ISSN: 0973-7510
E-ISSN: 2581-690X
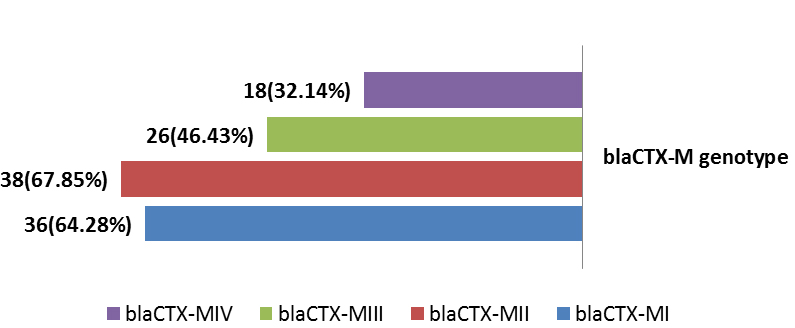
Urinary tract infection (UTI) is the second most common bacterial infection and important public health problem of human among all age groups from neonate to geriatric. Uropathogenic Escherichia coli (UPEC) accounts for approximately 85% of community acquired UTIs and 50% of hospital acquired UTIs. The current study aimed to investigate dominant CTX-M genotypes among local UPEC isolated from patients with cystitis. Phenotypic detection of Cefotaxime-Resistant Uropathogenic Escherichia coli (CRUPEC) were performed by Kirby-Bauer disk diffusion method (CLSI 2016). The DNA were extracted and investigation of four resistance genes blaCTX-M by a multiplex PCR assay using specific primer pairs. The results of phylogenetic subgrouping of UPEC using multiplex PCR to detect chuA (279bp), yjaA (211bp) and TspE4.C2 (152bp) revealed that among 56 CRUPEC 47(83.93%) were ExPEC (37(66.07%) for B23, 6(10.72%) for B22, 3(5.36%) for D1 and 1(1.78%) for D2). InPEC compile 9(16.07%). Genotypic investigation of cefotaxime resistance among 56 CRUPEC were performed using specific four primer pairs to detect the genotypes of blaCTX-M clusters (I, II, III and IV). Among 65 CRUPEC, 36 (64.28%) were positive for for blaCTX-MI, 38(67.85%) for blaCTX-MII 26(46.43%) for blaCTX-MIII and 18(32.14%) for blaCTX-MIV (Figure 9). Concern possessing of isolates for more than one blaCTX-M genotypes were also reported in this study. Our results revealed that blaCTX-M were present in 50 (89.29%) of CRUPEC. Co-existence of more than on genotypes were reported in 38(67.86%). The results displayed that 40(80%) of blaCTX-M positive CRUPEC were belong to group B2, 4(8%) for group D, 3(6%) for group A and 3(6%) for group B1. The current study conclude the presence of blaCTX-M clusters (I, II, III, IV) as a main mechanisim or cefotaxime resistance among CRUPEC and coexistance of multible cluters within same isolates that may leads to emergence of new hybrides of blaCTX-M.
CRUPEC, UTIs, blaCTX-M, Genotypes
Urinary tract infection (UTI) is the one of the most common bacterial infection in humans and a major cause of morbidity and represent an important public health problem of all ages from neonate to geriatric age group (Mazzariol et al., 2017). Among of the most common infectious diseases, second ranking after respiratory tract infection is urinary tract infection which involve about 250 million people in developing countries annually. (Piranfar et al., 2014) . It is classified to bladder infection (cystitis) and kidney infection (pyelonephritis), which can be either symptomatic or asymptomatic (Prakasam et al., 2012). Although diûerent causative agents can be responsible for UTIs, bacteria are the major cause being responsible for more than 95% of UTI cases, The Escherichia coli (E.coli) accounts for approximately 85% of community acquired UTIs and 50% of hospital acquired UTIs (Ahmad et al., 2015). Clermont and colleagues developed a triplex PCR assay to detect the genes chuA, yjaA, and TspE4.C2 in 2000. Regarding the presence/absence of these three genes, an E. coli strain could be classified into one of the main phylogroups intestinal pathogenic E. coli (InPEC) (include A, B1 group) while extraintestinal pathogenic E. coli (ExPEC) ( include B2, or D group) (Clermont et al., 2000). To increase the discrimination power of E. coli population analyses, it has been proposed the use of subgroups A0, A1, B1, B22, B23, D1 and D2, that are determined by the combination of the genetic markers (Escobar-Páramo et al., 2006).
Cefotaxime is one of third generation cephalosporins with broad spectrum activity and regards one of the WHO Model List of Essential Medicines, contains the medications considered to be most effective and safe to meet the most important needs in a health system (WHO, 2015). It is widely used as post-surgery infection prophylaxis and also used for UTIs. Cefotaxime is one of the extend spectrum cephalosporin that resist hydrolysis by b-lactamase but it is sensitive to one of the extended spectrum b-lactamases (ESBLs). The name ‘CTX’ is an abbreviation for ‘cefotaximase’ and refers to the potent hydrolytic activity of these enzymes against cefotaxime. CTX-M-type b-lactamases constitute a relatively small but growing group of ESBLs. Resistance to cefotaxime conferred by blaCTX-M (Bush class A b-lactamases) (Bush and Jacoby, 2010). CTX-M enzyme were common among community acquired and hospital acquired infection with E. coli which regards the most important pathogen producing these enzymes (Coque et al., 2008). Chromosome-encoded genes of intrinsic cefotaximases in Kluyvera spp. are proposed to be the progenitors of CTX-M family (Zhao and Hu, 2013). Most of CTX-Ms exhibit powerful activity against cefotaxime and ceftriaxone and some of them for ceftazidime. The family of CTX-M enzymes is grouped on the basis of similarities in amino acid sequences into four major phylogenetic groups: the CTX-M-I group (CTX-M-1, CTX-M-3, CTX-M-10, CTX-M-12, CTX-M-15, CTX-M-22, CTX-M-23, CTX-M-28, CTX-M-29, CTX-M-30 and CTX- M-32), the CTX-M-II group (CTX-M-2, CTX-M-4, CTX-M-5, CTX-M-6, CTX-M-7, CTX-20), the CTX-M-III group (CTX-M-8), and the CTX-M-IV group (CTX-M-9, CTX-M-13, CTX- M-14, CTX-M-16, CTX-M-17, CTX-M-18, CTX-M-19, CTX-M-21 and CTX-M-27) (Kiiru et al., 2012). The current study aimed to investigate dominant CTX-M genotypes among local UPEC isolated from patients with cystitis.
Bacterial Isolates
Fifty six UPEC isolates with confirmed resistance to cefotaxime (according to CLSI, 2016) were selected to study the phylogenetic subgroups using specific primer pairs to amplify chuA, yjaA and TspE2.C4 genes. Genotyping of blaCTX-M were performed using specific four pairs of primers for blaCTX-M-I, blaCTX-M-II, blaCTX-M-III and blaCTX-M-IV groups.
Extraction of Genomic DNA
Favor PrepTM Genomic DNA Mini Kit (Favorgen/Taiwan) was used to extract genomic DNA from E. coli isolates following the manufacturer’s protocol.
Polymerase Chain Reaction
Fifty-six isolates were screened for the resistance genes CTX-M by a multiplex PCR assay using specific primer pair (Table 1). PCR amplification reactions were performed in a volume of 20 ìl containing. The cycling parameters were as follows: an initial denaturation at 94°C for 2 min; followed by 30 cycles of 94°C for 30s, 55°C for 30s, and 72°C for 30s ; and with a final extension at 72°C for 5 min (Clermont et al., 2000; Kiiru et al., 2012). The amplified PCR products were subjected to electrophoresis at 1.5 % agarose gel in 0.5X TBE buffer.
Table (1):
Primer pairs with amplicon size for blaCTX-M genotyping.
| Target Gene | Primer sequence (5′-3′) | Product (bp) | Annealing (°C) | Ref. | |
|---|---|---|---|---|---|
| chuA | F | GACGAACCAACGGTCAGGAT | 279 | 59 | (Clermont et al., 2000) |
| R | TGCCGCCAGTACCAAAGACA | ||||
| yjaA | F | TGAAGTGTCAGGAGGCGCTG | 211 | 59 | |
| R | ATGGAGAATGCGTTCCTCAAC | ||||
| TSpE4.C2 | F | GAGTAATGTCGGGGCATTCA | 152 | 59 | |
| R | CGCGCCAACAAAGTATTACG | ||||
| bla CTX-M-I | F | GAC GAT GTC ACT GGC TGA GC | 499 | 55°C | (Kiiru et al., 2012 )
|
| R | AGC CG C CGA CGC TAA TAC A | ||||
| bla CTX-M-II | F | GCG ACC TGG TTA ACT ACA ATC C | 351 | 55°C | |
| R | CGG TAG TAT TGC CCT TAA GCC | ||||
| bla CTX-M-III | F | CGC TTT GCC ATG TGC AGC ACC | 305 | 55°C | |
| R | GCT CAG TAC GAT CGA GCC | ||||
| bla CTX-M-IV | F | GCT GGA GAA AAG CAG CGG AG | 474 | 62°C | |
| R | GTA AGC TGA CGC AAC GTC TG | ||||
The results of phylogenetic subgrouping of UPEC using multiplex PCR to detect chuA (279bp), yjaA (211bp) and TspE4.C2 (152bp) revealed that among 56 CRUPEC 47(83.93%) were ExPEC (37(66.07%) for B23, 6(10.72%) for B22, 3(5.36%) for D1 and 1(1.78%) for D2). InPEC compile 9(16.07%) (Table 2). The results in accordance with many similar studies, ExPEC compile 80%-94.82% (Lee et al., 2016; Ochoa et al., 2016; Lara et al., 2017; Al-Khaqani et al., 2017; Salehzadeh and Zamani, 2018). Predominance of B23 subgroup were stated by many studies (Alizade et al., 2014; Merza and Jubrael, 2015; Al-Khafaji and Al-Thahab, 2017).
Table (2):
Distribution of CRUPEC among phylogenetic subgroups.
| Total | No. (%) | chuA/ yjaA/TspE4.C2 | Phylogenic subgroup |
Phylogenic group |
|---|---|---|---|---|
| 9(16.07%) | 2(3.57%) | -ve /-ve/-ve | Subgroup A0 | Group A |
| 2(3.57%) | -ve /+ve/-ve | Subgroup A1 | ||
| 5(8.93%) | -ve /-ve/+ve | B1 | Group B1 | |
| 47(83.93%) | 6(10.72%) | +ve /+ve/-ve | Subgroup B22 | Group B2 |
| 37(66.07%) | +ve /+ve/+ve | Subgroup B23 | ||
| 3(5.36%) | +ve /-ve/-ve | Subgroup D1 | Group D | |
| 1(1.78%) | +ve /-ve/+ve | Subgroup D2 |
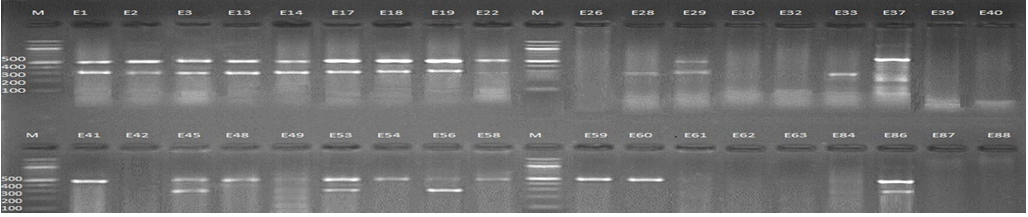
Fig. 1. 1.5% Agarose gel electrophoresis at 72 volt for 90 minutes of PCR to blaCTX-M-I amplicon (499bp) and blaCTX-M-III amplicon (305bp); lane M represent DNA marker size(100bp) while E1-E88 represent the isolates
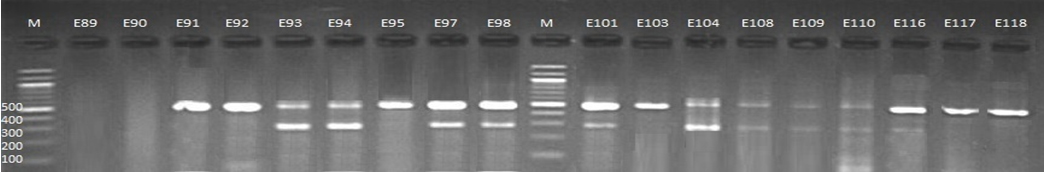
Fig. 2. 1.5% Agarose gel electrophoresis at 72 volt for 90 minutes of PCR to blaCTX-M-I amplicon (499bp) and blaCTX-M-III amplicon (305bp); lane M represent DNA marker size(100bp) while E89-E118 represent the isolates
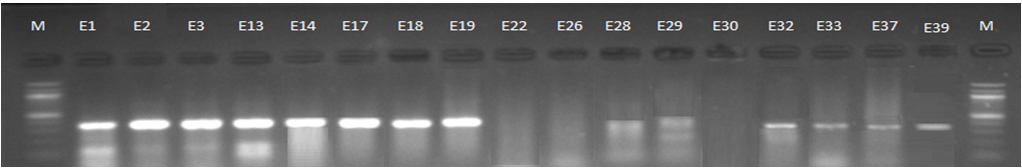
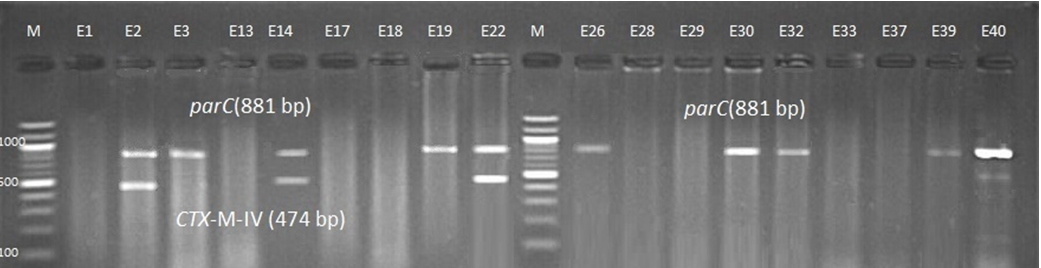
Fig. 3. 1.5% Agarose gel electrophoresis at 72 volt for 90 minutes of PCR to blaCTX-M-II amplicon (351bp); lane M represent DNA marker size(100bp) while E1-E39 represent the isolates
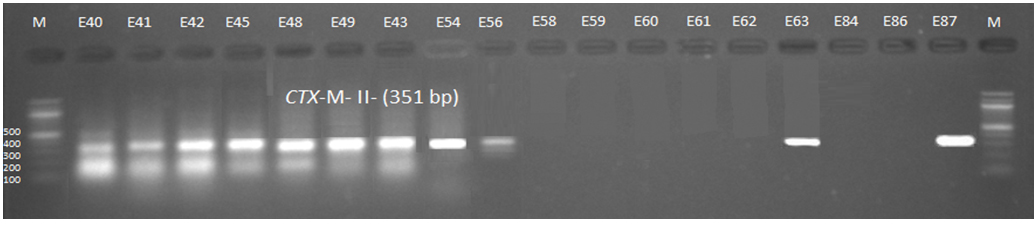
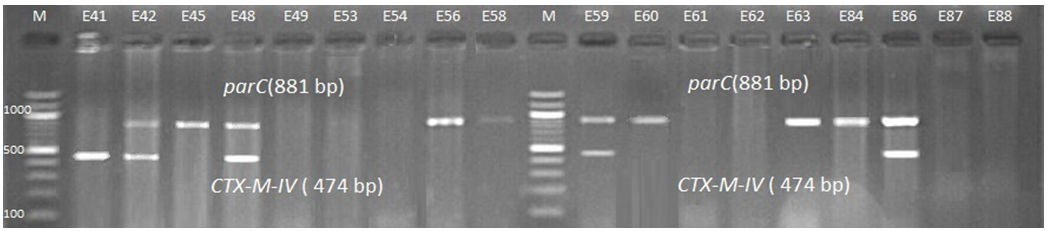
Fig. 4. 1.5% Agarose gel electrophoresis at 72 volt for 90 minutes of PCR to blaCTX-M-II amplicon (351bp); lane M represent DNA marker size(100bp) while E40-E87 represent the isolates
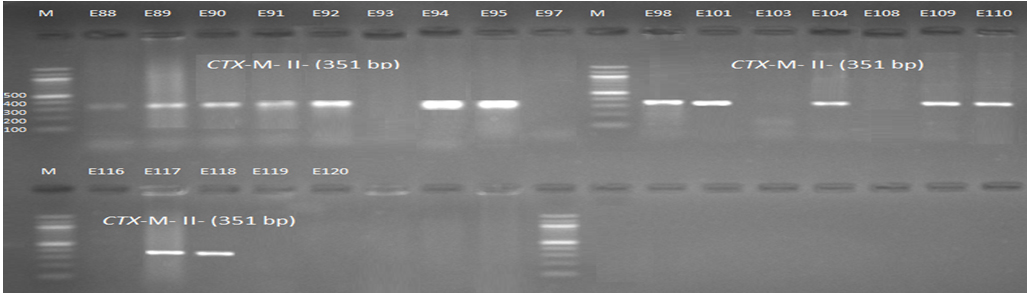
Fig. 5. 1.5% Agarose gel electrophoresis at 72 volt for 90 minutes of PCR to blaCTX-M-II amplicon (351bp); lane M represent DNA marker size(100bp) while E88-E120 represent the isolates
Fig. 6. 1.5% Agarose gel electrophoresis at 72 volt for 90 minutes of PCR to blaCTX-M-IV amplicon (474bp) lane M represent DNA marker size(100bp) while E1-E40 represent the isolates
Fig. 7. 1.5% Agarose gel electrophoresis at 72 volt for 90 minutes of PCR to blaCTX-M-IV amplicon (474bp) lane M represent DNA marker size(100bp) while E41-E88 represent the isolates
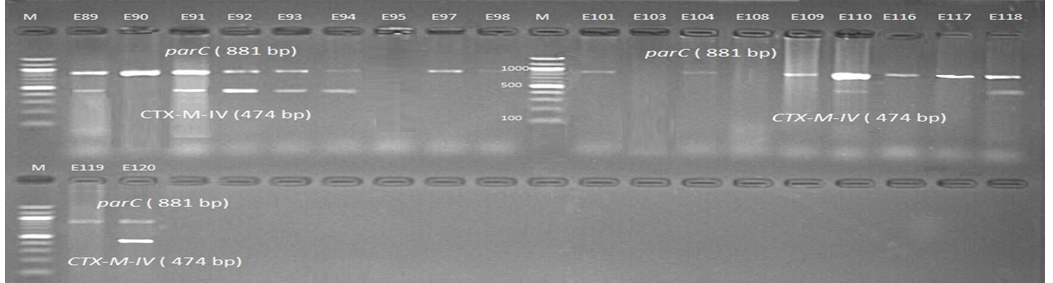
Fig. 8. 1.5% Agarose gel electrophoresis at 72 volt for 90 minutes of PCR to blaCTX-M-IV amplicon (474bp) lane M represent DNA marker size(100bp) while E89-E120 represent the iso-lates
Genotypic investigation of cefotaxime resistance among 56 CRUPEC were performed using specific four primer pairs to detect the genotypes of blaCTX-M groups. Multiplex-PCR were used to detect blaCTX-M-I (499bp) and blaCTX-M-III (305bp) while monoplex-PCR for blaCTX-M-II (351bp) and blaCTX-M-IV(474bp) (Figures 1-8). The results revealed existence of all blaCTX-M genotypes in different percentage: 36 (64.28%) for blaCTX-MI, 38(67.85%) for blaCTX-MII 26(46.43%) for blaCTX-MIII and 18(32.14%) for blaCTX-MIV (Figure 9). Concern possessing of isolates for more than one blaCTX-M genotypes were also reported in this study. Our results revealed that blaCTX-M were present in 50 (89.29%) of CRUPEC. Co-existence of more than on genotypes were reported in 38(67.86%) (Table 3). The results displayed that 40(80%) of blaCTX-M positive CRUPEC were belong to group B2, 4(8%) for group D, 3(6%) for group A and 3(6%) for group B1. Our results were roughly similar to those stated by other researcher. Mohajeri et al., (2014) found that UPEC that have blaCTX-M compile (93.3%). Occurrence of blaCTX-M ranged from 70-90% (Hernandez et al., 2014; Micenková et al., 2014; Poovendran and Ramanathan, 2015; Al-Mayahie and Al Kuriashy, 2016; Nojoomi et al., 2016; Padmavathy et al., 2016).
Table (3):
Distribution of cefotaxime resistance genotypes among UPEC.
| Cefotaxime Resistance Genotypes | Phylogenetic Subgroups | Total | No. (%) | ||||||
|---|---|---|---|---|---|---|---|---|---|
| A | B1 | B2 | D | ||||||
| A0 | A1 | B1 | B22 | B23 | D1 | D2 | |||
| CTX-MI/CTX-MII/CTX-MIII/CTX-MIV | 0 | 0 | 0 | 0 | 3 | 1 | 0 | 4 | 38(67.86) |
| CTX-MI/CTX-MIII/CTX-MIV | 0 | 0 | 0 | 0 | 3 | 1 | 0 | 4 | |
| CTX-MI/CTX-MII/CTX-MIII | 0 | 0 | 1 | 2 | 10 | 0 | 1 | 14 | |
| CTX-MI/CTX-MII/CTX-MIV | 0 | 0 | 0 | 0 | 3 | 1 | 0 | 4 | |
| CTX-MI/CTX-MII | 0 | 0 | 0 | 0 | 4 | 0 | 0 | 4 | |
| CTX-MI/CTX-MIII | 0 | 0 | 0 | 1 | 1 | 0 | 0 | 2 | |
| CTX-MI/CTX-MIV | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 1 | |
| CTX-MII/CTX-MIII | 0 | 0 | 1 | 0 | 1 | 0 | 0 | 2 | |
| CTX-MII/CTX-MIV | 0 | 0 | 0 | 1 | 2 | 0 | 0 | 3 | |
| CTX-MI | 0 | 1 | 1 | 1 | 0 | 0 | 0 | 3 | 12(21.43) |
| CTX-MII | 2 | 0 | 0 | 0 | 5 | 0 | 0 | 7 | |
| CTX-MIII | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | |
| CTX-MIV | 0 | 0 | 0 | 0 | 2 | 0 | 0 | 2 | |
| Negative | 0 | 0 | 2 | 0 | 4 | 0 | 0 | 6 | 6(10.71) |
| Total | 2 | 1 | 3 | 5 | 35 | 3 | 1 | ||
| Percentage % | 4 | 2 | 6 | 10 | 70 | 6 | 2 | 56(100) | |
The dominance of B23 phylosubgroup can be attributed to that most of ExPEC belong to group B2 and D while presence of isolated belong to intestinal phylogroups (A and B1) may be due to contamination and autoinoculation from feces in person with low personal hygiene (Cao et al., 2011; Luo et al., 2011; Merza and Jubrael, 2015; Abdullah and Lakshmidevi, 2016). BlaCTX-M is Extend spectrum beta-lactamase (ESBLs) with hydrolytic activity against cefotaxime and ceftriaxone with high susceptibility to tazobactam. The high prevalence of BlaCTX-M may attributed to the quick spread worldwide among both hospital and community acquired infections especially UTIs and so it called pandemic-BlaCTX-M (Cantón and Coque, 2006). The global spread of this enzyme may be due to high usage of third generation especially cefotaxime which leads to explosive of resistance. Also due to high spread of plasmid, transposon, intergron and insertion sequence which ac a carrier for blaCTX-M may responsible for their high prevalence (Cantón et al., 2012). The spread of blaCTX-M may be via international traveler (Arcilla et al.,2017). There are more than 25 variant of BlaCTX-M and emergence of these types may be resulted from recombination as a results of presence of the two different blaCTX-M types (Sun et al., 2017). Dominance of blaCTX-MI were reported in many studies and blaCTX-M15 is the commonly documented variant among UPEC (Park et al., 2012; Castanheira et al., 2014; Hasan et al., 2015; Bonger et al., 2016; Mshana et al., 2016; Abrar et al., 2017; Hashemizadeh et al., 2018). The low prevalence of blaCTX-M-III and blaCTX-M-IV were also reported in many studies from different geographical region (Tekiner and Özpýnar, 2016; Nairoukh et al., 2018).
Concern co-existence of multiple genotypes of blaCTX-M clusters, it is very risky and may leads to new variant of blaCTX-M. Our results revealed 38/56 CRUPEC isolates with multiple blaCTX-M clusters. He et al., 2016 found that as a results of recombination of blaCTX-MI cluster (blaCTX-M-15) and blaCTX-MIV cluster (blaCTX-M-14), the new variants with high stability and catalytic activity will results like blaCTX-M-64. Several stable and highly active hybrid including blaCTX-M-123, blaCTX-M-137, and blaCTX-M-132 were also resulted from recombination of blaCTX-M-15 and blaCTX-M-14 (Nagano et al., 2009; He et al., 2013; Tian et al., 2014; He et al., 2015; Liu et al., 2015).
The current study conclude the presence of blaCTX-M clusters (I, II, III, IV) as a main mechanisim or cefotaxime resistance among CRUPEC and coexistance of multible cluters within same isolates that may leads to emergence of new hybrides of blaCTX-M.
ACKNOWLEDGMENTS
I am warmly thanks Dr. Noor S.K. Al-Khafaji for kind cooperation and many thanks for Dr. Naeem R. Al-Jebori for assistance in isolate collection.
CONFLICT OF INTEREST
The authors declare that there is no conflict of interest.
- Abdullah, A.H.K.Y. and Lakshmidevi, N., Prevalence of Escherichia Coli sequence type 131 (ST131) among extra-intestinal clinical isolates in different phylogenetic groups. Health Sciences, 2016; 5(3), pp.90-94.
- Abrar, S., Vajeeha, A., Ul-Ain, N. and Riaz, S., Distribution of CTX-M group I and group III b-lactamases produced by Escherichia coli and klebsiella pneumoniae in Lahore, Pakistan. Microbial pathogenesis, 2017; 103, pp.8-12.
- Ahmad, W., Jamshed, F. and Ahmad, W., Freequncy of escherichia coli in patients with community acquired urinary tract infection and their resistance pattern against some commonly used anti bacterials. Journal of Ayub Medical College Abbottabad, 2015; 27(2), pp.333-337.
- Alizade, H., Ghanbarpour, R. and Aflatoonian, M.R., Virulence genotyping of Escherichia coli isolates from diarrheic and urinary tract infections in relation to phylogeny in southeast of Iran. Trop Biomed, 2014; 31(1), pp.174-82.
- Al-Khafaji, N.S. and Al-Thahab, A.A., Phylogenetic Study of Escherichia coli Isolatedfrom Clinical Samples in Hilla City, Iraq. Journal of Pure and Applied Microbiology, 2017; 11(4): p. 1777-1781.
- Al-Khaqani, M.M., Alwash, M.S. and Al-Dahmoshi, H.O., Investigation of phylogroups and some virulence traits among cervico-vaginal Escherichia coli (CVEC) isolated for female in Hilla City, Iraq. Malaysian Journal of Microbiology, 2017; 13(2), pp.132-138.
- Al-Mayahie, S. and Al Kuriashy, J.J., Distribution of ESBLs among Escherichia coli isolates from outpatients with recurrent UTIs and their antimicrobial resistance. The Journal of Infection in Developing Countries, 2016; 10(06), pp.575-583.
- Arcilla, M.S., van Hattem, J.M., Haverkate, M.R., Bootsma, M.C., van Genderen, P.J., Goorhuis, A., Grobusch, M.P., Lashof, A.M.O., Molhoek, N., Schultsz, C. and Stobberingh, E.E., Import and spread of extended-spectrum b-lactamase-producing Enterobacteriaceae by international travellers (COMBAT study): a prospective, multicentre cohort study. The Lancet Infectious Diseases, 2017; 17(1), pp.78-85.
- Bogner, C., Miethke, T., Wantia, N., Gebhard, F., Busch, D. and Hoffmann, R., 2016. Differences in ESBL Genes between E. coli, Klebsiella spp. and Enterobacter Cloacae Strains. International Journal of Clinical & Medical Microbiology, 2016.
- Bush, K. and Jacoby, G.A., Updated functional classification of b-lactamases. Antimicrobial agents and chemotherapy, 2010; 54(3): pp.969-976.
- Cantón, R. and Coque, T.M., The CTX-M b-lactamase pandemic. Current opinion in microbiology, 2006; 9(5), pp.466-475.
- Cantón, R., González-Alba, J.M. and Galán, J.C., CTX-M enzymes: origin and diffusion. Frontiers in microbiology, 2012; 3 :p.110.
- Cao, X., Cavaco, L.M., Lv, Y., Li, Y., Zheng, B., Wang, P., Hasman, H., Liu, Y. and Aarestrup, F.M., Molecular Characterization and Antimicrobial Susceptibility testing of Escherichia coli isolates from urinary tract infections in 20 Chinese hospitals. Journal of clinical microbiology, 2011; pp.JCM-02503.
- Castanheira, M., Farrell, S.E., Krause, K.M., Jones, R.N., and Sader, H.S., Contemporary Diversity of b-Lactamases among Enterobacteriaceae in the Nine U.S. Census Regions and Ceftazidime-Avibactam Activity Tested against Isolates Producing the Most Prevalent b-Lactamase Groups. Antimicrobial Agents and Chemotherapy, 2014; 58(2): 833–838.
- Clermont, O., Bonacorsi, S. and Bingen, E., Rapid and simple determination of theEscherichia coli phylogenetic group. Applied and environmental microbiology, 2000; 66(10), pp.4555-4558.
- CLSI. Performance Standards for Antimicrobial Susceptibility Testing. 26th ed. CLSI supplement M100S. Wayne, PA: Clinical and Laboratory Standards Institute; 2016.
- Coque, T.M., Baquero, F. and Canton, R., Increasing prevalence of ESBL-producing Enterobacteriaceae in Europe. Eurosurveillance, 2008; 13(47), p.19044.
- Escobar-Páramo, P., Le Menac’H, A., Le Gall, T., Amorin, C., Gouriou, S., Picard, B., Skurnik, D. and Denamur, E., Identification of forces shaping the commensal Escherichia coli genetic structure by comparing animal and human isolates. Environmental microbiology, 2006; 8(11), pp.1975-1984.
- Hasan, B., Olsen, B., Alam, A., Akter, L. and Melhus, Å., Dissemination of the multidrug-resistant extended-spectrum b-lactamase-producing Escherichia coli O25b-ST131 clone and the role of house crow (Corvus splendens) foraging on hospital waste in Bangladesh. Clinical Microbiology and Infection, 2015; 21(11), pp.1000-e1.
- Hashemizadeh, Z., Kalantar-Neyestanaki, D. and Mansouri, S., Clonal relationships, antimicrobial susceptibilities, and molecular characterization of extended-spectrum beta-lactamase-producing Escherichia coli isolates from urinary tract infections and fecal samples in Southeast Iran. Revista da Sociedade Brasileira de Medicina Tropical, 2018; 51(1), pp.44-51.
- He, D., Chiou, J., Zeng, Z., Chan, E.W.C., Liu, J.H. and Chen, S., Comparative characterization of CTX-M-64 and CTX-M-14: insights into the structure and catalytic activity of the CTX-M class of enzymes. Antimicrobial agents and chemotherapy, 2016; pp.AAC-00917.
- He, D., Chiou, J., Zeng, Z., Liu, L., Chen, X., Zeng, L., Chan, E.W.C., Liu, J.H. and Chen, S., Residues distal to the active site contribute to enhanced catalytic activity of variant and hybrid b-lactamases derived from CTX-M-14 and-15. Antimicrobial agents and chemotherapy, 2015; pp.AAC-04920.
- He, D., Partridge, S. R., Shen, J., Zeng, Z., Liu, L., Rao, L., … Liu, J.-H. CTX-M-123, a Novel Hybrid of the CTX-M-1 and CTX-M-9 Group b-Lactamases Recovered from Escherichia coli Isolates in China. Antimicrobial Agents and Chemotherapy, 2013; 57(8), 4068–4071. http://doi.org/10.1128/AAC.00541-13
- Hernandez, E., Araque, M., Millan, Y., Millan, B. and Vielma, S., Prevalence of beta-lactamase CTX-M-15 in phylogenetic groups of uropathogenic Escherichia coli isolated from patients in the community of Merida, Venezuela. Investigacion clinica, 2014; 55(1), pp.32-43.
- Kiiru, J., Kariuki, S., Goddeeris, B.M. and Butaye, P., Analysis of b-lactamase phenotypes and carriage of selected b-lactamase genes among Escherichia coli strains obtained from Kenyan patients during an 18-year period. BMC microbiology, 2012; 12(1), p.155.
- Lara, F., Nery, D.R., de Oliveira, P.M., Araujo, M.L., Carvalho, F.R., Messias-Silva, L.C., Ferreira, L.B., Faria-Junior, C. and Pereira, A.L., Virulence markers and phylogenetic analysis of Escherichia coli strains with hybrid EAEC/UPEC genotypes recovered from sporadic cases of extraintestinal infections. Frontiers in microbiology, 2017; 8: p.146.
- Lee, J.H., Subhadra, B., Son, Y.J., Kim, D.H., Park, H.S., Kim, J.M., Koo, S.H., Oh, M.H., Kim, H.J. and Choi, C.H., Phylogenetic group distributions, virulence factors and antimicrobial resistance properties of uropathogenic Escherichia coli strains isolated from patients with urinary tract infections in South Korea. Letters in applied microbiology, 2016; 62(1), pp.84-90.
- Liu, L., He, D., Lv, L., Liu, W., Chen, X., Zeng, Z., Partridge, S.R. and Liu, J.H., blaCTX-M-1/9/1 hybrid genes may have been generated from blaCTX-M-15 on an IncI2 plasmid. Antimicrobial agents and chemotherapy, 2015; pp.AAC-00501.
- Luo, C., Walk, S.T., Gordon, D.M., Feldgarden, M., Tiedje, J.M. and Konstantinidis, K.T., Genome sequencing of environmental Escherichia coli expands understanding of the ecology and speciation of the model bacterial species. Proceedings of the National Academy of Sciences, 2011; p.201015622.
- Mazzariol, A., Bazaj, A. and Cornaglia, G., Multi-drug-resistant Gram-negative bacteria causing urinary tract infections: a review. Journal of Chemotherapy, 2017; 29(sup1), pp.2-9.
- Merza, N.S. and Jubrael, J.M., Phylogenetic Grouping of Uropathogenic Escherichia coli Using Different Molecular Typing Methods in Kurdistan Region-Iraq. Int. J chem. BioMol. Sci, 2015; pp.51-55.
- Merza, N.S. and Jubrael, J.M., Phylogenetic Grouping of Uropathogenic Escherichia coli Using Different Molecular Typing Methods in Kurdistan Region-Iraq. Int. J chem. BioMol. Sci, 2015; pp.51-55.
- Micenková, L., Šišková, P., Bosák, J., Jamborová, I., Èernohorská, L. and Šmajs, D., Characterization of human uropathogenic ESBL-producing Escherichia coli in the Czech Republic: spread of CTX-M-27-producing strains in a university hospital. Microbial Drug Resistance, 2014; 20(6), pp.610-617.
- Mohajeri, P., Rostami, Z., Farahani, A. and Norozi, B., Distribution of ESBL producing Uropathogenic Escherichia coli and carriage of selected b-lactamase genes in Hospital and community isolates in west of Iran. Annals of Tropical Medicine and Public Health, 2014; 7(5), p.219.
- Mshana, S.E., Falgenhauer, L., Mirambo, M.M., Mushi, M.F., Moremi, N., Julius, R., Seni, J., Imirzalioglu, C., Matee, M. and Chakraborty, T., Predictors of bl a CTX-M-15 in varieties of Escherichia coli genotypes from humans in community settings in Mwanza, Tanzania. BMC infectious diseases, 2016; 16(1), p.187.
- Nagano, Y., Nagano, N., Wachino, J.I., Ishikawa, K. and Arakawa, Y., Novel chimeric b-lactamase CTX-M-64, a hybrid of CTX-M-15-like and CTX-M-14 b-lactamases, found in a Shigella sonnei strain resistant to various oxyimino-cephalosporins, including ceftazidime. Antimicrobial agents and chemotherapy, 2009; 53(1), pp.69-74.
- Nairoukh, Y.R., Mahafzah, A.M., Irshaid, A. and Shehabi, A.A., Molecular Characterization of Multidrug Resistant Uropathogenic E. Coli Isolates from Jordanian Patients. The open microbiology journal, 2018; 12, p.1.
- Nojoomi, F. and Ghasemian, A., Prevalence of ESBL phenotype, blaCTX-M-1, blaSHV and blaTEM genes among uropathogenic Escherichia coli isolates from 3 military hospitals of Tehran, Iran. Journal of Coastal Life Medicine, 2016; 4(8), pp.616-618.
- Ochoa, S.A., Cruz-Córdova, A., Luna-Pineda, V.M., Reyes-Grajeda, J.P., Cázares-Domínguez, V., Escalona, G., Sepúlveda-González, M., López-Montiel, F., Arellano-Galindo, J., López-Martínez, B. and Parra-Ortega, I., Multidrug-and extensively drug-resistant uropathogenic Escherichia coli clinical strains: phylogenetic groups widely associated with integrons maintain high genetic diversity. Frontiers in microbiology, 2016; 7: p.2042.
- Padmavathy, K., Padma, K. and Rajasekaran, S., Multidrug resistant CTX-M-producing Escherichia coli: a growing threat among HIV patients in India. Journal of pathogens. Journal of pathogens. 2016; 1(1):1-6.
- Park, S.H., Byun, J.H., Choi, S.M., Lee, D.G., Kim, S.H., Kwon, J.C., Park, C., Choi, J.H. and Yoo, J.H., Molecular epidemiology of extended-spectrum b-lactamase-producing Escherichia coli in the community and hospital in Korea: emergence of ST131 producing CTX-M-15. BMC infectious diseases, 2012; 12(1), p.149.
- Piranfar, V., Mirnejad, R. and Erfani, M., Incidence and Antibiotic Susceptibility Pattern of Most Common Bacterial Pathogen Causing Urinary Tract Infection (UTI) in Tehran, Iran, 2012-2013. Int J Enteric Pathog, 2014; 2(1), pp.1-4.
- Poovendran, P., Ramanathan, N., Molecular Characterization of Bla CTX-M, Bla TEM, Bla SHV- Beta Lactamase Produced by Uropathogenic Escherichia coli Isolates. International Journal of Microbiological Research, 2015; 6(2): pp.67-73.
- Prakasam, A.K.C., Kumar, K.D. and Vijayan, M., A cross sectional study on distribution of urinary tract infection and their antibiotic utilisation pattern in Kerala. Int J PharmTech Res, 2012; 4(3): pp.1310-6.
- Salehzadeh, A. and Zamani, H., Characterization of (Uropathogenic) E. coli isolated from urinary tract infections: phylogenetic typing and distribution of virulence-associated traits. British journal of biomedical science, 2018; 75(1), pp.40-42.
- Sun, P., Bi, Z., Nilsson, M., Zheng, B., Berglund, B., Lundborg, C.S., Börjesson, S., Li, X., Chen, B., Yin, H. and Nilsson, L.E., Occurrence of blaKPC-2, blaCTX-M and mcr-1 in Enterobacteriaceae from Well Water in rural China. Antimicrobial agents and chemotherapy, 2017; pp.AAC-02569.
- Tekiner, Ý.H. and Özpýnar, H., Occurrence and characteristics of extended spectrum beta-lactamases-producing Enterobacteriaceae from foods of animal origin. Brazilian Journal of Microbiology, 2016; 47(2), 444–451.
- Tian, G.B., Huang, Y.M., Fang, Z.L., Qing, Y., Zhang, X.F. and Huang, X., CTX-M-137, a hybrid of CTX-M-14-like and CTX-M-15-like b-lactamases identified in an Escherichia coli clinical isolate. Journal of Antimicrobial Chemotherapy, 2014; 69(8), pp.2081-2085.
- World Health Organization, 2015. The Selection and Use of Essential Medicines: Report of the WHO Expert Committee, 2015 (including the 19th WHO Model List of Essential Medicines and the 5th WHO Model List of Essential Medicines for Children) (No. 994). World Health Organization.
- Zhao, W.H. and Hu, Z.Q., Epidemiology and genetics of CTX-M extended-spectrum b-lactamases in Gram-negative bacteria. Critical reviews in microbiology, 2013; 39(1): pp.79-101.
© The Author(s) 2018. Open Access. This article is distributed under the terms of the Creative Commons Attribution 4.0 International License which permits unrestricted use, sharing, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.