Keratin is a fibrous and recalcitrant protein found in feathers, nails, horns, hooves, and the epidermis of the skin. The presence of the high degree of disulfide bonds, hydrogen bonds, and hydrophobic interactions makes them resistant to mechanical stress and are not degraded by common proteases such as trypsin, pepsin, and papain. Due to the slow degradation of keratinous protein, accumulation of solid wastes from the poultry, slaughterhouse, textile, and leather industries leads to solid waste problems and other environmental and health related problems. In this review, efficient biodegradation of keratinous wastes by microorganisms, as a low-cost, environmentally friendly strategy has been discussed. Keratinases are the microbial proteases and hydrolyze the hard keratin. The decomposition of keratin by keratinases maintains the original structure of the final products, including short peptides, amino acids, and organic nitrogen which are deteriorated when traditional or chemical method is implemented. In this article, the role of keratinases producing bacterial and fungal species and their attributes has been elaborated, along with the biochemical characteristics of keratinases, and further, protein engineering approaches has been discussed, with the prospects to enhance keratinases activity for their biotechnological applications.
Keratin, Solid Wastes, Biodegradation, Keratinolytic Enzymes
Microorganisms can be found everywhere and are the primary source of proteases, which have vast biotechnological implications. Additionally, protease can efficiently degrade various proteins. Keratin is a fibrous and resistant protein found in feathers, nails, horns, hooves, and the epidermis of the skin, among other tissues (Figure 1). 1-5 The durability and rigidity of these proteins is due to the high degree of disulfide bonds, hydrogen bonds, and hydrophobic interactions.4,6,7 Fibrous proteins are stable and resistant to degradation by proteolytic enzymes such as trypsin, pepsin, and papain due to their compact disulfide and intra-molecular-bond-based structural organization.8 Due to their slow biodegradation, keratin-based wastes wreak havoc on the environment. The poultry industry is one of the emerging industries due of the rise in global meat consumption, as chicken meat is a good protein sources.9 The major producers of these keratin-based solid wastes that are difficult to degrade are the poultry, leather, and textile industries.10 It is estimated that the annual production of keratinous wastes from chicken processing farms is in the millions of tons.6 They have become harmful contaminants in the environment because of their rigidity, high mechanical strength, recalcitrant property, and poor deposition of wastes.11 Traditional methods of keratin decomposition, such as incineration, burial, composting, and mechanical grinding, produce toxic gases that harm terrestrial and aquatic life. Chemical treatment of keratin wastes (acid/alkaline hydrolysis or treatment with oxidizing/reducing agents) is a costly and energy-intensive process that reduces the nutritional value of the amino acids obtained through keratin decomposition.10,12–14 The accumulation of solid waste reduces the land’s surface and contributes to land pollution. One of the major problem caused by the poultry industries in the solid waste problem originated from the slow degradation of hard keratin protein feather. Due to the presence of pathogenic microorganisms, improper disposal of poultry feathers has been linked to a number of human health issues, such as chlorosis and fowl cholera.15 Particulate matter (PM), greenhouse gas emissions (such as methane and sulfur dioxide), nitrogenous compounds (NH3, N2O, NOx), and volatile organic compounds (VOCs) are just some of the reported hazardous substances produced by the poultry industry. Ecosystems (via processes like soil acidification following atmospheric deposition) as well as human health are negatively affected by these threats. Bronchitis, asthma in youngsters, heart disease, lung illness, and cancer are some of the ailments that might develop in poultry farm workers.16–18 Poultry manure can also contain microorganisms and pharmaceuticals (such as antibiotics) used in poultry production, which can contaminate soil and water, leading to antimicrobial resistance, including multidrug resistance.17
Figure 1. Different sources of keratin in natural available form a) Nails b) Pig Bristle c) Chicken Feathers d) Tails e) Hairs f) Hoof g) Horn
The microorganisms have the potential to produce proteases. Keratinase is a protease produced by microorganisms that can break down the protein keratin to release amino acids. The degradation of keratin by microbial keratinases maintains the original structure of the final products, including short peptides, amino acids, and organic nitrogen.11 This method is cost-effective and environmentally friendly. The products resulting from the hydrolysis of feathers are suitable for use in fertilizers and as a source of valuable amino acids such as serine, cysteine, and proline.19 The ability of keratinase to withstand extreme conditions is advantageous for the development of green technology and sustainability. On the basis of the enzyme’s catalytic site, keratinases can be classified as serine or metallo-serine proteases.20
Keratinase-producing bacteria consist of Chryseobacterium sp. strain kr621, Stenotrophomonas maltophilia DHHj22, Bacillus subtilis SLC23, Bacillus megaterium SN124, Bacillus pumilus GRK25, Bacillus pumilus FH919, Bacillus licheniformis ALW112, Bacillus sp. Nnolim-K114, Ochrobactrum intermedium26, Deinococcus geothermalis27, Bacillus aerius NSMk228, Chryseobacterium cucumeris FHN16 etc and keratinase producing fungi includes Aspergillus oryzae29, Trichoderma atroviride F630, Aspergillus parasiticus31, Coriolopsis byrsina32, Aspergillus sp. DHE733. Since keratin is used in so many different industries (including textiles, leather, detergents, cosmetics, and bio fertilizer)6,34, it has become the prominent areas of research.
Structure and classification of keratin
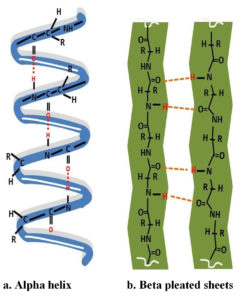
Keratins are intermediate superfamily proteins that are a major component of epidermal appendages such as nails, hair, feathers, and horn.35–37 Keratins are highly resistant proteins because of their compact structure. The structural classification of keratin is determined by its amino acid composition and polypeptide chain arrangement. The fibrous keratin is classified into α-keratin which is composed of α-helix and β-keratin is defined by β-sheets shown in Figure 2. Mammalian cells contain α-keratin, whereas avian and reptilian tissues contain β-keratin, with the exception of the pangolin, which contains both types of keratin in its skin8,36. Around 91 percent of the keratin in feathers is a β-keratin. α-helical structures are the coiled structures formed by α-helical-coils types I and II (basic/neutral) that form a helix filament and a fibril held together by an interchain linkage. α-helix structure is rigid due to the disulfide bond of cysteine and the hydrogen bond. The α-keratin is distinguished by its low sulfur content. The diameters of α- and β-keratin were determined to be 7-10 nm and 3-4 nm, respectively, using transmission electron microscopy. Molecular analyses of α-keratin revealed the helix chain, which causes the chain to twist and display a helical shape due to the presence of a disulphide bond. Stability in β-keratin is provided by intermolecular hydrogen bonds between the protein’s β-sheets, which are arranged in a parallel or antiparallel orientation.37 α-keratin has a size of 40–68 kDa, while β-keratin is 20–40 kDa (10-22 kDa). Additionally, keratins have been categorized into distinct groups, such as reptilian keratin, mammalian keratin, and avian keratin.38 Due to its insolubility in inorganic solvents and water, beta-keratin is resistant to the proteolytic effects of common enzymes such as trypsin and pepsin.9 γ-keratin, also known as, is a globular protein with a molecular weight of 15 kDa. It provides structural integrity via disulfide bond and is a structural component of the epidermis, fibril cortex, and globular fiber matrix. Based on the sulfur content, keratin are further classified into soft and hard keratin of which epidermal cells of skin constitute the soft keratin while feathers, nail, horn, hooves etc., are hard keratin.36
Mechanism of keratin degradation
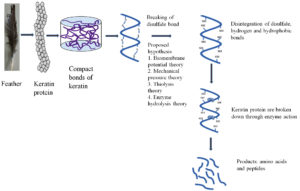
Keratin is a highly resistant and indigestible protein due to its complex structure. The mechanism of its decomposition is still unknown. Numerous hypotheses have been developed to illustrate the microbial breakdown of keratin. The biochemical mechanism of keratin disruption found that the degradation begins only after the denaturation of disulfide bond in the keratin followed by the action of extracellular proteases39. Similarly, Rajak et al. hypothesized that keratin degrades via sulfitolysis and keratinolysis.40 The mechanism begins with the cleavage of the keratin disulfide bond, followed by the unfolding of the active sites for microbial keratinases, which subsequently degrade the keratin into amino acids and peptides.41 Numerous theories illustrate the initial phrase denaturation or the destruction of the disulfide bond. The biomembrane potential theory illustrates the ability of the cells to produce sulfite or reducing substance in the medium which disintegrate the disulfide bond. The reduction of disulfide bond in feather Streptomyces pactum and found the production of reducing agent that disintegrate the disulfide bond and makes the protein chains available for cleavage by proteases.42 Similarly, the presence of disulfide reductase, which is responsible for the dissociation of disulfide linkages, was detected during the degradation of feathers by a bacterial cell, as inferred by using glutathione.43 Mycelia invading the structure’s epidermal layer and destroying the surface keratin protein are central to the mechanical pressure theory.40 Thiolysis theory explains how denaturation occurs when a microbe secretes sulfite, disulfide reductase, or enzymes, which then facilitates further hydrolysis of the protein by proteases.41 The keratinolytic enzyme aids in the further breakdown of resistant keratin, as proposed by the theory of enzyme hydrolysis. According to the theories, the dissolution of the disulfide bond is a prominent feature of keratin degradation. Along with provided theories of keratin degradation, the cooperative action of disulfide reductase and proteases in the breakdown of feather keratin. The rigid complex cysteine disulfide bond is shattered by disulfide reductase, and the action of keratinases loosens the keratin structure in conjunction with the formation of amino acids.44 To facilitate the breakdown of keratin in microorganisms, Peng et al. proposed a cascade of reactions involving cysteine catabolism. The microbial cell can convert cysteine to sulfite, which exits the cell and breaks the disulfide bond in feather keratin. Cysteine, a byproduct of feather keratin hydrolysis, enters the cell and the cycle is repeated.45 Keratin degradation begins with the hydrolysis of a cysteine bond, which is then catalyzed by microbial keratinases. The possible mechanism of keratin degradation by microbial keratinases is depicted in Figure 3, which depicts the initial phase, i.e. denaturation, and the subsequent phase, hydrolysis, which produces amino acids.
Characterization of keratinases
Different factors have a significant impact on the keratinases’ activity and output. The pH and the temperature stand out the most. According to the measured physical parameters, such as pH, keratinase activity increases from an acidic to an alkaline medium. Most of the keratinases from Bacillus megaterium SN1 are active at a pH of 324. Researchers found that keratinase activity was greatest between the pH range of 7 and 9, which is neutral to alkaline. The bacterial and fungal keratinases showing maximum activity under alkaline pH includes keratinases from Streptomyces gulbargensis,46 Bacillus pumilus FH919, Scopulariopsis brevicaulis,47 Paecilomyces marquandii and Doratomyces microspores,48 Aspergillus sp. DHE733, Bacillus licheniformis ALW112, Scopulariopsis brevicaulis47, Proteus vulgaris EMB-1449, Aspergillus sp. DHE733, Trichophyton sp. HA-230 etc. The alkaline medium facilitated the severing of the disulfide bond, resulting in the highest keratinase activity at alkaline pH. However, the keratinases from thermophilic bacterium Fervidobacterium islandicum AW-1 showed maximum activity in the range from 90 to 100°C.50 The optimal temperature range for most keratinases was 40°C to 60°C. According to a survey of published data on microbial keratinolytic proteases, their molecular weights can vary anywhere from 17 to 240 kDa12,14,31–33,48,51–53,19,22–26,29,30 Keratinases from S. maltophilia BBE11-154 and S. albidoflavus55 have low molecular weights of 17 and 18 kDa, respectively. In contrast, the keratinase produced by K. rosea was discovered to have a molecular weight of up to 240 kDa.56 The various biochemical characterizations of keratinases has been described in Table 1 and Table 2.
Table (1):
Biochemical characterization of keratinases from some fungal species.
Reported species |
Mol. weight |
Protease type |
Optimum pH |
Optimum Temp. |
Method of Keratinase production |
Inhibitors |
Stimulators |
Reference |
|---|---|---|---|---|---|---|---|---|
Aspergillus oryzae |
60kDa |
Metallo |
8.0 |
50 °C |
Submerged |
Hg2+,Cd2+ Pb2+ and EDTA |
Ca2+, Ba2+, Cu2+, Na2+, K+, & Mg2 |
29 |
Paecilomyces marquandii Doratomyces microspores |
33kDa 30kDa |
serine |
8.0 8.0 |
60-65 °C 60-65 °C |
Submerged Submerged |
PMSF PMSF |
DDT DDT |
48 |
Trichoderma atroviride F6 |
21kDa |
serine |
8.0-9.0 |
50°C |
Submerged |
Hg2+, Cu2, EDTA SDS |
Ca2+, Ba2+, Mn2+, Zn2+ |
30 |
Alternaria tenuissima K2 Aspergillus niduluns K7 |
— — |
7.5 7.5 |
35 °C 35 °C |
Submerged submerged |
NH4H2, KNO3, NH4Cl, pectin |
Starch, Maltose |
53 |
|
Asperigillus flavus K-03 |
31kDa |
serine |
8.0 |
45 °C |
submerged |
PMSF, EDTA iodoacetate |
Mn2+ Hg2+, Fe2+ |
92 |
Aspergillus parasiticus (MTCC 9164) |
36kDa |
serine |
7.0 |
50 °C |
submerged |
PMSF, chymostatin, leupeptin and pepstatin |
Ca2+, Mg2+, Mn2+,
DDT, β-ME |
31 |
Aspergillus niger 3T5B8 Aspergillus niger 9D80 |
14-130kDa 130kDa |
serine |
5.0 5.0 |
45 °C 45 °C |
Solid state fermentation Submerged |
PMSF |
— |
51 |
Aspergillus flavusS125 |
— |
— |
9.0 |
55 °C |
Solid state fermentation |
Dextrin, sucrose, starch, glucose, yeast, malt, urea |
— |
52 |
Coriolopsis byrsina |
— |
serine |
7-7.5 |
40-55°C |
submerged |
PMSF, EDTA |
— |
32 |
Aspergillus sp. DHE7 |
33kDa |
Serine-metalloprotease. |
60°C |
submerged |
PMSF, EDTA |
Ca2+, Mn2+, Zn2+ |
33 |
Abbreviations: EDTA- Ethylenediaminetetraacetic acid, SDS-sodium dodecyl sulfate, PMSF-phenylmethylsulfonyl fluoride, BSA- Bovine serum albumin DDT-Dichlorodiphenyltrichloroethane, β-ME-β- mercaptoethanol
Table (2):
Production and characterization of keratinases from bacterial species.
Reported species |
Mol. weight |
Protease type |
Optimum pH |
Optimum temp. |
Method of Keratinase production |
Inhibitors |
Stimulator |
Reference |
|---|---|---|---|---|---|---|---|---|
Chryseobacterium sp. strain kr6 |
— |
Metallo-protease |
7.5 |
55°C |
submerged |
EDTA, 1-10 phenanthroline, Hg2+, Cu2+ |
Ca2+, Triton X-100 |
21 |
Stenotrophomonas maltophilia DHHj |
35.2kDa |
serine |
7.8 |
40°C |
submerged |
PMSF, Zn2+, Pb2+, Cd2+, Hg2+, |
Ca2+, Ba2+, Cu2+, Na+, K+and Mg2+ |
22 |
Bacillus subtilis SLC |
serine |
8 |
60°C |
submerged |
PMSF |
23 |
||
Bacillus megaterium SN1 |
30kDa |
3 |
70°C |
submerged |
Hg2+, Ba2+ |
Co2+, Mn2+, Mg2+, |
24 |
|
Bacillus pumilus GRK |
— |
serine |
10 |
37°C |
submerged |
PMSF β- ME Hg2+ |
Ca2+ Mg2+ |
25 |
Bacillus pumilus FH9 |
9 |
60°C |
Submerged |
Zn2+, EDTA, Co2+ and Hg2+ |
Ca2+ Mg2+ |
19 |
||
Bacillus aerius NSMk2 |
— |
7.5 |
35°C |
submerged |
— |
— |
28 |
|
Bacillus licheniformis ALW1 |
— |
8.0 |
65C |
submerged |
— |
— |
12 |
|
Ochrobactrum intermedium. |
— |
serine |
7.0 |
40°C |
submerged |
PMSF, EDTA |
Ca2+ |
26 |
Bacillus sp. Nnolim-K1 |
metalloprotease |
8.0 |
60°C |
submerged |
EDTA, 1,10-phenanthroline, Zn2+, Hg2+, Cu2+ |
Ca2+ |
14 |
|
Bacillus haynesii ALW2 |
— |
8.0 |
70°C |
submerged |
— |
— |
93 |
|
Proteus vulgaris EMB-14 |
49kD |
9.0 |
60°C |
submerged |
Cu2+, Pb2+, β- ME |
Ca2+ Mg2+, |
49 |
|
Bacillus zhangzhouensis |
42 kDa |
Serine |
9.5 |
60°C |
submerged |
PMSF |
Ca2+, Mn2+, Na+, K+ |
94 |
Deinococcus gobiensis |
6-8 |
60°C |
Cr3+ |
K+, Li+, Mg2+ and Co2+ |
95 |
Abbreviations: EDTA- Ethylenediaminetetraacetic acid, SDS-sodium dodecyl sulfate, PMSF-phenylmethylsulfonyl fluoride, BSA-bovine serum albumin, DDT-Dichlorodiphenyltrichloroethane, β-ME-β- mercaptoethanol.
Strategies to improve keratinases for keratinases production
Protein Engineering Techniques
There are numerous applications for microbial keratinases in biotechnological industries, but their poor tolerance to temperature, pH, organic salts, and detergent, as well as their low catalytic activity, precludes their use. Because the processes of degrading feathers and dehairing leather took place in an alkaline environment, the high performance of alkaline keratinase may be especially significant. One of the most important characteristics of an enzyme for its potential use in industrial processes is its resistance to salt, also known as halotolerance. Protein engineering methods were used to develop superior enzymes with enhanced catalytic efficiency, thermostability, and substrate specificity.34 To achieve the desired yield, scientists use protein engineering techniques like signal pro-peptide engineering, domain swapping, truncation, etc. to improve the protein’s activity, thermostability, and catalytic efficiency. Analysis of keratinase amino acid sequences reveals that the different domains of the enzymes are largely responsible for substrate specificity, thermostability, alkalinity, feather degrading efficiency, etc., as well as other effects of organic compounds and detergents. Keratinases from Stenotrophomonas maltophilia BBE11-1 have been deduced to have three distinct domains based on their amino acid sequences: an N-propeptide domain, a catalytic domain, and a PPC (Pre-peptidase C-terminal) domain.57 When each domain was altered, specific results were observed. Substrate specificity was found to be controlled by a C-terminal domain. Changes in the N-terminal domain exchange between the isolated keratinases KerSMF and KerSMD formed a mutant FDD with enhanced catalytic and feather degradation activity.58
The variant V355 from Keratinase KerSMD of Stenotrophomonas maltophilia was created by partial truncation of the PPC domain, and it exhibited improved secretion of keratinase, enzyme stability, salt tolerance, surfactant stability, and other properties that make it prominent for use in laundry detergents and leather treatments.59,99 The enhancement of protein or the enzymes can be achieved by two approaches which are as follows:
Rational protein engineering
The rational protein design can effectively predict and modify the residues responsible for the enzyme’s substrate and co-factor binding sites, thermostability, and other specific functional properties by comparing the target sequence (predicted structure) to fully characterized enzymes.60 Site directed mutagenesis which is one of rational approach in protein engineering is used for modifying enzyme substrate specificity. Site directed mutagenesis was studied in mutant FDD keratinases from by the N-terminal domain exchange between the isolated keratinases KerSMF and KerSMD from Stenotrophomonas sp.57 The keratinase variant FDD thus engineered through site directed mutagenesis and the two variants (Y94F and Y215F) showed higher extracellular keratinolytic activity (about 3-fold increase).58
Directed evolution
Directed evolution tries to accelerate up the natural evolution of biological molecules and it is based on random mutagenesis and/or gene recombination to generate a library of variants and screening/selection of variants with enhanced phenotype.61 This strategies has been used to make enzymes with a wide range of traits, such as substrate specificity, resistance to organic solvents, stability at high temperatures, and the best working pH.61–63 The method was used to engineered keratinases (KerPA) from Pseudomonas aeruginosa CCTCC AB2013184 and found that the variant of keratinases (Y21pBpF,Y70pBpF and Y114pBpF) thus obtained showed enhanced activity and thermostability.64 Similarly, Zhang et al., found that keratinase mutant (KerBp) obtained from using directed evolution technology showed enhanced enzyme activity from 1150 to 8448 U/mL and feather degradation rate increases from 49 to 88% when trypsin was added.65
The outcomes of the various protein engineering techniques used to improve enzyme performance are summarized in the table 3 that follows.
Table (3):
Different molecular Strategies to improve keratinases production and its possible outcomes.
Microorganism |
Gene |
Expression host |
Engineering strategy |
Variant/Mutant |
Effects/outcome |
Reference |
|---|---|---|---|---|---|---|
Brevibacillus parabrevis CGMCC 10798 |
KERBP |
E.coli |
site-directed mutagenesis and combinatorial mutagenesis |
T218S, S236C and N181D |
Improved thermostability and catalytic efficiency, higher dehairing capacity |
96 |
Stenotrophomonas maltophilia BBE11-1 |
KerSMD |
E.coli |
Domain exchange with KerSMF |
FDD and DDF |
Higher catalytic activity towards casein, feather degradation and higher keratinolyic activity |
57 |
Stenotrophomonas maltophilia BBE11-1 |
Keratinase FDD |
E.coli |
Site-directed mutagenesis |
Y94F, Y187V, Y215F, A218G and A218S |
Higher keratinolytic activity, better Thermostability(70°C) |
58 |
Stenotrophomonas maltophilia BBE11-1 |
Keratinase DDF |
E.coli |
C-terminus fusion |
DDFD |
Higher keratinolytic activity, better Thermostability(70°C) |
58 |
Streptomyces fradiae |
keratinase Sfp2 |
plasmid p Bluescript SK |
site-directed mutagenesis of its N-terminal pro-sequence |
L(-1)D,L(-1)FL(-1)G, L(-1)H, K(-2)E, K(2)L |
Improved thermostability |
97 |
Bacillus subtilis WB600 |
kerBv |
Multi-enzyme cascade pathway |
— |
Wound healing, tissue engineering |
98 |
|
Pseudomonas aeruginosa CCTCC AB2013184 |
KerPA |
E. coli DH5α/ pET22b to |
Directed evolution with non-canonical amino acids (ncAAs) using genetic code expansion |
Y21pBpF,Y70pBpF and Y114pBpF |
Enhanced activity and thermostability |
64 |
Stenotrophomonas maltophilia |
KerSMD |
E. coli |
Partial truncation of pre-peptidase C-terminal PPC domain |
V435, V415, V395, V380, V370, and V355 showed |
Can be used for laundry detergent and leather treatment |
99 |
Stenotrophomonas maltophilia |
KerSMD |
E. coli |
N-terminal propeptide replacement and site-directed mutagenesis in S1 pocket |
S180G/ Y215S |
Higher enzyme production |
100 |
Bacillus licheniformis BBE11‐1 |
KerZ1 |
Bacillus subtilis WB600 |
Pro‐peptide engineering (using the three‐codon mutation principle) |
2‐D12 |
High feather degradation ability along with sulfite |
101 |
Pseudomonas aeruginosa |
Ker P |
E.coli HB101 |
N-Terminal Pro-sequence |
KerP F1, KerP F2, KerP F3, and Ker P F4 |
Showed higher catalytic activity and thermostability |
102 |
Bacillus sp. LCB12 |
B. subtilis SCK6, pMA0911 |
signal peptide optimization and site-directed mutagenesis |
M123L,V149I and A242N |
Higher extracellular keratinase activity |
103 |
|
Bacillus licheniformis Bacillus pumilus |
(Ker BL) (Ker BP) |
E. coli HB101-pEZZ18 |
Swapping of pro-sequences between Ker BL and Ker BP |
Ker ProBP–BL and Ker ProBL–BP |
Higher thermostability and substrate efficiency |
104 |
Stenotrophomonas maltophilia |
(KerSMD) |
E.coli |
Truncation of PPC domain |
V456, V445, V435, V415, V395, V380 ,V370 and V355 |
1.7 fold increase keratinolytic activity, increased thermostability from 50°C to 60°C |
99 |
Mutagenesis
UV mutagenesis
The effect of non-ionizing radiation on mutagenesis is due to the excitation of DNA molecules, which results in the formation of thymine base pairs between adjacent bases in the DNA molecule. The strain’s keratinase production is changed by UV light incubation. Lateef et al. discovered that the mutant (89.2 U/mL) from Bacillus safensis LAU 13 produced more keratinase than that of the wild type (57.4 U/mL) and, when the strain was pre-incubated with UV light of 254 nm, the production increased further. It was observed that the resulting mutant possessed dehairing capabilities.66 The Bacillus cereus group with isolates (S1, S13, S15, S26, and S39) were introduced with UV radiation and found the mutants S13uv and S26uv showed high keratinase activity showed higher keratinase production of after 72 hours of incubation.67 Similarly, the purified proteases obtained from the mutant RS 1 obtained through UV irradiation of Bacillus sp. resulted in 44-fold increase in specific activity compared with the crude protease of the wild type Bacillus sp. RS168.
Chemical Mutagenesis
Enzyme production is significantly impacted by the presence of chemical agents like ethyl methyl sulfonate (EMS), N-methyl-N-nitro-N-nitrosoguanidine (MNNG), and ethidium bromide in the culture medium. A 2011 study by Duarte et al. found that incubating the Candida parapsilosis strain in 3% ethyl methanesulfonate (EMS) resulted in greater keratinase activity in the mutant strain J5(140 U/mL) than in the wild type strain (80 U/mL).69 The bacterial cereus isolates (S1, S13, S15, S26, and S39) were treated with EMS and the mutant thus formed showed increased keratinase activity from 1.5–3.7-fold compared with the wild isolates.67
Site-Directed mutagenesis
Site-directed mutagenesis entails the modification of a gene via substitutions, targeted base insertions, or deletions. The process is primarily concerned with increasing the activity, yield, efficiency, and thermostability of enzymes, as well as their resistance to acidic to alkaline environments.70 Various molecular strategies applied for obtaining higher keratinases yield along with the possible outcomes were tabulated in table 3.
Biotechnological applications of keratinases
Keratinases are essential enzymes for the degradation of numerous keratin-based substances. Keratinases provide numerous environmental and agricultural benefits. The eco-friendly nature of the microbial degradation of resistant hard keratin by keratinases is of utmost importance, as it prevents the loss of nutritionally significant amino acids that can be damaged by chemical treatment. Several applications of keratinases are described below: –
Animal feed additives
Keratin can be broken down by microbial keratinases, which can then be used by the microbes as a source of carbon and nitrogen. Essential amino acids like cysteine, threonine, phenylalanine, leucine, valine, and isoleucine are produced as a byproduct of keratinous waste and can be used to strengthen animal feed.71,72 Odetallah et al. investigated the effects of adding keratinases to the chickens’ diet on their overall body weight. Supplementing corn-soy based broiler starter diets with a keratinase was found to boost chick growth performance and increase body weight. It was deduced that the young chickens’ increased body mass was the result of their consuming the crude protein, which served as a substrate for the enzymes and released the free amino acids that the chicks then consumed. When added to low protein diets on day one, keratinase increased chicken body weight by day 21.73 Aina et al. conducted a study in which Wickerhanomyces Anomalus keratinase was introduced into chicken feed. It was found that when enzymes were introduced into the feed, the total protein content and nitrogen content both increased.74 The study done by Xu et al. in 2022 showed that when keratinase (200,000 U/kg) was added to a corn-soybean-feather meal-based diet (BD) for 6 weeks, the body weight of broiler chicken increases from by 3.6% to 4.3% and also improves the meat quality of the chicken.75 Using Ross 308 chicks, Ayanwale et al. did an experiment to compare the growth performance and meat quality of broiler chickens fed on keratinase-treated and untreated feather meal-based diets. The broiler chickens fed a diet with 16% enzyme-treated feather meal (T6) had the highest average final body weight (2113.00 g), compared to the untreated feather meal-based diets that had average final body weight (1935.70g).76 Therefore, the supplementation of keratinase could be used as feed additives to enhance growth performance, total protein content and meat quality in the poultry industries.
Leather Industry
In developing nations, the leather trade is seen as a promising new sector. Chemicals like sodium sulfite, lime, and calcium carbonate are used in the dehairing process, and they lower the quality of the leather and pollute the air with their byproducts. When compared to chemical dehairing, which damages fibers and yields low-quality leather, the use of microbial proteases as an alternative is both environmentally friendly and cost-effective. Keratinolytic proteases are more effective for depilation and show no damage to leather when the process is optimized, in comparison to common alkaline proteases.70,77 Depilation requires an extremely alkaline environment and the use of detergents like sodium dodecyl sulfonate, inactivate most of the proteases. In contrast, keratinases are recalcitrant enzymes that can withstand high concentrations of alkali and detergents like SDS etc. Keratinases are used in the dehairing process to remove hair follicles, leaving the skin smooth and hairless which infers good quality leather.70 Chaturveti et al. found that all of the hairs were removed when goat hairs were incubated with 40 U/ml of crude keratinase for 20 hours. Skin treated with keratinase was soft, swollen, and white in color after removal of hairs, while skin treated with sodium sulfite and calcium carbonate was hard, dark, and brown in color after shrinking.78 SEM analysis confirmed that both the crude and purified keratinases from Bacillus sp. NKSP-7 were able to completely dehair goat skin while leaving it otherwise undamaged.79 Goat, rabbit, bovine, and sheep skins were completely dehaired when treated with the recombinant keratinase rKERDZ isolated from Actinomadura viridilutea DZ50. Scanning electron microscopy (SEM) analyses corroborated the findings, showing that the leather’s quality was unaltered from the control sample. Leather processing using chemicals raises concentrations of harmful byproducts like COD and BOD, which is bad for the environment. As a result, enzymatic techniques may have biotechnological and ecological significance. Bioremediation with the help of the microbial product has significant economic value and advantages over chemical treatment in the leather industry. Utilizing purified or recombinant keratinase may ameliorate the process more effectively than would be possible with crude enzymes.80
Detergent industry
Some keratinases are considered thermoactive, which provides them with a high degree of tolerance against any solvent, surfactant, or detergent. In addition to aiding in the removal of scarves and blood stains from cotton garments in hospitals, they have extensive application in the detergent industry.81 Researchers have analyzed keratinases for their potential use in the detergent industry due to their enzymatic washing properties. On various fabrics stained with blood, egg yolk, and chocolate, the crude enzyme (76 U/ml) supplemented with inactive detergent was used to observe its cleansing effects. Protein stains like blood and egg yolk were found to be easily removed by the enzyme, while the chocolate stain was not completely removed.82 Both the wild type (57.4 U/mL) and the mutant strain (89.2 U/mL) created by UV mutagenesis were able to efficiently remove the blood stain from the fabric without damaging the fibers. Keratinases from Bacillus sp. NKSP-7 were found to be stable and detergent-compatible. When incubated with blood-stained fabrics, both the purified enzyme and the crude enzyme demonstrated cleaning efficacy.79 Another cleansing properties was observed when the cloth stained with coffee and blood was incubated with keratinases from Stenotrophomonas maltophillia.83 The washing capacity of the egg stained was shown by Citrobacter diversus-derived keratinases which was tolerant to detergent mixture which can implemented in detergent formulations.84 Therefore, the use of keratinases plays a significant role in the detergent industry, as it can give a natural means of cleaning without compromising the fabric quality, which is negatively affected by the use of detergents.
Other Applications
Keratinases can be utilized as both biopesticides and plant growth factor. Root knot nematodes (Meloidogyne incognita), which cause damage to plants, can be destroyed by treating with purified alkaline keratinases obtained from Bacilllus sp., which digest the nematode’s keratin and collagen cuticle. Keratinases’ ability to degrade the cuticle layer of nematodes lead to the dead of the nematodes.85 The role of keratinases in growth is investigated in Vigna radiate. The plant treated with feather hydrolysate obtained from Stenotrophomonas maltophilia showed higher growth in plant compared to untreated one.83 The application of keratinases can also be observed in transdermal drug delivery systems. In mice infected with Staphylococcus aureus, keratinases from Bacillus cereus were used as a drug delivery agent alongside fusidic acid by Shalaby et al. When a keratinases/fusidic acid mixture was used, rather than fusidic acid alone, S.aureus growth was greatly reduced. It was hypothesized that keratinase could break down the stratum corneum, allowing the drug to penetrate the skin and speed up the mice’s recovery. The research laid the groundwork for future investigations into the medical application of keratinases as delivery systems.86
The current scope and future challenges in keratinases research
Assessment of research publications accumulated over a while on a topic of interest is a very reliable method to decipher the relevance, viability, aspects, national/ international cooperation, funding pattern by the different funding agencies, and existing lacunae on the concerned research topic. Such studies not only give an insight into the researcher’s involvement but also inherently depicts and dissect the focal themes and trends of research.87 A recent study conducted by Nnolim and Nwodo based on bibliometric data of three decades (1990 – 2019) on keratinase research by 20 top most researchers in this area highlighted that the focus of research has largely been associated with different aspects of keratinase production, characterization, and keratinous biomass degradation. The study also revealed that India scaled the summit among the top twenty most productive countries on keratinase research, ahead of China and Brazil, by achieving not only ~ 26% of the total research publications but also the highest number of citations (1533 times with average article citation of 18.04) documented during this 3-decade period.88
The ultimate aim needs to focus on collective efforts through national/ international collaborations for addressing the unsolved and most challenging research areas on keratinases to make the study more profound and innovative that in turn would contribute more to the discovery sciences rather than settling for just mundane findings. International collaborations not only bring about cross-pollination of ideas to enhance productivity and innovation but also foster scientific capacity.89 While some of the seminal research work conducted during the past 3-decades has now eventually established excellent platforms to cement the academia-industry linkages and collaborations for harnessing the laboratory-based keratinase research to accrue economic benefits out of the same, there remains considerable scope to explore and exploit new vistas in this arena. One of the major limitations encountered by numerous researchers involved in keratinase research is towards identifying the peptidases possessing keratinase activity. The impediment to the identification of the potential keratinase is compounded by the challenge of predicting the substrate specificity of the peptidase with keratinolytic activity.90 The establishment of efficient analytical methods is fundamental, both for the production as well as its application to environmental waste management.91
Although it is imperative to have the stimulus of appropriate S&T infrastructure, adequate research funding, and sound collaborative partnerships for qualitative and quantitative research outputs, it is even more crucial to define the research niches on keratinases and the expected outputs from those researches before framing any research proposal. This assumes even greater significance as currently, due to overall constraints of R&D funding globally, efforts are directed towards resource optimization and restrictions in funding proposals that project only incremental advances in domain knowledge in any particular area of S&T research. Hence, to secure grants from different funding agencies, the different aspects of research on Keratinases that needs to be conceptualized may revolve around at least one or more factors such as: i) research that contributes to value addition to discovery sciences, ii) research that caters to generating more academia-industry link-ups ensuring economic returns, iii) research having a significantly high societal impact, and iv) research that addresses grand challenges by aligning with one or more of the National Missions related to sustainable habitats and green energy.
Microbial proteases play a prominent role in biotechnological as well as in agro-industries which are growing at a tremendous rate. The microbial keratin degradation is advantageous over traditional chemical methods as its eco-friendly, cost-effective, and recovers the valuable amino acid. Due to the vast applicability of keratinases in various biotechnological industries, the demands for enzymes are also simultaneously increasing. Various molecular strategies like a heterologous expression of keratinase genes, codon mutagenesis, and site-directed mutagenesis used for the production of a recombinant or mutant keratinase gene. The recombinant keratinase were found to have a better degradation percentage, and dehairing properties compared with crude or purified keratinases. The involvement of keratinases in drug delivery system may have immense important and it gives the pavement for further research of keratinases in different fields of science.
ACKNOWLEDGMENTS
The authors would like to thank GBP-National Institute of Himalayan Environment and Assam University, Silchar, Assam, for providing the facilities.
CONFLICT OF INTEREST
The authors declare that there is no conflict of interest.
AUTHORS’ CONTRIBUTION
SK conceptualized and designed the study. BS performed data curation. IG, SM, AnB, PP, ArB, SK wrote the manuscript. SM, AnB, PP, ArB, SK, BS reviewed the manuscript. BS and SK edited the manuscript. SK approved the final manuscript for publication.
FUNDING
This study was funded by GBP-National Institute of Himalayan Environment under IERP research grant no- GBPI/IERP/18-19/04.
DATA AVAILABILITY
All datasets generated or analyzed during this study are included in the manuscript.
ETHICS STATEMENT
Not applicable.
- Huang Y, Łężyk M, Herbst FA, Busk PK, Lange L. Novel keratinolytic enzymes, discovered from a talented and efficient bacterial keratin degrader. Sci Rep. 2020;10(1):1-11.
Crossref - Kang D, Huang Y, Nesme J, et al. Metagenomic analysis of a keratin-degrading bacterial consortium provides insight into the keratinolytic mechanisms. Sci Total Environ. 2021;761.
Crossref - Lange L, Huang Y, Busk PK. Microbial decomposition of keratin in nature—a new hypothesis of industrial relevance. Appl Microbiol Biotechnol. 2016;100(5):2083-2096.
Crossref - McKittrick J, Chen PY, Bodde SG, Yang W, Novitskaya EE, Meyers MA. The structure, functions, and mechanical properties of keratin. Jom. 2012;64(4):449-468.
Crossref - Qiu J, Barrett K, Wilkens C, Meyer AS. Bioinformatics based discovery of new keratinases in protease family M36. N Biotechnol. 2022;68:19-27.
Crossref - Hendrick Q, Nnolim NE, Nwodo UU. Chryseobacterium cucumeris FHN1 keratinolytic enzyme valorized chicken feathers to amino acids with polar, anionic and non-polar imino side chain characteristics. Biocatal Agric Biotechnol. 2021;35:102109.
Crossref - Huang W, Restrepo D, Jung JY, et al. Multiscale Toughening Mechanisms in Biological Materials and Bioinspired Designs. Adv Mater. 2019;31(43):1901561.
Crossref - Hassan MA, Abol-Fotouh D, Omer AM, Tamer TM, Abbas E. Comprehensive insights into microbial keratinases and their implication in various biotechnological and industrial sectors: A review. Int J Biol Macromol. 2020;154:567-583.
Crossref - Parinayawanich S, Sittipol D, Ajingi YS, Rodpan S, Pattanapanyasat K, Jongruja N. Application of recombinant hyperthermostable keratinase for degradation of chicken feather waste. Biocatal Agric Biotechnol. 2021;36:102146.
Crossref - Łaba W, Żarowska B, Chorążyk D, et al. New keratinolytic bacteria in valorization of chicken feather waste. AMB Express. 2018.
Crossref - Yadav S, Bumbra P, Laura JS, Khosla B. Optimization of nutritional and physical parameters for enhancing the keratinase activity of Bacillus cereus isolated from soil of poultry dump site in Gurugram , Haryana. Bioresour Technol Reports. 2022;18:101108.
Crossref - Abdel-fattah AM, El-gamal MS, Ismail SA, Emran MA, Hashem AM. Biodegradation of feather waste by keratinase produced from newly isolated Bacillus licheniformis ALW1. J Genet Eng Biotechnol. 2018;16(2):311-318.
Crossref - Li ZW, Liang S, Ke Y, et al. The feather degradation mechanisms of a new Streptomyces sp. isolate SCUT-3. Commun Biol. 2020;3(1):1-13.
Crossref - Nnolim NE, Mpaka L, Okoh AI, Nwodo UU. Biochemical and molecular characterization of a thermostable alkaline metallo-keratinase from bacillus sp. Nnolim-k1. Microorganisms. 2020;8(9):1-24.
Crossref - Anbesaw MS. Bioconversion of Keratin Wastes Using Keratinolytic Microorganisms to Generate Value-Added Products. Int J Biomater. 2022;1-24.
Crossref - Dróżdż D, Wystalska K, Malińska K, Grosser A, Grobelak A, Kacprzak M. Management of poultry manure in Poland – Current state and future perspectives. J Environ Manage. 2020;264.
Crossref - Gržinić G, Piotrowicz-Cieślak A, Klimkowicz-Pawlas A, et al. Intensive poultry farming: A review of the impact on the environment and human health. Sci Total Environ. 2023;858(Pt 3).
Crossref - Mitroi R, Stoian O, Covaliu CI, Manea D. Pollutants resulting from intensive poultry farming activities and their impact on the environment. E3S Web Conf. 2021;286:0-5.
Crossref - Abdel-naby MA, El-refai HA, Ibrahim MHA. Structural characterization , catalytic , kinetic and thermodynamic properties of Keratinase from Bacillus pumilus FH9. Int J Biol Macromol. 2017;105:973-980.
Crossref - Gurung SK, Adhikari M, Kim SW, et al. Discovery of Two Chrysosporium Species with Keratinolytic Activity from Field Soil in Korea. Mycobiology. 2018;46(3):260-268.
Crossref - Riffel A, Lucas F, Heeb P, Brandelli A. Characterization of a new keratinolytic bacterium that completely degrades native feather keratin. Arch Microbiol. 2003;179(4):258-265.
Crossref - Cao ZJ, Zhang Q, Wei DK, et al. Characterization of a novel Stenotrophomonas isolate with high keratinase activity and purification of the enzyme. J Ind Microbiol Biotechnol. 2009;36(2):181-188.
Crossref - Cedrola SML, de Melo ACN, Mazotto AM, et al. Keratinases and sulfide from Bacillus subtilis SLC to recycle feather waste. World J Microbiol Biotechnol. 2012;28(3):1259-1269.
Crossref - Agrahari S, Wadhwa N. Isolation and characterization of feather degrading enzymes from Bacillus megaterium SN1 isolated from Ghazipur poultry waste site. Appl Biochem Microbiol. 2012;48(2):175-181.
Crossref - Ramakrishna Reddy M, Sathi Reddy K, Ranjita Chouhan Y, Bee H, Reddy G. Effective feather degradation and keratinase production by Bacillus pumilus GRK for its application as bio-detergent additive. Bioresour Technol. 2017;243:254-263.
Crossref - Sharma I, Kango N. Production and characterization of keratinase by Ochrobactrum intermedium for feather keratin utilization. Int J Biol Macromol. 2021;166:1046-1056.
Crossref - Tang Y, Guo L, Zhao M, et al. A novel thermostable keratinase from Deinococcus geothermalis with potential application in feather degradation. Appl Sci. 2021;11(7).
Crossref - Bhari R, Kaur M, Singh RS, Pandey A, Larroche C. Bioconversion of chicken feathers by Bacillus aerius NSMk2: A potential approach in poultry waste management. Bioresour Technol Reports. 2018;3:224-230.
Crossref - Farag AM, Hassan MA. Purification, characterization and immobilization of a keratinase from Aspergillus oryzae. Enzyme Microb Technol. 2004;34(2):85-93.
Crossref - Cao L, Tan H, Liu Y, Xue X, Zhou S. Characterization of a new keratinolytic Trichoderma atroviride strain F6 that completely degrades native chicken feather. Lett Appl Microbiol. 2008;46(3):389-394.
Crossref - Anitha TS, Palanivelu P. Purification and characterization of an extracellular keratinolytic protease from a new isolate of Aspergillus parasiticus. Protein Expr Purif. 2013;88(2):214-220.
Crossref - Duffeck CE, de Menezes CLA, Boscolo M, da Silva R, Gomes E, da Silva RR. Keratinases from Coriolopsis byrsina as an alternative for feather degradation: applications for cloth cleaning based on commercial detergent compatibility and for the production of collagen hydrolysate. Biotechnol Lett. 2020;42(11):2403-2412.
Crossref - El-ghonemy DH, Ali TH. Effective bioconversion of feather-waste Keratin by Thermo-Surfactant Stable Alkaline Keratinase produced from Aspergillus sp . DHE7 with promising biotechnological application in detergent formulations. Biocatal Agric Biotechnol. 2021;35:102052.
Crossref - Su C, Gong JS, Qin A, et al. A combination of bioinformatics analysis and rational design strategies to enhance keratinase thermostability for efficient biodegradation of feathers. Sci Total Environ. 2022;818(1800):151824.
Crossref - Feroz S, Muhammad N, Ranayake J, Dias G. Keratin – Based materials for biomedical applications. Bioact Mater. 2020;5(3):496-509.
Crossref - Qiu J, Wilkens C, Barrett K, Meyer AS. Microbial enzymes catalyzing keratin degradation : Classification , structure , function. Biotechnol Adv. 2020;44:107607.
Crossref - Wang B, Yang W, McKittrick J, Meyers MA. Keratin: Structure, mechanical properties, occurrence in biological organisms, and efforts at bioinspiration. Prog Mater Sci. 2016;76:229-318.
Crossref - Schweizer J, Bowden PE, Coulombe PA, et al. New consensus nomenclature for mammalian keratins. J Cell Biol. 2006;174(2):169-174.
Crossref - Kunert J. Biochemical mechanism of keratin degradation by the actinomycete Streptomyces fradiae and the fungus Microsporum gypseum: A comparison. J Basic Microbiol. 1989;29(9):597-604.
Crossref - Rajak RC, Parwekar S, Malviya H, Hasija SK. Keratin degradation by fungi isolated from the grounds of a gelatin factory in Jabalpur, India. Mycopathologia. 1991;114(2):83-87.
Crossref - Peng Z, Zhang J, Du G, Chen J. Keratin Waste Recycling Based on Microbial Degradation: Mechanisms and Prospects. ACS Sustain Chem Eng. 2019;7(11):9727-9736.
Crossref - Böckle B, Müller R. Reduction of disulfide bonds by Streptomyces pactum during growth on chicken feathers. Appl Environ Microbiol. 1997;63(2):790-792.
Crossref - Yamamura S, Morita Y, Hasan Q, et al. Characterization of a new keratin-degrading bacterium isolated from deer fur. J Biosci Bioeng. 2002;93(6):595-600.
Crossref - Yamamura S, Morita Y, Hasan Q, Yokoyama K, Tamiya E. Keratin degradation: A cooperative action of two enzymes from Stenotrophomonas sp. Biochem Biophys Res Commun. 2002;294(5):1138-1143.
Crossref - Peng Z, Xu P, Song Y, Du G, Zhang J, Chen J. Cysteine-Mediated Cyclic Metabolism Drives the Microbial Degradation of Keratin. ACS Sustain Chem Eng. 2021;9(29):9861-9870.
Crossref - Syed DG, Lee JC, Li WJ, Kim CJ, Agasar D. Production, characterization and application of keratinase from Streptomyces gulbargensis. Bioresour Technol. 2009;100(5):1868-1871.
Crossref - Anbu P, Gopinath SCB, Hilda A, Lakshmi T, Annadurai G. Purification of keratinase from poultry farm isolate- Scopulariopsis brevicaulis and statistical optimization of enzyme activity. 2005;36:639-647.
Crossref - Gradišar H, Friedrich J, Križaj I, Jerala R. Similarities and Specificities of Fungal Keratinolytic Proteases: Comparison of Keratinases of Paecilomyces marquandii and Doratomyces microsporus to Some Known Proteases. Appl Environ Microbiol. 2005;71(7):3420.
Crossref - Babalola MO, Ayodeji AO, Bamidele OS, Ajele JO. Biochemical characterization of a surfactant-stable keratinase purified from Proteus vulgaris EMB-14 grown on low-cost feather meal. Biotechnol Lett. 2020;42(12):2673-2683.
Crossref - Kang E, Jin H, La JW, Park S, Kim W, Lee W. Identi fi cation of keratinases from Fervidobacterium islandicum AW-1 using dynamic gene expression profiling. 2020;13(2):442-457.
Crossref - Maria A, Couri S, Damaso MCT, Beatriz A. Degradation of feather waste by Aspergillus niger keratinases : Comparison of submerged and solid-state fermentation. Int Biodeterior Biodegradation. 2013;85:189-195.
Crossref - Mini K, George S, Mathew J. Keratinase Production by Solid State Fermentation and Optimization of Conditions of SSF and Formulation of Low Cost Medium for the Production of Keratinase. Int J Curr Microbiol App Sci. 2015;4(9):535-548. https://www.ijcmas.com/vol-4-9/K.D.%20Mini,%20et%20al.pdf
- Saber WIA, El-Metwally MM, El-Hersh MS.Keratinase Production and Biodegradation of Some Keratinous Wastes by Alternaria tenuissima and Aspergillus nidulans. Journal of Microbiology. 2010;5:21-35.
- Fang Z, Zhang J, Liu B, Du G, Chen J. Biochemical characterization of three keratinolytic enzymes from Stenotrophomonas maltophilia BBE11-1 for biodegrading keratin wastes. Int Biodeterior Biodegradation. 2013;82:166-172.
Crossref - Bressollier P, Letourneau F, Urdaci M, Verneuil B. Purification and characterization of a keratinolytic serine proteinase from Streptomyces albidoflavus. Appl Environ Microbiol. 1999;65(6):2570-2576.
Crossref - Bernal C, Cairó J, Coello N. Purification and characterization of a novel exocellular keratinase from Kocuria rosea. Enzyme Microb Technol. 2006;38(1-2):49-54.
Crossref - Fang Z, Zhang J, Liu B, Du G, Chen J. Enhancement of the catalytic efficiency and thermostability of Stenotrophomonas sp. keratinase KerSMD by domain exchange with KerSMF. Microb Biotechnol. 2016;9(1):35-46
Crossref - Fang Z, Zhang J, Du G, Chen J. Rational protein engineering approaches to further improve the keratinolytic activity and thermostability of engineered keratinase KerSMD. Biochem Eng J. 2017;127:147-153.
Crossref - Fang Z, Zhang J, Liu B, Jiang L, Du G, Chen J. Cloning, heterologous expression and characterization of two keratinases from Stenotrophomonas maltophilia BBE11-1. Process Biochem. 2014;49(4):647-654.
Crossref - Vidmar B, Vodovnik M. Microbial keratinases: Enzymes with promising biotechnological applications. Food Technol Biotechnol. 2018;56(3):312-328.
Crossref - Wang Y, Xue P, Cao M, Yu T, Lane ST, Zhao H. Directed Evolution: Methodologies and Applications. Chem Rev. 2021;121(20):12384-12444.
Crossref - Johannes TW, Zhao H. Directed evolution of enzymes and biosynthetic pathways. Curr Opin Microbiol. 2006;9(3):261-267.
Crossref - Xiao H, Bao Z, Zhao H. High Throughput Screening and Selection Methods for Directed Enzyme Evolution. Ind Eng Chem Res. 2014;54(16):4011-4020.
Crossref - Pan X, Yang J, Xie P, et al. Enhancement of Activity and Thermostability of Keratinase From Pseudomonas aeruginosa CCTCC AB2013184 by Directed Evolution With Noncanonical Amino Acids. Front Bioeng Biotechnol. 2021;9:1-15.
Crossref - Zhang J, Su C, Kong XL, et al. Directed evolution driving the generation of an efficient keratinase variant to facilitate the feather degradation. Bioresour Bioprocess. 2022;4.
Crossref - Lateef A, Adelere IA, Gueguim-Kana EB. Bacillus safensis LAU 13: A new source of keratinase and its multi-functional biocatalytic applications. Biotechnol Biotechnol Equip. 2015;29(1):54-63.
Crossref - Almahasheer AA, Mahmoud A, El-Komy H, et al. Novel feather degrading keratinases from bacillus cereus group: Biochemical, genetic and bioinformatics analysis. Microorganisms. 2022;10(1):1-27.
Crossref - Karn N, Karn SK. Evaluation and Characterization of Protease Production by Bacillus Sp . Induced By UV-Mutagenesis. 2014;3(1):3-7.
Crossref - Duarte TR, Oliveira SS, Macrae A, et al. Increased expression of keratinase and other peptidases by Candida parapsilosis mutants. Braz J Med Biol Res. 2011;44(3):212-6.
Crossref - Fang Z, Yong YC, Zhang J, Du G, Chen J. Keratinolytic protease: a green biocatalyst for leather industry. Appl Microbiol Biotechnol. 2017;101(21):7771-7779.
Crossref - Bertsch A, Coello N. A biotechnological process for treatment and recycling poultry feathers as a feed ingredient. Bioresour Technol. 2005;96(15):1703-1708.
Crossref - Nie L, Zhang R, Zhang L, et al. Mutations in the regulatory regions result in increased streptomycin resistance and keratinase synthesis in Bacillus thuringiensis. Arch Microbiol. 2021;203(9):5387-5396.
Crossref - Odetallah NH, Wang JJ, Garlich JD, Shih JCH. Keratinase in starter diets improves growth of broiler chicks. Poult Sci. 2003;82(4):664-670.
Crossref - Aina A, Ezeamagu CO, Akindele ST, Aleshinloye AO. Purification of Wickerhamomyces anomalus Keratinase and Its Prospective Application in Poultry Feed Industries. Fountain J Nat Appl Sci. 2021;10(1):1-12.
Crossref - Xu KL, Gong GX, Liu M, et al. Keratinase improves the growth performance, meat quality and redox status of broiler chickens fed a diet containing feather meal. Poult Sci. 2022;101(6):101913.
Crossref - Ayanwale BA, Egwim EC, Alemede IC, et al. Growth performance and meat quality of broiler chickens on diets containing Keratinase-treated and untreated feather meal-based diets. Acta Agric Scand A Anim Sci. 2023;1-11.
Crossref - Brandelli A, Daroit DJ, Riffel A. Biochemical features of microbial keratinases and their production and applications. Appl Microbiol Biotechnol. 2010;85(6):1735-1750.
Crossref - Chaturvedi V, Bhange K, Bhatt R, Verma P. Production of kertinases using chicken feathers as substrate by a novel multifunctional strain of Pseudomonas stutzeri and its dehairing application. Biocatal Agric Biotechnol. 2014;3(2):167-174.
Crossref - Akram F, Haq I ul, Jabbar Z. Production and characterization of a novel thermo- and detergent stable keratinase from Bacillus sp. NKSP-7 with perceptible applications in leather processing and laundry industries. Int J Biol Macromol. 2020;164:371-383.
Crossref - Ben Elhoul M, Zaraî Jaouadi N, Bouacem K, et al. Heterologous expression and purification of keratinase from Actinomadura viridilutea DZ50: feather biodegradation and animal hide dehairing bioprocesses. Environ Sci Pollut Res. 2020.
Crossref - Rajput R, Sharma R, Gupta R. Biochemical characterization of a thiol-activated, oxidation stable leratinase from Bacillus pumilus KS12. Enzyme Res. 2010.
Crossref - Paul T, Das A, Mandal A, et al. An efficient cloth cleaning properties of a crude keratinase combined with detergent: Towards industrial viewpoint. J Clean Prod. 2014;66:672-684.
Crossref - Parmar BN, Trivedi SH. Production of microbial keratinase using newly isolated strain of Stenotrophomonas maltophilia its characterization and applications. Kuwait J Sci. 2021;48(4):1-14.
Crossref - Duffeck CE, de Menezes CLA, Boscolo M, da Silva R, Gomes E, da Silva RR. Citrobacter diversus-derived keratinases and their potential application as detergent-compatible cloth-cleaning agents. Brazilian J Microbiol. 2020;51(3):969-977.
Crossref - Yue XY, Zhang B, Jiang DD, Liu YJ, Niu TG. Separation and purification of a keratinase as pesticide against root-knot nematodes. World J Microbiol Biotechnol. 2011;27(9):2147-2153.
Crossref - Shalaby MM, Samir R, Goma FAZM, Rammadan MA. Enhanced fusidic acid transdermal delivery achieved by newly isolated and optimized Bacillus cereus Keratinase. Biotechnol Reports. 2021.
Crossref - Tripathi M, Kumar S, Sonker SK, Babbar P. Occurrence of author keywords and keywords plus in social sciences and humanities research : A preliminary study. Collnet J Sci Inf Manag. 2018;12(2):215-232.
Crossref - Nnolim NE, Nwodo UU. Microbial keratinase and the bio-economy: a three-decade meta-analysis of research exploit. AMB Express. 2021;11(1).
Crossref - Chinchilla-Rodríguez Z, Miao L, Murray D, Robinson-García N, Costas R, Sugimoto CR. A Global Comparison of Scientific Mobility and Collaboration According to National Scientific Capacities. Front Res Metrics Anal. 2018;3:1-14.
Crossref - Martinez JPDO, Cai G, Nachtschatt M, et al. Challenges and opportunities in identifying and characterising keratinases for value-added peptide production. Catalysts. 2020;10(2).
Crossref - Gopinath SCB, Anbu P, Lakshmipriya T, et al. Biotechnological Aspects and Perspective of Microbial Keratinase Production. Biomed Res Int. 2015;1-10.
Crossref - Kim J-D. Purification and Characterization of a Keratinase from a Feather-Degrading Fungus, Aspergillus flavus Strain K-03. Mycobiology. 2010;35(4):219.
Crossref - Emran MA, Ismail SA, Abdel-Fattah AM. Valorization of feather via the microbial production of multi-applicable keratinolytic enzyme. Biocatal Agric Biotechnol. 2020;27:101674.
Crossref - Moridshahi R, Bahreini M, Sharifmoghaddam M, Asoodeh A. Biochemical characterization of an alkaline surfactant-stable keratinase from a new keratinase producer, Bacillus zhangzhouensis. Extremophiles. 2020;24(5):693-704.
Crossref - Meng Y, Tang Y, Zhang X, Wang J, Zhou Z. Molecular Identification of Keratinase DgokerA from Deinococcus gobiensis for Feather Degradation. Appl Sci. 2022;12(1):1-12.
Crossref - Zhang RX, Gong JS, Su C, et al. Recombinant expression and molecular engineering of the keratinase from Brevibacillus parabrevis for dehairing performance. J Biotechnol. 2020;320(1800):57-65.
Crossref - Li J, Chen D, Yu Z, Á PÁS. Improvement of expression level of keratinase Sfp2 from Streptomyces fradiae by site-directed mutagenesis of its N -terminal pro-sequence. Biotechnology Letters. 2013:743-749.
Crossref - Ye JP, Gong JS, Su C, et al. Fabrication and characterization of high molecular keratin based nanofibrous membranes for wound healing. Colloids Surfaces B Biointerfaces. 2020;194:111158.
Crossref - Fang Z, Zhang J, Du G, Chen J. Improved catalytic efficiency, thermophilicity, anti-salt and detergent tolerance of keratinase KerSMD by partially truncation of PPC domain. Sci Rep. 2016;6(October 2015):1-11.
Crossref - Fang Z, Sha C, Peng Z, Zhang J, Du G. Protein engineering to enhance keratinolytic protease activity and excretion in Escherichia coli and its scale-up fermentation for high extracellular yield. Enzyme Microb Technol. 2019;121:37-44.
Crossref - Peng Z, Zhang J, Song Y, Guo R, Du G, Chen J. Engineered pro-peptide enhances the catalytic activity of keratinase to improve the conversion ability of feather waste. Biotechnol Bioeng. 2021;118(7):2559-2571.
Crossref - Sharma R, Murty NAR, Gupta R. Molecular characterization of N-terminal pro-sequence of keratinase ker P from Pseudomonas aeruginosa: Identification of region with chaperone activity. Appl Biochem Biotechnol. 2011;165(3-4):892-901.
Crossref - Tian J, Long X, Tian Y, Shi B. Enhanced extracellular recombinant keratinase activity in: Bacillus subtilis SCK6 through signal peptide optimization and site-directed mutagenesis. RSC Adv. 2019;9(57):33337-33344.
Crossref - Rajput R, Tiwary E, Sharma R, Gupta R. Swapping of pro-sequences between keratinases of Bacillus licheniformis and Bacillus pumilus: Altered substrate specificity and thermostability. Enzyme Microb Technol. 2012;51(3):131-138.
Crossref
© The Author(s) 2023. Open Access. This article is distributed under the terms of the Creative Commons Attribution 4.0 International License which permits unrestricted use, sharing, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.