ISSN: 0973-7510
E-ISSN: 2581-690X
Polar microbiology remains as the most fascinating area of research which mainly focuses on exploration of psychrophilic organisms for having their cold-active enzymes of biotechnological potential. In this study, we have explored a culturable bacterial community and isolated 27 bacterial isolates with a different morphology from an unexplored site of Arctic region, for the possibility of identifying various active biomolecules. Screening of various isolates in a culture dependent manner helped us to identify strains capable of producing extracellular enzymes. The optimal growth parameters of most of the isolates are ranges between 18-22°C temperature, 3-5 days of incubation, 6-9 pH, and 3-5% (w/v) NaCl in LB media. It has also been found that among these isolates, 63% are able to produce lipase, 17% amylase, 7% xylanase and 7% isolates have responded for phosphatase activity but there are no isolates found for gelatinase and cellulase production ability. In addition, few isolates can also produce secretory protease, urease, β-galactosidase, etc. 16SrRNA gene sequence-based phylogeny revealed that the isolates belong to the genera of Psychrobacter, Planococcus, Halomonas, Arthrobacter, Oceanisphaera, Marinbacter, Pseudomonas, Algoriphagus. Strikingly, none of the Arctic isolates showed resistance towards commonly used antibiotics which indicates that the unexplored habitat is devoid of antibiotic exposure and so does the rise of antimicrobial resistance. The structure-function relationship of the isolated bioactive compounds from these isolates are the major focus of future research.
Psychrotrophs, Arctic, Community Analysis, Polar Microbiology, Lipase, Pigment, Cold-active enzymes
The cold ecosystems cover almost three-fourth of the earth’s surface, and this has been colonized by low temperature loving extremophilic microbes.1 It is estimated that only 1% of the total bacterial biodiversity is culturable in the laboratory using different media. Diverse cold-adapted microorganisms i.e., psychrophiles have recently been found in different low-temperature environments, and their enzymes have attracted the interest of the scientists for their potentials in modern biotechnology.2 Thereby, contributing economic and ecological benefits like energy saving, minimizing undesirable chemical reactions that occur at high temperature, cold-active enzymes which can be neutralized easily by heating when required. Enzymes that are active in low temperatures show a key role in psychrophiles to adapt to lower temperatures and remain as the primary importance to scientists owing to their application potential in industries.3 Psychrophiles can produce cold-active enzymes4 that are structurally flexible and thus have high complementarity between the active site and the substrate resulting in high catalytic activity. Lipases are ubiquitously found among prokaryotes and eukaryotes and are studied extensively, but less consideration has been paid to psychrotrophic bacterial lipases.3 Lipase produced by cold-loving bacteria can be used for their applications in food processing, bioremediation in cold environment, etc.5 These are also useful as detergent additives for cold washing to protect delicate fabrics.6 Pigments are another class of important biomolecules produced by psychrotrophs. The use of bio-natural pigments is much safer, as these are harmless, biodegradable and noncarcinogenic.7 The global trends are rapidly shifting towards the usage of daily commodities which are natural and eco-friendly. The application of natural dyes is rising gradually among nutraceuticals, pharmaceuticals, cosmetics and dye industries. Excitingly, various environmental waste-materials have also been used for such pigment production, in a better sustainable manner.8 Natural pigments of bacterial origin are preferable over the pigments of plant origin due to higher solubility, stability and easy production by culturing the respective bacteria.9 Pigments are not just coloring agents; they also possess different chemical structures with diverse biological activities.10 Other than bacteria, many micro-algae and fungi remain as the alternatives for production of such natural pigments.8,9,11-13 Other life forms in hostile cryosphere remain impossible in past but there are diverse microbial communities present for a long time and remain as the major candidate to build the glacial ecosystem.14,15 However the microbial communities living in such cold environments are exposed all the time with various radiations, low temperatures, higher salinity, nutritional deficiency and repeated freeze-thaw cycles. Thus, the pigments can also be able to protect the producing organisms from different ecological stresses. These can well be used as a source of energy16 and also are the tools to fight against stress,17 oxidants, extreme temperature and desiccation,18 and for the safety against UV irradiation.19 Sometimes such molecules can be used as anti-microbials to protect from the neighboring organisms and sometimes as a protective shield to evade antibiosis. There are many reports on diverse group of pigment molecules produced from microorganisms residing in glaciers, ice cores, and marine surface waters and found to be essential to the microorganisms to overcome environmental stress.20-23 Psychrophiles can undergo morphological change and produce various metabolites for their protection while growing in the hostile environment. Carotenoid is one of such examples that is produced by many organisms as a protective mechanism for their life in cryospheric environments.24 In this study, we screened psychrotophic bacterial strains isolated from NyAlesund (78˚55″N, 11˚54”E), Svalbard, Norway for exploration of bacterial diversity in focus of few novel isolate identification which are industrially important for their active biomolecule production.
Collection, pre-treatment and assessment of soil sample
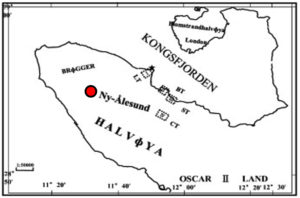
Collection of the soil samples has been done from different spots of Ny Alesund (latitude: 78˚55″N, longitude: 11˚54”E), Norway (Fig. 1). Soil samples were stored at -20˚C for analyzing their physiochemical attributes in future. The estimation of electrical conductivity,25 pH, organic carbon, total organic matter following titration method,26 available phosphorous has been carried out following the procedure explained in Sarkar and Haldar.27 Flame photometer has been implemented to measure the total potassium available.27 Available soil potassium was extracted by using CH3COONH4 and filtered by the Whatman paper. Based on the principle of emission spectrometry the sample was analysed using the flame photometer.
Isolation of bacteria
Soil samples were diluted up to 10-5 and 0.1 mL of this was spread on LA (in 1 L, tryptone 10 g, sodium chloride 10 g, yeast extract 5 g, agar 15 g, pH 7.0); R2A medium (for 1 L medium, agar 20 g, pH 7.3) and Zobell Marine agar medium (for 1 L, agar 15 g, pH 8.1) with 50 µg/mL of nystatin or cycloheximide supplementation as antifungal agent.28 Inoculated plates were incubated for 3-5 days at 18-22°C. Individual colony appeared upon incubation were selected by their morphology and/or developed colour and to get pure culture streaked on fresh plate.
Characterization of bacteria
Morphological and cultural characteristics
Gram-staining properties and the colony morphology based on the colony shape, size, margin, texture, optical properties were investigated after incubating each culture in LA medium for 4 days. For scanning electron microscopy, the freshly grown colonies were placed on cover-slip and fixed for 5-20 min with 0.1 M cacodylate buffer with the pH 7.2 at 4˚C having 0.5% glutaraldehyde. This further has been washed for 30 min with 0.1 M cacodylate buffer, and the colonies were postfixed in 1 % OsO4 for 2-3 min at 4˚C29 followed by dehydration with alcohol at ambient temperature. The morphology of these isolates was visualized through ZEISS EVO 18 scanning electron microscopy. For negative staining, transmission electron microscopy the water-suspended bacterial colonies were transferred on copper grids and with the same volume of an aqueous solution of 0.5 % w/v uranyl acetate, it has been stained negatively.30 In room temperature the sample were air dried and examined under electron microscope (JEOL JEM 2100). The optimal growth parameters of the isolates were determined in the range of temperature (4°C to 30°C), pH (4 to 10), and NaCl (3% to 15%).
Analysis of biochemical characters of the isolates
Biochemical characterization of each isolates were determined based on their mixed acid fermentation (methyl red, Voges-Proskauer), ability to produce indole, utilization of citrate, catalase production abilities and also on abilities to reduce nitrate, formation of H2S, ability to utilize different carbon-sources (ribose, glucose, lactose, sucrose, pyruvate, D-fructose, D-xylose, D-maltose, D-galactose, D-arabinose, D-mannose, D-adonitol, D-raffinose, D-melizitose, L-rhamnose, melibiose, inositol, and dulcitol) following standard protocol.31,32 The utilization ability of nitrogenous source (L-asparagines, L-arginine, and L-alanine) of these bacteria were checked following standard protocol.33,34
Assessment for biomolecule production ability
The bacterial isolates were tested for their capability to produce exoenzymes like lipase, amylase, cellulose and xylanase. Xylan and cellulose degradation were detected by using congo-red overlay method35 after streaking the pure colonies on solid media that contains either xylan or cellulose (1% xylan or 1% CM cellulose supplemented with 0.3% beef extract and also 2% agar in order to detect the xylanase or cellulase). To perform this, the respective plates were submerged with 0.1% congo red aqueous solution for about 10 min and then washed with 1M NaCl. The lipid degrading ability were assayed using tributyrin or olive oil (10 mL/L) containing agar media.36 Degradation of xylan, cellulose, and lipid was visualized by the clear-zone that appeared around the developed bacterial colony. Similarly, the gelatinase assay was performed on the nutrient-gelatin agar. Urease function was tested in medium containing urea in it. The modified starch agar media (consist of 0.3% beef-extract, 1% starch and 2% agar) has been used for checking the starch degradation ability confirmed by using the solution of Gram’s iodine. The skim-milk containing agar media has been used to test the caseinase secretion ability.34 Pikovakaya’s agar medium (Himedia) was used for identifying the bacteria which have phosphate solubilizing ability.
Genomic DNA isolation
Individual bacterium grew independently in 5 mL Luria broth for 3-4 days at 20°C in shaking conditions. Genomic DNA from all the bacterial culture were extracted by phenol-chloroform method.37
16S rDNA gene sequence based phylogenetic analysis
16S rDNAs of each isolate were PCR amplified using the 8F (5’-AGAGTTTGATCCTGGCTCAG-3’) and 1492R (5’-ACGGTTACCTTGTTACGACTT-3’)38 universal primer. After purification of the 16S rDNA PCR amplicon, they were sequenced following Sanger’s method of DNA sequencing. Sequence of both the DNA stands were aligned and arranged by using the Bioedit software (http://bioedit.software.informer.com) and recovered the 16S rDNA complete gene sequence of each isolate. The sequences were uploaded at EZbiocloud server (https://www.ezbiocloud.net/) to get the information of closely related strains. The output file has also been compared through the BLASTn program of NCBI (https://blast.ncbi.nlm.nih.gov). The sequences of the type strains were collected from the EZbiocloud server39 and multiple sequence alignments were performed using the clustal W program of Mega7.40 Through the neighbor joining (NJ) method,41 the phylogenetic tree were built and the evolutionary distances were calculated by Kimura 2 parameter model.42 Bootstrap values have been derived from the resampling at 1000 times.
Assay for antibiotic sensitivity
Widely used standard antibiotic have been used to screen the sensitivity of the isolated strains towards each antibiotic. Antibiotics included in this study were inhibitors of bacterial cell-wall like Penicillin G (10 U), Carbenicillin (10 µg), Amoxocillin/Clavulinic acid (20/10 µg), Aztreonam (30 µg), Cefazolin (30 µg), Cefoperazone (75 µg), Cefoxitin, Cefuroxime (30 µg) and Cefepime (30 µg), Mezlocillin (75 µg), Vancomycin (30 µg); outer membrane inhibitor like Polymyxin B (30 µg) and Colistin (10 µg); nucleic acid inhibitors: Ciprofloxacin (5 µg), Norfloxacin (10 µg), Levofloxacin (5 µg), Ofloxacine (5 µg), Co-trimoxazole (25 µg), Novobiocine (30 µg), Rifampicin (30 µg), and Nitrofurantoin (30 µg); protein synthesis inhibitor: Clindamycin (2 µg), Tobramycin (10 µg), Oleandomycin (15 µg), Lincomycin (15 µg), Chloramphenicol (30 µg), Tetracyclin (30 µg), Gentamycin (10 µg), Amikacin (30 µg) and Streptomycin (25 µg). On Muller Hinton agar plate 100 µL of fresh inoculum were spreaded using a spreader and incubated at 20°C for 3-4 days after placing the antibiotic discs on each. Sensitivity to any antibiotic was determined by the diameter of the zone of inhibition. Bacterial isolates were assigned either as resistant or sensitive towards used antibiotics.
Accession numbers of isolates
The sequences of 16S rRNA gene of 24 isolates were deposited in NCBI and the accession numbers are mentioned in Table 1.
Table (1):
List of culture dependent strains isolated from the Arctic soil.
Strain isolated |
Accession numbers |
Optimal growth temperature (℃) |
Appearance of visible colonies (days) |
|---|---|---|---|
RSAP1 |
MH348974 |
20-22 |
5 |
RSAP6 |
MH348975 |
4-10 |
4 |
RSAP7 |
MH348976 |
4-10 |
4 |
RSAP8 |
MH348977 |
15-18 |
5 |
RSAP9 |
MH348978 |
15-18 |
3 |
RSAP10 |
MH348979 |
15-18 |
3 |
RSAP11 |
MH348980 |
10-15 |
3 |
RSAP12 |
MH348981 |
15-18 |
3 |
RSAP13 |
MH348982 |
18-20 |
4 |
RSAP14 |
MH348983 |
18-20 |
4 |
RSAP15 |
MH348984 |
15-18 |
3 |
RSAP18 |
MH348985 |
15-18 |
3 |
RSAP19 |
MH348986 |
10-15 |
4 |
RSAP21 |
MH348987 |
15-18 |
3 |
RSAP23 |
MH348988 |
10-15 |
4 |
RSAP24 |
MH348989 |
20-22 |
4 |
RSAP25 |
MH348990 |
20-22 |
3 |
RSAP26 |
MH348991 |
20-22 |
3 |
RSAP27 |
MH348992 |
10-15 |
2 |
RSAP17 |
MK391379 |
20-22 |
3 |
RSAP22 |
MK391380 |
20-22 |
4 |
RSAP16 |
MK391381 |
20-22 |
5 |
RSAP8 |
MK391382 |
15-18 |
3 |
RSAP2 |
MW346644 |
20-22 |
5 |
Description of sample collection site
The sample collection sites are from Arctic region which is absolutely free from anthropogenic disturbances. The blackish-coloured soils are having moderate moisture with granular forms i.e., not muddy or dough. General characters of the Arctic samples, where from the bacterial isolates have been cultured, are found to be alkaline (pH 8.1). The soil conductivity is 0.06528 ds/m which indicates it as a non-saline soil. The total organic substance, organic carbon, total potassium and phosphorous were 0.9076%, 0.526%, 58 ppm, 0.4775 kg ha-1, respectively (Table 2) indicating the presence of relatively low organic carbon, phosphorus and potassium content in the soil sample. In general, as there is a very low level of organic matter present, the diversity of microbial community might be affected due to very high competition among organisms for the limited source of carbon. Thus, the possibility of prevalence of fastidious organisms and diversity in the bacterial community are also under challenge.
Table (2):
Characteristic of soil and its elemental composition.
Soil characters |
Description |
|---|---|
Soil color |
Blackish |
Moisture content |
Moderate |
pH |
8.1 |
Soil conductivity |
0.6528 ds/m |
Organic C (%) |
0.526 |
Total organic matter (%) |
0.9076 |
Total phosphorous |
0.4775 kg ha-1 |
Total Potassium |
58 ppm |
Bacterial community analyses
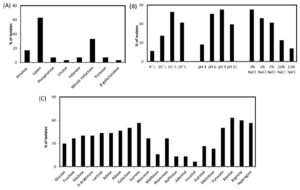
Demand of cold-adaptive enzymes for probable industrial applications are always high. Drive on the discovery of new enzymes from psychrophilic or psychtrophic bacteria are even more promising. Hence for searching novel bacterial isolates from such unexplored region increase the possibility to get new enzymes and useful bioactive-molecules. Among those isolated strains few are able to produce pigment also. A total of 27 bacteria were isolated. Among the used media for culturing these isolates, the most diversities in colony morphology were found in LB media (Fig. 2). Analysis based on the 16S rRNA sequence predicted that among the total isolates, 5 are from the genus Psychrobacter, 3 from genus Planococcus, 2 from genus Halomonas, 2 from genus Arthrobacter, 2 from genus Oceanisphaera, 2 from genus Marinbacter, 2 from genus Pseudomonas, and 2 from the genus Algoriphagus. Among the 27 isolates, 5 colonies exhibited pigment while growing on LA medium (Fig. 2). Many isolates have potential secretory exoenzymes and they could essentially open significant industrial application. Most of the isolates showed the secretion of lipase, cellulase and amylase while some showed the ability to produce substantial urease and xylanase enzyme. From primary screening, it has been found that out of 27 isolates, none of the isolates represents any anti-microbial activity. Out of 27 isolates, 20 isolates produce lipase enzyme and 10, 5, 2, 2, 2,1 and 1 isolates produce nitrate reductase, amylase, xylanase, protease, phosphatase, β-galactosidase and urease, respectively. We have found that there are 5 isolates of which are capable to produce pigments while grown on respective mediums. Furthermore, the carbon and nitrogen utilization patterns in these isolates are different (Fig. 3C). Most of the isolates are sensitive towards almost all the antibiotics we have tested. Once again it indicates that the environment which is devoid of any disturbance of human civilization and has not been exposed to the antibiotics are having bacterial flora that are sensitive to most of the existing antibiotics. The rise of antimicrobial resistance is almost nil in microbes of such environment and this is a clear indication that the rise of antimicrobial resistance is aggravated among microbes which are exposed to various antibiotics. 23 isolates are able to grow in 15°C, 12 isolates in 10°C, and 18 isolates can grow in 20°C however the optimum temperature of their growth has been recorded as 20°C±2°C. The level of tolerance of 24 isolates is 3% NaCl, 6 isolates up to 15% NaCl and 18 isolates up to 7% NaCl (Fig. 3B). The isolates can even grow in absence of NaCl, and also the majority of these can survive in as high as 3% NaCl and rationalize themselves as mild halotolerant. The total 8 isolates are able to grow in pH 4. However, most isolates favor to grow under the media condition of neutral to alkaline pH 6 to 9 (Fig. 3B).
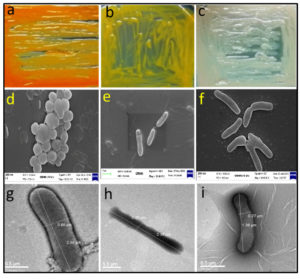
Fig. 2. Morphological appearance of random representative isolates. Upper panel represents the appearance of representative bacterial colonies on LA medium, middle panel represents the scanning electron micrograph and the lower panel represents the transmission electron micrograph. (a) RSAP19, (b) RSAP10, (c) RSAP25, (d) RSAP19, (e) RSAP21, (f) RSAP1, (g) RSAP22, (h) RSAP17 and (i) RSAP16.
Fig. 3. Production ability of biomolecules and growth parameters of the isolates. (a) Production of different enzymes by isolated bacteria; (b) Physiological parameter of isolated bacteria; (c) Carbon and nitrogen utilization pattern of isolated bacteria.
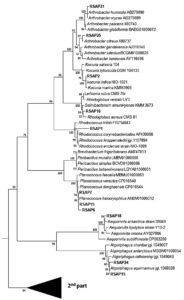
The BLASTn analysis with 16S rDNA sequence revealed that the isolates are mostly between the range of 98-100% sequence similarity. Five isolates were found to have 99% sequence similarity with the genus Psychrobacter, three isolate with Planococcus, 2 isolates are 99% similar to Arthrobacter and Halomonas, 2 isolates are having 98% similarity with Marinobacter and Algoriphagus sp. About 63% of these isolates are able to produce lipase enzymes. Three isolates have the potential to be novel species as these are showing unique morphological, biochemical and molecular characteristics. As for example, on isolate shows less than 16S rDNA sequence 98.7% similarity and can be claimed as new species following the modern trend of bacterial systematics.43 Whereas 3 isolates which showed 16S rDNA sequence similarity between 98.7 -99 % also likely to be a new bacterial species which needs to be validated upon exploring their genome sequence and chemotaxonomic features. Most of the isolates fit within a monophyletic group (which shows close-similarity when compared with their nearest ancestors) based on both maximum likelihood (ML) and neighbour joining (NJ) phylogeny. The 16S rDNA linear sequence-based neighbour joining tree were shown in Fig. 4 where only >50% bootstrap values (from 1000 replicates) are mentioned at the branch nodes.
Demand of novel enzymes, pigments and different biomolecules are always high. The new enzymes and pigment discovery always remained a primary thrust area for scientists. Thus, the search for novel bacteria from the under-explored Arctic region should have a higher probability to obtain new enzymes and bioactive molecules from them. 27 bacteria were isolated using three media and most diverse strains with enzyme and pigment produce were found in LA media. Among the isolates, 20 isolates produce lipase enzyme; 10, 5, 2, 2, 2 and 1 isolates produce nitrate reductase, amylase, xylanase, protease, phosphatase and urease, respectively. Each of these enzymes is supposed to be the cold-active one as the source organisms survived in the hostile environment of the Arctic region. The phylogenetic analysis, based on the 16S rRNA gene sequence, indicated that among the culturable bacteria we isolated, 5 species of Psychrobacter, 3 Planococcus, 2 Halomonas, 2 Arthrobacter, 2 Oceanisphaera, 2 Marinbacter, 2 Pseudomonas, 2 species of Algoriphagus from LB media. The detailed understanding of few isolates were found to be unique based on their morphological, biochemical and molecular characterization. Among these novel isolates, RSAP17 is a potent lipase producer hence its lipase enzyme production, purification and characterization have further been studied. Lipases have the potential for application in the biotechnological industry in diverse fields of food processing, detergent additives, biodegradation of unwanted wastes from cold environments. Another isolate has been identified as Kocuria sp. RSAP2, produces profuse yellow colour pigment. The 16S rDNA-phylogeny and the morpho-biochemical investigation also predicts the strain RSAP2 under the genus Kocuria. We would further analyze the pigment production from the isolate RSAP2 using different media with a varied range of temperatures, pH, concentration of NaCl and time of incubation in order to optimize the production of the pigment. Cold active extracellular enzymes and other non-enzymatic biomolecules produced by different psychrotrophs are important for their possible industrial applications. Moreover, the knowledge database helps to understand the eco-physiological role of these bacteria in their extreme habitat.
ACKNOWLEDGMENTS
The authors are grateful to Dr. Pulak Kumar Maiti for his suggestion on analyzing the phylogeny of the strains.
CONFLICT OF INTEREST
The authors declare that there is no conflict of interest.
AUTHORS’ CONTRIBUTION
RU performed all the experiments, interpreted data and prepared the manuscript. PR helped in procuring sample and edited the manuscript. SM conceptualized the research, designed the experiments, wrote and edited the manuscript.
FUNDING
None.
ETHICS STATEMENT
This article does not contain any studies with human participants or animals performed by any of the authors.
AVAILABILITY OF DATA
The datasets generated and analysed during the current study are available from the corresponding author on reasonable request.
- Feller G, Gerday C. Psychrophilic enzymes: hot topics in cold adaptation. Nat Rev Microbiol. 2003;1(3):200-208.
Crossref - Cavicchioli R, Charlton T, Ertan H, Omar SM, Siddiqui KS, Williams TJ. Biotechnological uses of enzymes from psychrophiles. Microb Biotechnol. 2011;4(4):449-460.
Crossref - Salwoom L, Raja Abd Rahman RNZ, Salleh AB, et al. Isolation, Characterisation, and Lipase Production of a Cold-Adapted Bacterial Strain Pseudomonas sp. LSK25 Isolated from Signy Island, Antarctica. Molecules. 2019;24(4):715.
Crossref - Srinivas TNR, Rao SSSN, Reddy PVV, et al. Bacterial diversity and bioprospecting for cold-active lipases, amylases and proteases, from culturable bacteria of Kongsfjorden and Ny-A°lesund, Svalbard, Arctic. Curr Microbiol. 2009;59:537-547.
Crossref - Maiangwa J, Ali MS, Salleh AB, Rahman RN, Shariff FM, Leow TC. Adaptational properties and applications of cold-active lipases from psychrophilic bacteria. Extremophiles. 2015;19(2):235-247.
Crossref - Joseph B, Ramteke PW, Thomas G. Cold active microbial lipases: some hot issues and recent developments. Biotechnol Adv. 2008;26(5):457-470.
Crossref - Cristea D, Vilarem G. Improving light fastness of natural dyes on cotton yarn. Dyes and Pigments. 2006;70(3):238-245.
Crossref - Joshi VK, Attri D, Bala A, Bhushan S. Microbial pigments. Indian J Biotechnol. 2003;2:362-369.
- Narsing Rao MP, Xiao M, Li WJ. Fungal and Bacterial Pigments: Secondary Metabolites with Wide Applications. Front Microbiol. 2017;8:1113.
Crossref - Kim SB, Nedashkovskaya OI, Mikhailov VV, et al. Kocuria marina sp. nov., a novel actinobacterium isolated from marine sediment. Int J Syst Evol Microbiol. 2004;54(5):1617-1620.
Crossref - Choi SY, Yoon KH, Lee JI, Mitchell RJ. Violacein: Properties and Production of a Versatile Bacterial Pigment. Biomed Res Int. 2015;2015:465056.
Crossref - Pandey N, Jain R, Pandey A, Tamta S. Optimisation and characterisation of the orange pigment produced by a cold adapted strain of Penicillium sp. (GBPI_P155) isolated from mountain ecosystem. Mycology. 2018;9(2):81-92.
Crossref - Ramesh C, Vinithkumar NV, Kirubagaran R. Marine pigmented bacteria: a prospective source of antibacterial compounds. J Nat Sc boil Med. 2019;10:104-113.
Crossref - Anesio AM, Lutz S, Chrismas NAM, Benning LG. The microbiome of glaciers and ice sheets. NPJ Biofilms Microbiomes. 2017;3:10.
Crossref - Anesio AM, Laybourn-Parry J. Glaciers and ice sheets as a biome. Trends Ecol Evol. 2012;27(4):219-225.
Crossref - Madigan MT, Martinko JM, Stahl DA, Clark DP. Brock biology of microorganisms, 13th edn. Pearson Education, Inc, San Francisco. 2012.
- Martin-Cerezo ML, Garcia-Lopez E, Cid C. Isolation and Identification of a Red Pigment from the Antarctic Bacterium Shewanella frigidimarina. Protein Pept Lett. 2015;22(12):1076-1082.
Crossref - Wada N, Sakamoto T, Matsugo S. Multiple roles of photosynthetic and sunscreen pigments in cyanobacteria focusing on the oxidative stress. Metabolites. 2013;3(2):463-483.
Crossref - Becker-Hapak M, Troxtel E, Hoerter J, Eisenstark A. RpoS dependent overexpression of carotenoids from Erwinia herbicola in OXYR deficient Escherichia coli. Biochem Biophys Res Commun. 1997;239(1):305-309.
Crossref - Foght J, Aislabie J, Turner S, et al. Culturable bacteria in subglacial sediments and ice from two Southern Hemisphere glaciers. Microb Ecol. 2004;47(4):329-340.
Crossref - Lu Y, Wang L, Xue Y, et al. Production of violet pigment by a newly isolated psychrotrophic bacterium from a glacier in Xinjiang, China. Biochemical Engineering Journal. 2009;43:135-141.
Crossref - Agogue H, Casamayor EO, Bourrain M. A survey on bacteria inhabiting the sea surface microlayer of coastal ecosystems. FEMS Microbiol Ecol. 2005;54(2):269-280.
Crossref - Shen L, Liu Y, Wang N, Jiao N, Xu B, Liu X. Variation with depth of the abundance, diversity and pigmentation of culturable bacteria in a deep ice core from the Yuzhufeng Glacier, Tibetan Plateau. Extremophiles. 2018;22(1):29-38.
Crossref - Vila E, Hornero-Mendez D, Azziz G, Lareo C, Saravia V. Carotenoids from heterotrophic bacteria isolated from Fildes Peninsula, King George Island, Antarctica. Biotechnol Rep. 2019;21:e00306.
Crossref - Jackson ML. Soil Chemical Analysis. Prentice Hall of India Private Limited, New Delhi.1973.
- Walkley A, Black JA. An Estimation of Digestion Method for Determining Soil Organic Matter and a Proposed Modification of Chromic Acid Titration method. Soil Science. 1934;37(1):29-38.
Crossref - Sarkar DK, Haldar A. Physical and Chemical Methods of Soil Analysis. New Age International Publishers, India. 2005.
- Maiti PK, Mandal S. Majority of actinobacterial strains isolated from Kashmir Himalaya soil are rich source of antimicrobials and industrially important biomolecules. Advan Microbiol. 2019;9(3):220-238.
Crossref - Mandal M, Paul S, Uddin MR, Mondal MA, Mandal S, Mandal V. In vitro antibacterial potential of Hydrocotyle javanica Thunb. Asian Pac J Trop Dis. 2016;6(1):54-62.
Crossref - Bozzola JJ, Russell LD. Electron Microscopy: Principles and Techniques for Biologists. Jones and Bartlett, Boston. 1999;670.
- Shirling EB, Gottlieb D. Methods for characterization of Streptomyces species. Int J Sys Bacteriol. 1966;16(3):313-340.
Crossref - Maiti PK, Das S, Sahoo P, Mandal S. Streptomyces sp. SM01 isolated from Indian soil produces a novel antibiotic picolinamycin effective against multi drug resistant bacterial strains. Sci Rep. 2020;10(1):10092.
Crossref - Williams ST, Davies FL. Use of a scanning electron microscope for the examination of Actinomycetes. J Gen Microbiol. 1967;48(2):171-177.
Crossref - Williams ST, Goodfellow M, Alderson G, Wellington EM, Sneath PH, Sackin MJ. Numerical classification of Streptomyces and related genera. J Gen Microbiol. 1983;129(6):1743-1813.
Crossref - Anand AAP, Vennison SJ, Sankar SG, et al. Isolation and characterization of bacteria from the gut of Bombyx mori that degrade cellulose, xylan, pectin and starch and their impact on digestion. J Insect Sci. 2010;10:107.
Crossref - Kumar D, Kumar L, Nagar S, Raina C, Parshad R, Gupta VK. Screening, Isolation and Production of Lipase/Esterase Producing Bacillus sp. Strain DVL2 and Its Potential Evaluation in Esterification and Resolution Reactions. Archives of Applied Science Research. 2012;4:1763-1770.
- Marmur J. A procedure for the isolation of deoxyribonucleic acid from micro-organisms. J Mol Biol. 1961;3(2):208-218.
Crossref - Turner S, Pryer KM, Miao VP, Palmer JD. Investigating deep phylogenetic relationships among cyanobacteria and plastids by small subunit rRNA sequence analysis. J Eukaryot Microbiol. 1999;46(4):327-338.
Crossref - Yoon SH, Ha SM, Kwon S, et al. Introducing EzBioCloud: a taxonomically united database of 16S rRNA gene sequences and whole-genome assemblies. Int J Syst Evol Microbiol. 2017;67(5):1613-1617.
Crossref - Kumar S, Stecher G, Tamura K. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger data sets. Mol Biol Evol. 2016;33(7):1870-1874.
Crossref - Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987;4(4):406-425.
Crossref - Kimura M. A Simple Method for Estimating Evolutionary Rates of Base Substitutions through Comparative Studies of Nucleotide Sequences. J Mol Evol. 1980;16(2):111-120.
Crossref - Stackebrandt E, Ebers J. Taxonomic Parameters Revisited: Tarnished Gold Standards. Microbiol Today. 2006;33:152-155.
© The Author(s) 2022. Open Access. This article is distributed under the terms of the Creative Commons Attribution 4.0 International License which permits unrestricted use, sharing, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.