ISSN: 0973-7510
E-ISSN: 2581-690X
https://dx.doi.org/10.22207/JPAM.13.2.31 | © The Author(s). 2019
Pathogenic bacteria are constantly adapted against antimicrobial drugs by arising new traits of drug resistance. The genes of such resistance are mostly possessed by pathogens due to the random use of antibiotics and commonly held on plasmid DNA. Plasmid-mediated antimicrobial resistance genes are easily transferred horizontally from one bacterial cell to another in an epidemic manner causing an increasing and serious challenge to clinicians to overcome the infectious pathogens. UTI is the most common infection caused by bacteria which has become hard to be treated due to the emerging problem of antibiotic resistance. The current study correlates plasmid DNA diversity and antibiotic resistance in E. coli; the common causative agent of UTI. Obtaining plasmid restriction maps provides a clear view about how DNA could be diversified in one bacteria. The result showed a clear DNA polymorphism in plasmids purified from E. coli. There has been 10 forms of different restriction plasmid profiles among 63 sample of E. coli. The 10 forms of plasmid profiles have been classified into two groups: highly diversified and lowly diversified profiles. Strains of highly diversified plasmid profiles showed significantly more resistance toward antibiotics than strains of less diversified plasmid profiles. Measuring plasmid DNA diversity together with the antibiotic resistance indicates an epidemic transfer and acquisition of different plasmids in pathogenic E. coli as a result of antibiotic random treatment.
Plasmid, Mapping, UTI, Antibiotic, Resistance.
E. coli is Gram-negative rod-shaped coliform bacterium which is facultative anaerobic and normally found harmlessly as normal flora in the lower intestinal part of warm blood animals1. However, serious infections could be caused by E. coli such as gastroenteritis due to food poisoning, burn infections, meningitis, and urinary tract infection (UTI)2,3.
There are approximately 150 million person who develops UTI annually word wide that impacts significantly on national economies in terms of treatment and management4. The incidence of UTI has been estimated to occur at least once during the life span of about 40% of females of all ages, while 11% of females of above 18 years old develop UTI in the USA only5,6.
Uropathogenic E. coli (UPEC), which is part of the intestinal normal flora, has been recorded as the most popular pathogenic bacteria isolated from 80% to 90% of the patients suffering from community acquired UTI6. This is due to the bacterium ability to invade both lower and upper parts of the urinary tract causing bladder and kidney infections respectively7,8. The virulence factors of structural adhesive (such as pili, flagella, and fimbriae) and toxic secreted elements qualify UPEC to be the primary cause of UTI9. Other bacterial pathogens which were also isolated from UTI involve Pseudomonas aeruginosa, Klebsiella pneumonia, Enterobacter cloacae, Proteus mirabilis, Streptococcus bovis, and Enterococcus faecalis. Fungal causative agents including Candida albicans was recorded as well10-12.
In spite of antibiotic treatment, it has been recorded that UTI could be recurred with the same pathogenic bacteria after 6-12 months of the patient remedy13. The UTI has been shown to recurred even after two years in about 33% of urinary tract-infected one year old Children, while 20% to 30% of women who are over 18 years old has been noticed to experience UTI recurrence after 3 to 4 months of the first infection14. The challenging pathogenic E. coli which develop antibiotic resistance and cause the infection to occur repeatedly could lead to surgical intervention performed by physicians to patient to manage the UTI 15.
Antimicrobial treatment of UTI must be prescribed after performing a precise diagnosis and drug sensitivity identification of the causative uropathogenic bacteria in order to avoid the emerging antibiotic resistance resulting from the arbitrary antibiotics prescription16. Pharmaceutical antibiotic industry has been significantly developed to produce drugs of good choice to manage the UTIs but still UPEC is challenging pathogen persisting high levels of antibiotic resistance17.
For the sake of avoiding the drawbacks (such as antibiotic resistance) resulting from the complete dependence on antibiotics treatment, the Urology Association in Europe has recently recommended patients to perform changes in their life style to minimize the symptoms of UTI without taking any antibiotics for a certain period of time, then antibiotics could be given when the no antibiotic treatment has not worked18.
Records obtained between 2007-2010 in the United States showed that 31.3% of UPEC isolated from hospitalized patients were resistant toward fluoroquinolones antibiotics, while in Europe, UPEC appeared to be less resistant to fluoroquinolones (22.3%) which were less active than 3rd generation cephalosporins that appeared 11% resistance by UPEC9,19. Such clinical studies highlight the serious problem of decreased drugs of choice used against resistant UPEC16.
Antimicrobial resistance in bacteria is an inherited trait which is expressed by pathogenic bacteria that carry the genetic material for such traits. Antibiotic resistance genes are not considered as essential genes for microbial growth and metabolism but an adaptive genes that applied to bacteria as a result of stressful adaptive selection derived by exposure to antibiotics. This adaptive genetic traits are mostly not carried on the bacterial chromosome but on a luxurious DNA molecules called plasmids20,21.
Plasmids are extra-chromosomal circular DNA molecules which are able to replicate inside the cell independently of the chromosomal DNA and transferred to other bacterial cells by conjugation22. Having a unique origin of replication, a given plasmid will not only be able to replicate itself but also determine its copy number inside the bacterial cell. Other genetic systems of toxin-antitoxin working mechanisms, for example, that ensure a 100% maintenance of plasmids in the bacterial offspring regardless the adaptive selection have also been detected on plasmid molecules23.
Studying plasmid DNA content in pathogenic bacteria constitutes a vital step toward the understanding of antibiotic resistance which is the major problem of treating bacterial infectious disease nowadays24. One of the approaches of understanding the plasmid DNA content is to measure its diversity or poly-morphism by restriction mapping which is the objective of the current study. Applying restriction enzymes will cut DNA molecules specifically from certain restriction sites. These restriction cutting sites are assumed by us to be distributed differently in their numbers and/or positions among the different plasmid DNA molecules producing unique patterns of DNA bands on agarose gel. Each pattern is related to a certain group of plasmid.
The aim of the present study is measuring the polymorphism of plasmid DNA content and correlating it with antimicrobial resistance profiles of E. coli isolated from UTI. This provides an indication on how antibiotic are being used randomly and ineffectively in Iraq toward this pathogen which will derive the pressure of stressful selection in bacteria to acquire and maintain a wide variety of plasmid DNA molecules, that carry antibiotic resistance traits, in order to survive the stress.
Samples collection
Urine samples of patients were collected from hospitals laboratories using sterile container and directly taken to the laboratory for culture on the same day.
Samples culture and Bacteria Identification
Each Urine sample was transferred into sterile centrifuge tubes and centrifuged at 5000 rpm for 10 minutes. The supernatant was discarded. The pellet was cultured on 5 ml brain heart infusion broth and incubated overnight at 37°C in order to activate bacteria. Samples which show turbidity in the brain heart infusion broth were sub-cultured on freshly made plates of blood and Macconkey agars. Lactose fermenting bacterial colonies were picked for and sub-cultured separately for the purpose of performing distinguishing biochemical test to diagnose E. coli according to the manuals of25.
Antibiotic sensitivity test
The antibiotic resistance ability of pathogenic E.coli has been measured by disc diffusion method on Muller-Hinton agar plates 25. Discs of 9 different antibiotics, which are commonly used in Iraq to treat UTI, were used in this study.
Plasmid DNA Isolation
The obtained E. coli isolates were activated by culturing on 15 ml of brain heart infusion broth and incubated overnight at 37°C. Bacterial cells are then precipitated by centrifugation at 6000 rpm for 15 minutes. The plasmid content of the precipitated bacterial cells were isolated by alkaline lyses method using plasmid extraction kit from Qiagen®. The manual provided with kit were followed for the plasmids isolation.
Plasmid DNA restriction digestion
The Plasmid content isolated from each E. coli isolate were digested in a double digestion reaction using high fidelity BamHI and HindIII restriction enzymes from New England Biolab®. Instruction manuals of the company regarding these enzymes were followed in performing restriction digestion.
Agarose Gel Electrophoresis and Visualization
After restriction digestion completed, 4 ml of 6X DNA loading dye (from New England Biolab®) were mixed with each 20 ml of the restriction reaction and applied into the wells of 1% Agarose gel prepared by adding 1 gram of agarose to 100 ml of Tris Acetic acid EDTA buffer in addition to 5µl ethidium bromide. The loaded gel were immersed in TAE in the electrophoresis tank and a current of 90 Volt was applied for 1.5 hour. DNA standard ladder from Bioline® were also loaded on the gel. The gels were read using computerized ultra violate transilluminator.
Urinary tract infection is the most commonly occurred infection in the community worldwide26-28, therefore, the current study involves isolation and identification of pathogenic E. coli from patients suffering from UTI and studying the plasmids polymorphism.
One hundred and twelve urine samples were collected from hospitals in Al-Najaf and Karbala. After centrifugation and culture on blood and Macconkey, 21 samples showed no bacterial growth, 91 samples showed bacterial growth due to UTI. Entrobacteriaceae was the common cause of UTI accounting for 89%, most of them (63 sample) were identified to be E. coli (78%).
It has been shown by28 that community-acquired UTI is caused mostly by entero-bacteriaceae (88%), 80% of them was diagnosed as E. coli. The recurrence of UTI was seen to be higher in females compared with males patients due to the anatomic difference of urogenital tract between male and female. This goes to agreement with the finding of29,30.
The treatment of bacterial infections including UTI has become a main problem due to the random use of antibiotics which induces the adaptive processes in pathogenic bacteria to resist such unconcerned antibiotic treatment. Enzymatic resistance by extended spectrum beta-lactamases is the main adaptation tool which has evolved in pathogenic bacteria converting such pathogents to be multidrug resistant and more challenging to physicians day by day31,32.
The emerging new genes of antibiotic resistance has become more challenging not only in the bacteria under the stress of antibiotic treatment but also to other bacteria of different species and genera due to the ability of such adaptive genes to be held on plasmids (mobile DNA elements) which are then easily moved horizontally to other bacteria making it ready and resistant to any suspected antibiotic treatment33,34.
Enterobacteriaceae is well known of plasmid-mediated antibiotic resistance which could be spread easily across its genera24,35. This means easily-treated E. coli could be converted to a highly resistant (virulent) pathogen due to plasmid acquisition from other virulent bacteria such as Klebsiella spp. for instance.
It has been thought that studying the plasmid types in pathogenic bacteria is an important step toward the well understanding of antibiotic resistance as plasmids are commonly found in bacteria to hold genes of drug resistance. Restriction mapping is one of the precise methods to study the genetic polymorphism depending on the specific DNA digestion from certain loci36,37.
The number and positions of certain restriction site are different among the different plasmid DNA molecules. In other words, the restriction nucleotide sequence of certain endonuclease is variant in its frequency among the plasmid DNA variants. Therefore, treating such plasmid variants with one restriction endonuclease, for instance, will produce groups of DNA segments of different sizes which will show unique patterns or finger print of DNA bands on agarose each pattern (finger print) is related to its original plasmid.
The results revealed 10 different plasmid patterns among E.coli isolates (fig. 1). These 10 patterns were divided into two groups according to their diversity which are highly and lowly diversified respectively. The more the number of DNA bands the more the diversity of plasmid content and vice versa. This indicates the clear variation of plasmid content in pathogenic bacteria of the same species which is isolated from only one type of infection. Different plasmid restriction maps in such single pathogen confirm the ability of the latter to accept wide variety of plasmids in order to resist antibiotics. As mentioned previously that plasmids are acquired by pathogenic bacteria when its under stress of antibiotic treatment, especially when antibiotics are used randomly.
In other words, an epidemic spread of different plasmid types from different origins is most likely to be a common phenomenon simultaneously with the random use of antimicrobial drug treatment20.
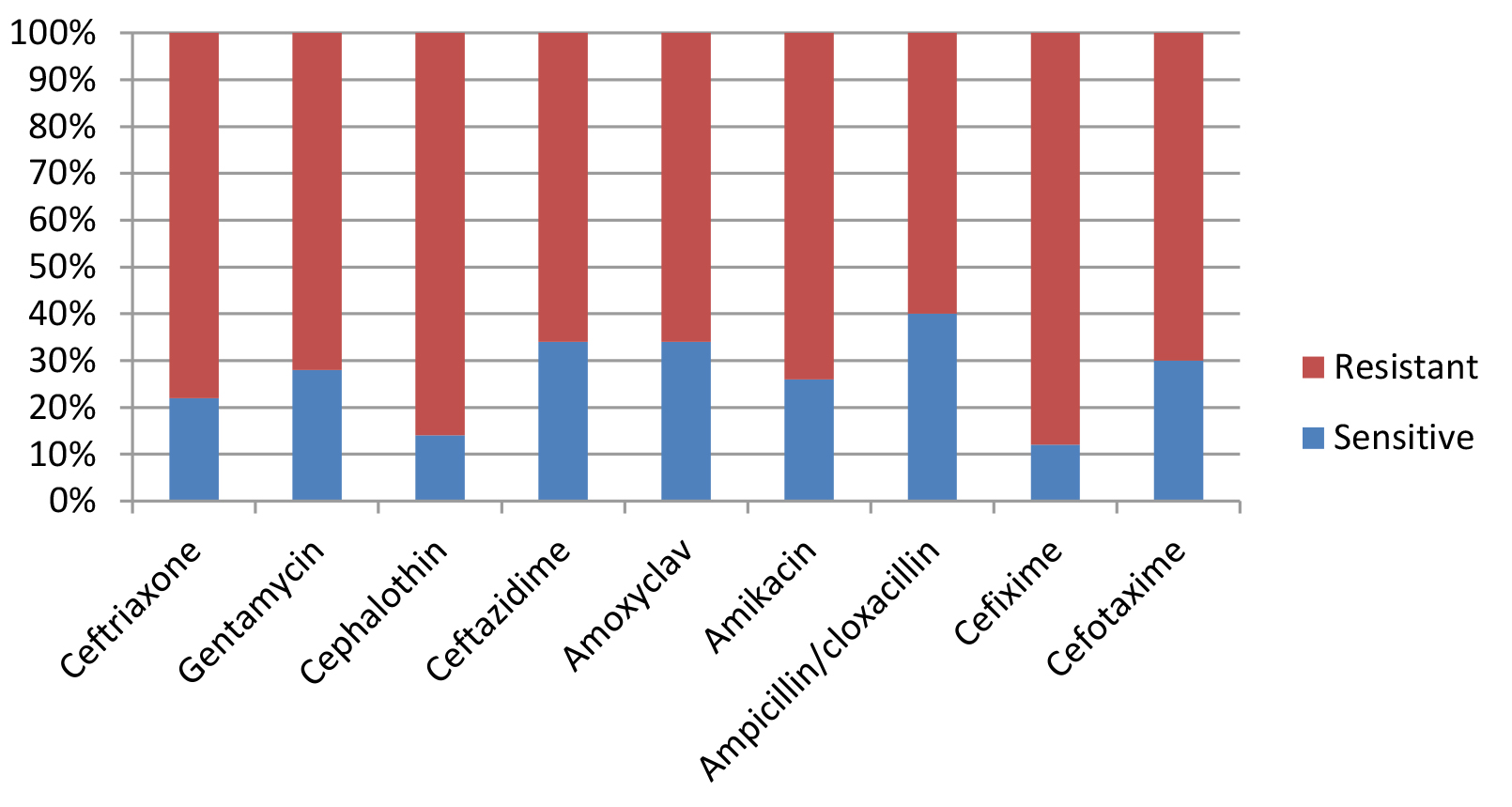
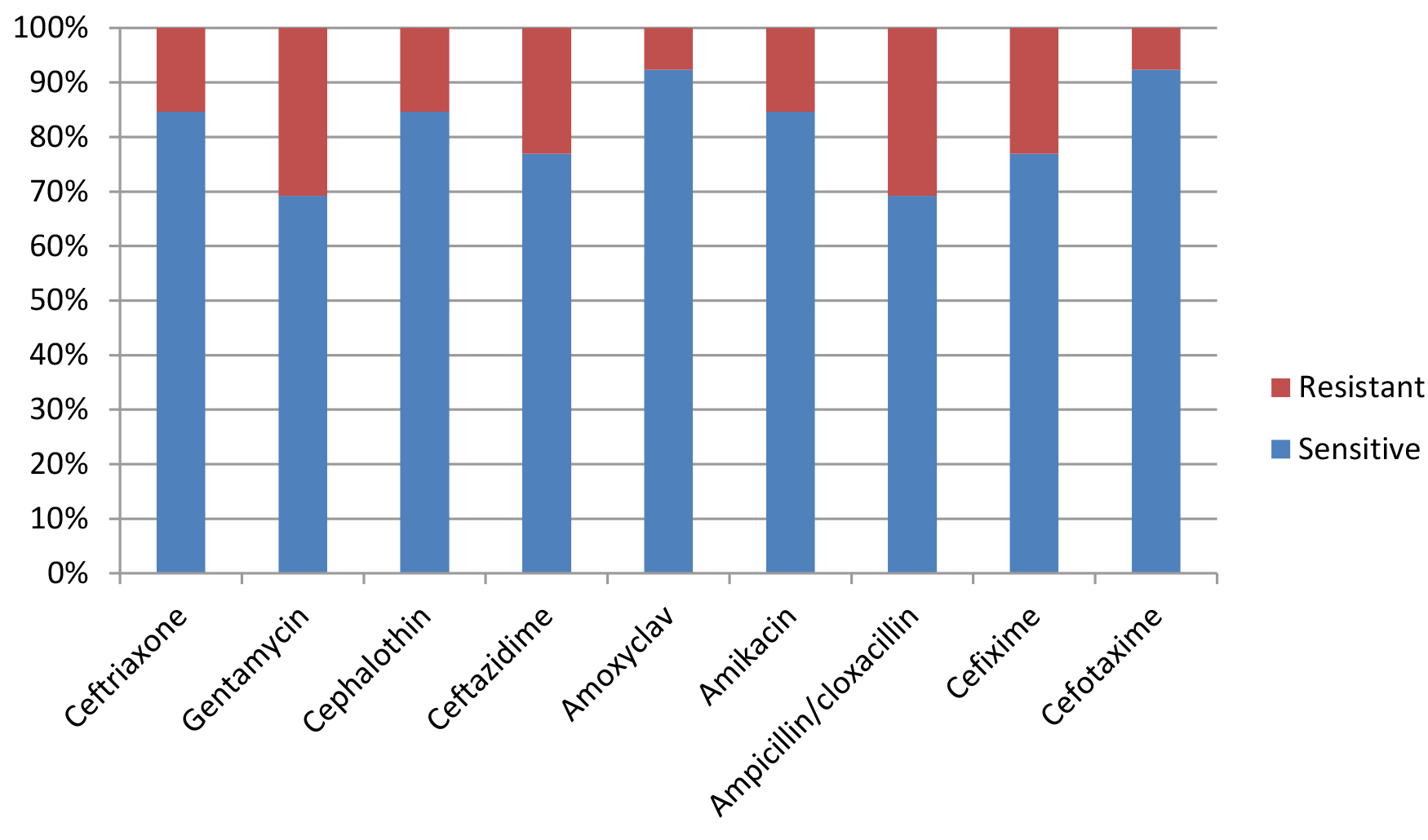
Most E. coli isolates (50 isolates 79.4%) in this study appeared with highly diversified plasmid DNA (Fig. 1, lanes 2,3,4,5,6, and 9), while 20.6% (13 isolates ) showed less diversity of plasmid content (Fig. 1, lanes 1,7,8, and 10). Isolates of highly diversified plasmids appeared significantly more resistance toward antibiotics (Table 1, Fig. 2), while isolates of less diversified plasmid content showed less resistance (Table 2, Fig. 3). This indicates an epidemic widespread of plasmids in relation with antibiotic resistance which is considered as a clear evidence of antibiotics random usage in the Iraqi community.
Table (1):
Antibiotic sensitivity of isolates with highly diversified plasmid DNA (50 samples).
| Antibiotic
|
Ceftriaxone
|
Gentamycin
|
Cephalotdin
|
Ceftazidime
|
Amoxyclav
|
Amikacin
|
Ampicillin/cloxacillin
|
Cefixime
|
Cefotaxime
|
|||||||||
| Sensitivity | s | r | s | r | s | r | s | r | s | r | s | r | s | r | s | r | s | r |
| Numbers | 11 | 39 | 14 | 36 | 7 | 43 | 17 | 33 | 17 | 33 | 13 | 37 | 20 | 30 | 6 | 44 | 15 | 35 |
|
Percentage |
22 % |
78% % |
28 % |
72 % |
14 % |
86 % |
34 % |
66 % |
34 % |
66 % |
26 % |
74 % |
40 % |
60 % |
12 % |
88 % |
30 % |
70 % |
S= Sensitive, R= Resistance
In the current study, the plasmids purified from UTI-causing pathogenic E.coli were treated by two different endonucleases which are BamHI and HindIII which recognize and digest plasmid DNA molecules at every 5′-GGATCC-3′ and 5′-AAGCTT-3′ respectively. The more endo-nucleases used in restriction mapping the more precise DNA finger print will be produced provided that endonucleases are not sensitive toward DNA methylation, therefore, BamHI and HindIII were used in double digestion reaction for this purpose. These two restriction enzymes are highly active and not blocked by dam, dcm, and CpG methyltransferases.
Table (2):
Antibiotic sensitivity of isolates with less diversified plasmid DNA (13 samples).
| Antibiotic
|
Ceftriaxone
|
Gentamycin
|
Cephalotdin
|
Ceftazidime
|
Amoxyclav
|
Amikacin
|
Ampicillin/cloxacillin
|
Cefixime
|
Cefotaxime
|
|||||||||
| Sensitivity | s | r | s | r | s | r | s | r | s | r | s | r | s | r | s | r | s | r |
| Numbers | 11 | 2 | 9 | 4 | 11 | 2 | 10 | 3 | 12 | 1 | 11 | 2 | 9 | 4 | 10 | 3 | 12 | 1 |
|
Percentage |
84.6 % |
15.4 % |
69.2 % |
30.8 % |
84.6 % |
15.4 % |
77 % |
23 % |
92 % |
8 % |
84.6 80 % |
15.4 % |
69.2 % |
30.8 % |
77 % |
23 % |
92 % |
8 % |
There are many types of methods used in plasmid typing most of them rely on amplifying specific sequences or genes which could be present only in certain types of plasmids. Such methods are considered to investigate the presence or absence of the target plasmids among the bacteria by locating some characteristic genes related to such plasmids. For example, replicon typing method which is based on PCR (PBRT method) developed by38 is one of the methods used to distinguish IncI1 and ColE plasmids by locating relaxases-encoding MOBp and MOBf genes using PCR39. While in this study, restriction mapping distinguishes extremely short DNA sequence repeats (restriction sites) of six base pair length which are not hard to be found in a wide range of different plasmids. Therefore, the assumption of random distribution of BamHI and HindIII restriction sequences among the plasmid content of pathogenic E.coli was easy to be estimated here providing us with clear view of plasmid DNA polymorphism.
There are 28 different plasmids (such as IncA/C, IncL, IncI, IncN, IncF, and IncH) identified in Enterobacteriaceae recently which are mostly related with antimicrobial drug resistance24. The plasmids are fixed in their sizes but they are modified by means of high frequency recombination (Hfr); a process by which plasmids could be recombine with the chromosomal DNA leading to insertion of plasmid genes into the chromosome or insertion of chromosomal genetic materials into plasmids40. This could give a molecular explanation to the result of the current study of getting different plasmid restriction maps. Therefore it is not surprising to find certain genes (such as antibiotic resistance genes) of chromosomal origins held on the plasmids.
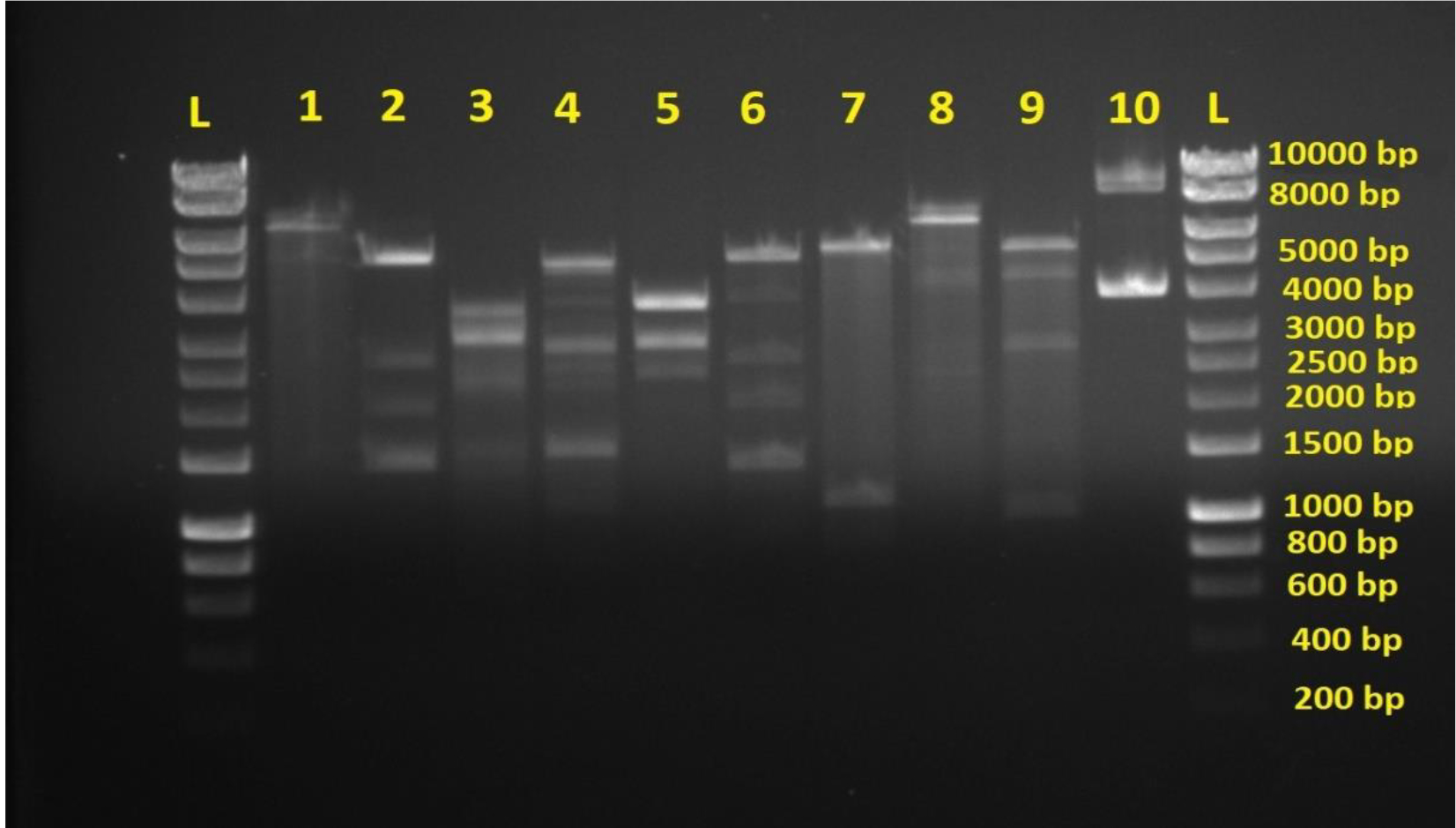
Fig. 1. Agarose gel electrophoresis of plasmids digested by BamHI and HindIII endonucleases. L is hyperladderI. Lane 1-10 are double digested plasmid samples from different E.coli isolates showing DNA variations. Lanes 2,3,4,5,6 and 9 show more DNA bands indicating more DNA diversity. Lanes 1,7,8, and 10 show less DNA bands indicating less DNA diversity.
Acknowledgments
None
Conflicts Of Interest
The authors declare that there are no conflicts of interest.
Authors’ Contribution
All authors have made substantial, direct and intellectual contribution to the work and approved it for publication.
Funding
None.
Data Availability
All datasets generated or analyzed during this study are included in the manuscript.
Ethics Statement
This article does not contain any studies with human participants or animals performed by any of the authors.
- Singleton P. Bacteria in biology, biotechnology and medicine: John Wiley & Sons; 1997.
- Murphy F.A. Emerging zoonoses. Emerg Infect Dis. 1998; 4(3):429.
- Vogt R.L., Dippold L. Escherichia coli O157: H7 outbreak associated with consumption of ground beef, June–July 2002. Public health reports., 2005; 120(2): 174-178.
- Flores-Mireles A.L., Walker J.N., Caparon M., Hultgren S.J. Urinary tract infections: epidemiology, mechanisms of infection and treatment options. Nat. Rev. Microbiol., 2015; 13(5): 269.
- Foxman B., Brown P. Epidemiology of urinary tract infections: transmission and risk factors, incidence, and costs. Infectious disease clinics of North America, 2003; 17(2): 227-241.
- Foxman B. Urinary tract infection syndromes: occurrence, recurrence, bacteriology, risk factors, and disease burden. Infectious disease clinics of North America, 2014; 28(1): 1-13.
- Hilbert D.W. Uropathogenic Escherichia coli: the pre-eminent urinary tract infection pathogen: Nova Publishers; 2013.
- Lim J.Y., Yoon J.W., Hovde C.J. A brief overview of Escherichia coli O157: H7 and its plasmid O157. Journal of microbiology and biotechnology, 2010; 20(1):5.
- Terlizzi M.E., Gribaudo G., Maffei M.E. Uro-Pathogenic Escherichia coli (UPEC) infections: virulence factors, bladder responses, antibiotic, and non-antibiotic antimicrobial strategies. Frontiers in microbiology, 2017; 8: 1566.
- Parish A., Holliday K. Long-term care acquired urinary tract infections’ antibiotic resistance patterns and empiric therapy: a pilot study. Geriatric nursing (New York, N.Y.), 2012; 33(6): 473-478.
- Palou J., Angulo J.C., Ramon de Fata F., et al. [Randomized comparative study for the assessment of a new therapeutic schedule of fosfomycin trometamol in postmenopausal women with uncomplicated lower urinary tract infection]. Actas urologicas espanolas, 2013; 37(3): 147-155.
- Hof H. [Candiduria! What now? : Therapy of urinary tract infections with Candida. Der Urologe. Ausg. A.,2017; 56(2): 172-179.
- Fiore DC, Fox CL. Urology and nephrology update: recurrent urinary tract infection. FP essentials, 2014; 416: 30-37.
- Nuutinen M., Uhari M. Recurrence and follow-up after urinary tract infection under the age of 1 year. Pediatric nephrology (Berlin, Germany), 2001; 16(1): 69-72.
- Tolg C., Bagli D.J. Uropathogenic Escherichia coli infection: potential importance of epigenetics. Epigenomics, 2012; 4(2): 229-235.
- Bartoletti R., Cai T., Wagenlehner F.M., Naber K., Johansen T.E.B. Treatment of urinary tract infections and antibiotic stewardship. European Urology Supplements, 2016; 15(4): 81-87.
- Blango M.G., Mulvey M.A. Persistence of uropathogenic Escherichia coli in the face of multiple antibiotics. Antimicrobial agents and chemotherapy, 2010; 54(5): 1855-1863.
- Vahlensieck W., Perepanova T., Johansen T.E.B., Tenke P., Naber K.G., Wagenlehner F.M. Management of uncomplicated recurrent urinary tract infections. Euro Urol Supplements, 2016; 15(4): 95-101.
- Edelsberg J., Weycker D., Barron R., et al. Prevalence of antibiotic resistance in US hospitals. Diagn. Microbiol. Infect. Dis., 2014; 78(3): 255-262.
- Carattoli A. Resistance plasmid families in Enterobacteriaceae. Antimicrobial agents and chemotherapy, 2009; 53(6): 2227-2238.
- Thomas C.M., Nielsen K.M. Mechanisms of, and barriers to, horizontal gene transfer between bacteria. Nature reviews microbiology, 2005; 3(9): 711.
- Lawley T., Frost L.S., Wilkins B.M. Bacterial conjugation in gram-negative bacteria. Plasmid biology: American Society of Microbiology, 2004: 203-226.
- Hayes F. Toxins-antitoxins: plasmid maintenance, programmed cell death, and cell cycle arrest. Science, 2003; 301(5639): 1496-1499.
- Rozwandowicz M., Brouwer M., Fischer J., et al. Plasmids carrying antimicrobial resistance genes in Enterobacteriaceae. J Antimicrob Chemother, 2018; 73(5): 1121-1137.
- Mac Faddin J. Biochemical Tests for Identification of Medical. Bacteria. Williams and Wilkins. London, 2000.
- Rodrםguez-Bano J., Navarro M.D., Romero L., et al. Epidemiology and clinical features of infections caused by extended-spectrum beta-lactamase-producing Escherichia coli in nonhospitalized patients. J Clinic Microbiol, 2004; 42(3): 1089-1094.
- Azap ײ., Arslan H., erefhanoנlu K., et al. Risk factors for extended-spectrum ג-lactamase positivity in uropathogenic Escherichia coli isolated from community-acquired urinary tract infections. Clin. Microbiol. Infect., 2010; 16(2): 147-151.
- Park J.J., Seo Y.B., Lee J. Antimicrobial susceptibilities of Enterobacteriaceae in community-acquired urinary tract infections during a 5-year period: a single hospital study in Korea. Infection & chemotherapy, 2017; 49(3): 184-193.
- Al-Shamarti M.J., Hussein A.A., AL-Luhaiby A.I. The Relationship Between the Type of Infection and Antibiotic Resistance. J Pure Appl Microbiol, 2018; 12(2): 845-854.
- Al-Shamarti M.J. molecular evaluation of B-lactam resistance genes in klebsiella spp isolated from clinical cases in Al-Najaf province Master Thesis. University of Kufa: Department of Biology, University of Kufa 2010.
- Pitout J.D., Nordmann P., Laupland K.B., Poirel L. Emergence of Enterobacteriaceae producing extended-spectrum b-lactamases (ESBLs) in the community. J. Antimicrob. Chemother.. 2005; 56(1): 52-59.
- Rodriguez-Bano J., Navarro M. Extended-spectrum b-lactamases in ambulatory care: a clinical perspective. Clin. Microbiol. Infect., 2008;14: 104-110.
- Bennett P. Plasmid encoded antibiotic resistance: acquisition and transfer of antibiotic resistance genes in bacteria. Br. J. Pharmacol., 2008; 153(S1): S347-S357.
- San Millan A., Santos-Lopez A., Ortega-Huedo R., Bernabe-Balas C., Kennedy S.P., Gonzalez-Zorn B. Small-plasmid-mediated antibiotic resistance is enhanced by increases in plasmid copy number and bacterial fitness. Antimicrobial agents and chemotherapy, 2015; 59(6): 3335-3341.
- ALshamarti M.J., AL-Muhnna A. Molecular detection of AmpC Gene encoding antibiotic resistance among Klebsiella SPP. isolated from different infections. Al-Kufa Journal for Biology, 2011; 3(1):1-9.
- Kuspa A., Loomis W.F. Tagging developmental genes in Dictyostelium by restriction enzyme-mediated integration of plasmid DNA. Proceedings of the National Academy of Sciences, 1992; 89(18): 8803-8807.
- Roberts R.J. How restriction enzymes became the workhorses of molecular biology. Proceedings of the National Academy of Sciences, 2005; 102(17): 5905-5908.
- Carattoli A., Bertini A., Villa L., Falbo V., Hopkins K.L., Threlfall E.J. Identification of plasmids by PCR-based replicon typing. J Microbiol Methods, 2005; 63(3): 219-228.
- Garcillבn-Barcia M.P., Francia M.V., De La Cruz F. The diversity of conjugative relaxases and its application in plasmid classification. FEMS Microbiol Reviews, 2009; 33(3): 657-687.
- Adelberg E.A., Pittard J. Chromosome transfer in bacterial conjugation. Bacteriological reviews, 1965; 29(2): 161.
© The Author(s) 2019. Open Access. This article is distributed under the terms of the Creative Commons Attribution 4.0 International License which permits unrestricted use, sharing, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.