ISSN: 0973-7510
E-ISSN: 2581-690X
https://dx.doi.org/10.22207/JPAM.13.1.27 | © The Author(s). 2019
The Group B Streptococcus (Streptococcus agalactiae) (GBS) is one of the natural flora bacteria in the female reproductive system. In the recent years, GBS has become the major bacterial infections throw the perinatal period causing many troubles. This study was undertaken to determine the prevalence and antibiotic genes of GBS colonization in pregnant women of obstetrics hospital in Taif, Saudi Arabia. Fourteen Streptococcus agalactiae isolates obtained after screening about 134 swabs samples from genitourinary tract specimens of women patients from obstetrics hospital in Taif governorate, Saudi Arabia. These isolates were studied for antibiotic resistance and virulence genes. All obtained isolates were identified as Streptococcus agalactiae by the 16S rDNA gene sequence. These strains were found by Disc diffusion method sensitive against Meropenem, Cefotaxime, Cefepime, Amoxicillin, Penicillin G, Daptomycin, Chloramphenicol, Linezolid and Levofloxacin. The highest resistance was for Tetracyclin (85.7%) whereas the lowest resistance was found for Vancomycin (21.4%). Resistance against Erythromycin and Clindamycin was 71.5%, and 28.5% respectively. PCR based detection revealed 50% of isolates were carrying the tetT genes, while 92.8% of isolates were carrying tetO and tetM genes associated with Tetracycline resistance. All isolates were harboring genes that associated with Erythromycin resistance like ErmB1, ErmB2, and Erm(A|TR) genes, but only 28.5% of isolates were carrying ErmTR gene. Molecular detection of virulence associated genes revealed that out of fourteen isolates of S. agalactiae, only one isolate was carrying the LinB gene, while all isolates were positive for MreA and VanA genes respectively. We can conclude that the GBS isolates were found sensitive to many antibiotic while most isolates were resistance to Tetracycline due to the existence of tetO and tetM genes. Resistance against Erythromycin and Clindamycin was 71.5%, and 28.5% respectively. The two erythromycin resistance genes (ErmB and ErmTR) were found in all isolates, while, the third erythromycin resistance gene Erm(A|TR) was found only in 25.8% of the isolates.
Streptococcus agalactiae group B, antibiotic resistance, virulence genes, Saudi Arabia.
Group B streptococci (GBS)are a component of intestinal bacteria and human genitourinary microflora. These types are often associated with the disease found in new borns, affecting life-threatening diseases such as meningitis and sepsis. Also, these strains are linked to complications during pregnancy and postpartum1. GBS colonization of the Umayyad reproductive tract was considered the most important risk factor for the development of neonatal disease2,3. Moreover, GBS is a reason for infection in older infants and non-pregnant adults, particularly older people or those with any primary medical conditions. Clinical manifestations include urinary tract infections, skin infections or soft tissue, osteomyelitis, and pericardial infarction3,4. Territories of concern incorporate an expanded recurrence of perinatal disease and a higher death rate in grown-ups than in the neonatal group5. GBS was reliably defenseless against penicillin and other b-lactams. But, antimicrobial resistance utilized as an elective treatment, particularly macrolides, lincosamides and fluoroquinolones, has been recorded in various countries6-8.The most widely recognized macrolide resistance mechanisms in streptococcus is the change of the ribosome by methylase, which is encoded by Erm genes. These enzymes also give resistance to the formation of resistance to lycosamides and streptopogen B, which characterizes macrolide-linuxamidestry-ptoline B (MLSB) phenotypes. Another common mechanism is the flow of drugs via membrane-specific protein encoded by the MEF gene, which is linked with M phenotype9. Protection from fluoroquinolones in GBS was first depicted in 2003 and is related with point mutations in gyrA and parC10.In spite of the clinical impact of GBS disease and expanded protection from some antimicrobial, there is a limited number of reports indicating antibiotic resistance among the GBS strains in Saudi Arabia7,11, even from non-symptomatic colonization or clinical contamination12-15. In present study, fourteen streptococcus agalactiae of 134 strains, isolated from genitourinary tract specimens of women patients from obstetrics hospital in Taif, Saudi Arabia from December 2017 to August 2018, were tested for antimicrobial susceptibility and macrolide resistance genes.
Streptococcus strains
Fourteen S. agalactiae isolates obtained after screening about 134 swabs samples from genitourinary tract specimens of women patients from obstetrics hospital in Taif, Saudi Arabia. The bacterial species were identified using the fully automated VITEK-2 COMPACT microbiology system (Bio Mיrieux, Inc., Durham, NC, USA). Purchased from strain control S. agalactiae ATCC® BAA1138™) from ATCC, USA.
Antibiotic Susceptibility Testing
Using the recommended clinical standard (CLSI) standard, World Health Organization method, the susceptibility of S. agalactiae against 16 antibiotics was determined11,16.
The 16S rDNA gene sequencing
The genomic DNA was isolated using DNA extraction kit (Gena Bioscience, Germany) from all S. agalactiae isolates accordance the manufacturer’s instructions. For each isolate and as per previously described methods, one fragment of the DNA (about 1,400 barrels) was amplified from the 16S rDNA gene11. The pieces were punctuated using the QIA quick PCR purification kit (QIAGEN, Valencia, CA, USA) and sequenced in DNA Analyzer 3146 Applied Bioscience (Applied Biosystems, USA). The sequencing texts were edited and compiled using DNASTAR software (Laser gene, Madison, WI, USA). Using the NCBI server, BLAST searches were performed (http://www.ncbi.nlm.nih.gov/blast/Blast.cgi).
Detection of Antimicrobial Resistance Genes
The antimicrobial resistance genes (ThekpsII,ermB1ermB2,ermBA/TR, mreA, ,ermTR, linB, mefA, vanAtetT, tetO and tetT) were detected in the S. agalactiae strains17,18. Antibiotic resistance and virulence genes were detected in multiple-drug resistant enterococcal isolates by PCR with primers synthesized by the macrogen Co., Ltd. (Seoul, Korea) (Table 1).
Table (1):
PCR primers for detection of some antibiotic resistance and virulence genes in Streptococcus agalactiae group B isolates.
Primers name |
Primer sequence (5′→3′) |
Size (bp)
|
|---|---|---|
Erm(B1) |
GAA AAG GTA CTC AAC CAA AGT
GGT ACT TAA ATT GTT TAC |
640 |
Erm(B2) |
CATTTAACGACGAAACTGGC
GGAACATCTGTGGTATGGCG |
420 |
Erm(TR) |
GAA GTT TAG CTT TCC TAA GCT TCA GCA CCT GTC TTA ATT GAT |
495 |
Erm(A/TR) |
TCA GGA AAA GGA CAT TTT ACC
ATA CTT TTT GTA GTC CTT CTT |
425 |
MreA |
AGA CAC CTC GTC TAA CCT TC
TCT GCA GGT AAG TAA GTG CG |
498 |
LinB |
CCT ACC TAT TGT TTG TGG AA
ATA ACG TTA CTC TCC TAT TC |
925 |
MefA |
AGT ATC ATT AAT CAC TAG TGC
TTC TTC TGG TAC TAA AAG TGG |
328 |
Van-A |
GTAGGCTGCGATATTCAAAGC CGATTCAATTGCGTAGTCCAA |
231 |
KpsII |
GCGCAT TTGCTGATA CTGTTG
CAT CCA GAC GAT AAG CAT GA |
272 |
Tet(M) |
GTMGTTGCGCGCTATATTCC
GTGAAMGRWAGCCACCTAA |
696 |
Tet(O) |
GCGGAACATTGCATTTGAGGG
CTCTATGGACAACCCGACAGAAG |
538 |
Tet(T) |
CAGTGGGAATATAAGGACACGTC
CAAGCCTTCTCTACAGCATC |
644 |
Ethics Statement
This study was accepted by the ethics council of obstetrics hospital in King Faisal complex, Taif, Saudi Arabia, (1-439-6050). All subjects gave written informed consent before their consideration in the study.
Enterococcus isolates identification
Out of 134 isolates, fourteen strains (10.4%) were identified as Streptococcus agalactiae by the using a fully automated VITEK-2 COMPACT microbiology system (BioMיrieux, Inc., Durham, NC, USA). These results were confirmed by 16S rDNA sequencing (data not shown). All isolates were compared to the partial sequences of the 16S rRNA genes of S. agalactiae that deposited in the Gen Bank database and were found to be similar to the obtained sequence. Ribosomal operons are greatly relevant for the study of bacterial evolution and phylogeny. When re-constructing phylogenetic relationships of microorganisms, sequencing of 16S rDNA has been widely used. Phylogenetic analysis of the partial 16S rDNA sequences from the studied S. agalactiae strains, together with related sequences deposited in GenBank were positioned Streptococcus isolates into Streptococcus agalactiae group B (Fig. 1).
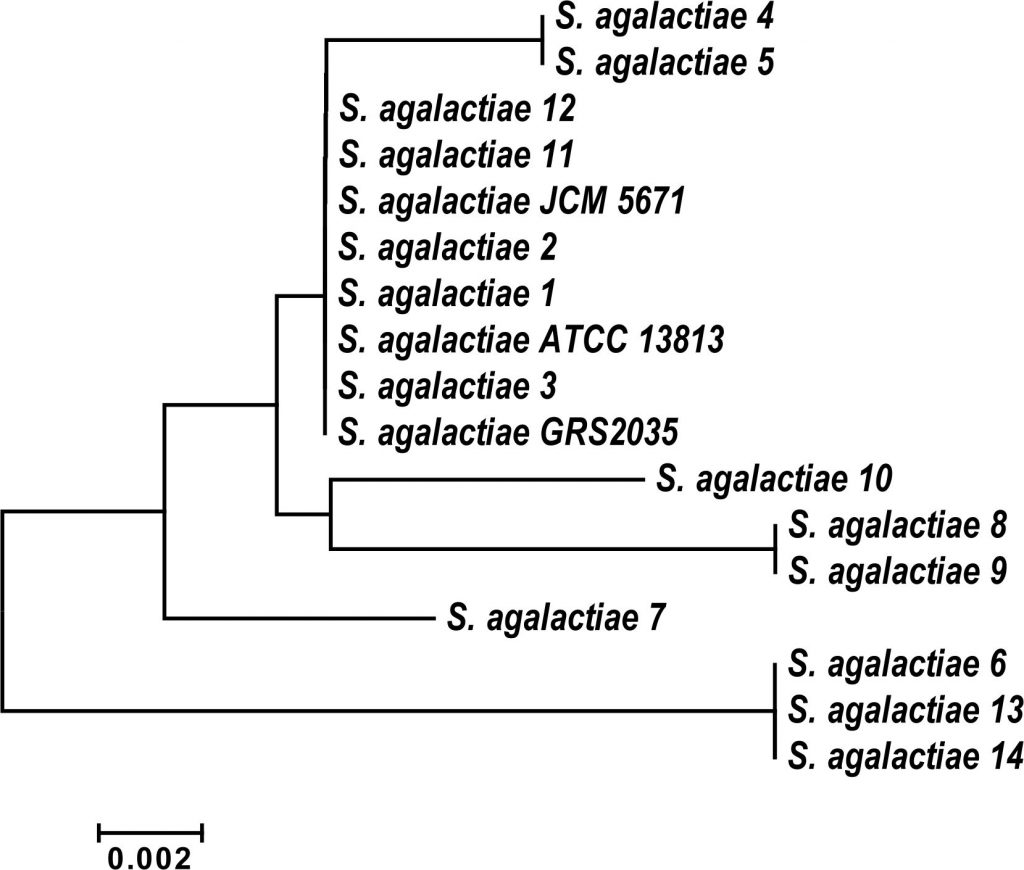
Fig. 1. Phylogenetic relationship of Streptococcus agalactiae group B isolates and related genera based on full size16SrDNA sequences. The tree was constructed using neighbor joining algorithm with Kimura 2 parameter distances in MEGA 7.1 software. The bar indicates the Juke-Cantor evolutionary distance.
Streptococcus specie sensitivity to antibiotics
All the fourteen Streptococcus strains were sensitive to Meropenem, Cefotaxime, Cefepime, Amoxicillin, Pencillin G, Daptomycin, Chloramphenicol, Lienezolid and Levofloxacin. The highest resistance was for Tetracyclin (85.7%) whereas the lowest resistance was found for Vancomycin (21.4%). Resistance against Erthromycin and Clindamycin was 71.5%, and 28.5% respectively (Fig. 2).
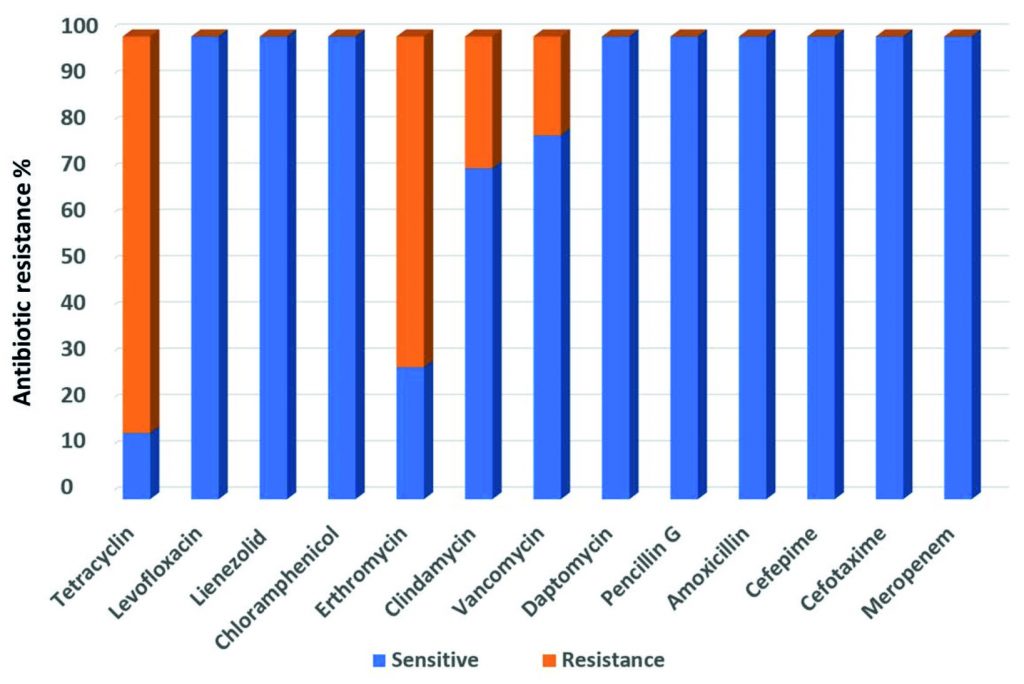
Fig. 2. Antimicrobial resistance profiles of nineteen multi-Drug resistance Streptococcus agalactiae group B against fourteen antibiotics.
Occurrence of antimicrobial resistance genes
The kpsII, ermB1ermB2, ermBA/TR, mreA, and vanA genes were detected in all Streptococcus isolates, while, ermTR, linB, mefA, and tetT genes were detected in 28.5, 7.1, 64.2, 50, and 50% of Streptococcus isolates, respectively. While, both tetM and tetO that associated with Tetracycline resistance were detected in 92.8 % of Streptococcus isolates. Table 2 shows the detection of these antimicrobial resistance genes in all isolates. The emeB1, ermB2 and ermA/TR genes were found in all isolates. In addition to drug efflux, such ermTR gene that were present in 28.5% of the erythromycin-sensitive enterococci and 7.1% of the linezolid-sensitive Streptococcus, indicating no expression of the emeBgene in some Streptococcus isolates. Further, the emeB gene occurred at a greater significance in the fluoroquinolone-resistant enterococci than that in the fluoroquinolone-sensitive Streptococcus. This indicates that the distribution of the emeBgene was related to the resistance to the three fluoroquinolones in the Streptococcus species (Fig. 3).
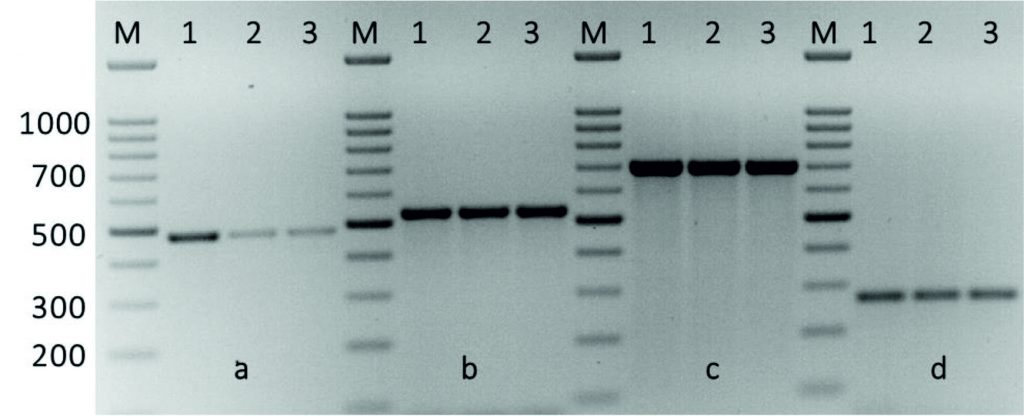
Fig. 3. Amplification of some specific genes producing in some Streptococcus agalactiae group B isolates by single PCR. (A) MreA gene with size about of 498 bp. (B)Tet(O)gene specific for tetracycline with size about of 538 bp. (C) Tet(M)gene specific also for tetracycline resistance with size about of 696 bp. (D) KpsII gene specific for capsule formationgene with size about of 272 bp. First lane on each panel is 100 bp molecular weight markers
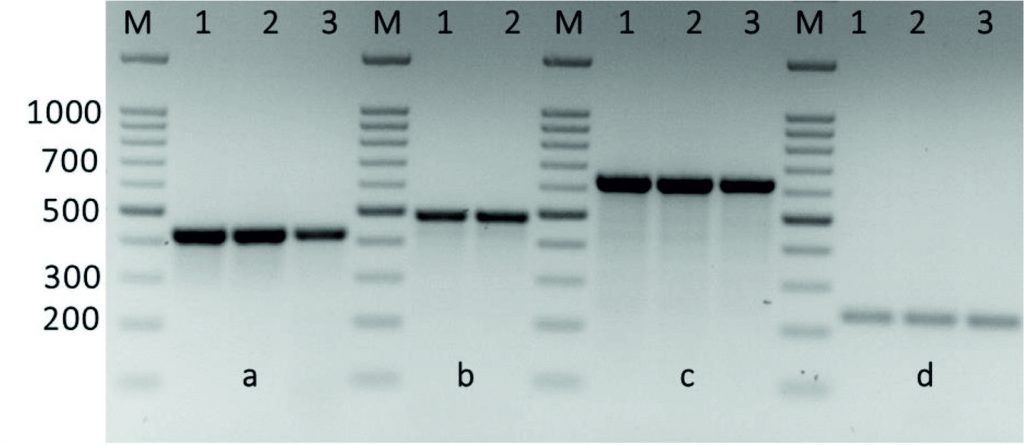
Fig. 4. Amplification of some specific genes producing in some Streptococcus agalactiae group B isolates by single PCR. (A) Erm(A/TR) gene specific for erythromycin resistance with size about of 425 bp. (B) Erm(TR)gene specific for erythromycin with size about of 495 bp. (C) Erm(B1) gene specific also for erythromycin resistance with size about of 640 bp. (D) Van-A gene specific for vancomycin resistance with size about of 231 bp. First lane on each panel is 100 bp molecular weight markers.
Table (2):
Molecular characterization of virulence and Antibiotic genes in Streptococcus agalactiae group B.
| Strain |
Antibiotic resistance genes | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| ErmB1 | ErmB2 | ErmTR | ErmA/TR | MreA | LinB | MefA | Van-A | TetM | TetO | TetT | |
| S-1 | + | + | ــ | + | + | ــ | + | + | + | + | + |
| S -2 | + | + | ــ | + | + | ــ | + | + | + | ــ | ــ |
| S -3 | + | + | + | + | + | ــ | + | + | + | + | + |
| S -4 | + | + | ــ | + | + | ــ | ــ | + | + | + | + |
| S -5 | + | + | ــ | + | + | ــ | ــ | + | ــ | + | ــ |
| S -6 | + | + | ــ | + | + | ــ | ــ | + | + | + | ــ |
| S -7 | + | + | ــ | + | + | + | + | + | + | + | + |
| S -8 | + | + | + | + | + | ــ | + | + | + | + | + |
| S -9 | + | + | ــ | + | + | ــ | + | + | + | + | ــ |
| S -10 | + | + | + | + | + | ــ | + | + | + | + | + |
| S -11 | + | + | ــ | + | + | ــ | + | + | + | + | ــ |
| S -12 | + | + | ــ | + | + | ــ | + | + | + | + | ــ |
| S -13 | + | + | + | + | + | ــ | ــ | + | + | + | ــ |
| S -14 | + | + | ــ | + | + | ــ | ــ | + | + | + | + |
Protection from erythromycin, clinda-mycin and tetracycline antibiotics was seen among the considered Streptococcus isolates. The resistance frequency of S. agalactiae group B to such antibiotics have wide-ranging depending on the region and time assessed, as reported in experiments accompanied in various countries8,9. In Saudi Arabia, available recorded data about S. agalactiae group Bantimicrobial susceptibility were obtained from studies performed in different governorates12,13,15,19,20. In this investigation, the general occurrence of GBS colonization amongst pregnant ladies was observed to be 10.4%. Comparative discoveries have been accounted for in other African countries, for example, in Malawi, Egypt, Zimbabwe, Gambia, and Tanzania; the prevalence of GBS in these countries ranges from 16.5 to 23%21-23. Nevertheless, low colonization rate was stated in a report directed in Ethiopia (9%) and some Latin American countries, for example, Peru (6%)24,25. Diverse examinations directed in Saudi Arabia indicated results higher than present investigation, 27.6% at Riyadh governorate12. On other hand, other reports showed results lower than present investigation as documented in 3-5% carriage rate in term of pregnant women at Aseer governorate13 and 4.76% at Abha Maternity Hospital14. As shown in our results, Tetracycline and clindamycin resistance rates were fundamentally the same as those saw in past investigations directed in differentarea26,27. A point of worry is the discovery of an Erthromycin-resistant isolate (Figure 2). GBS extremely resistant to Erthromycin was first depicted in Japan10,and from that point forward, it found in different countries7,11. Here, we report for the first time, the existence of Erthromycin resistance among S. agalactiae group B in Saudi patients at Taif governorate which it be recognized by molecular techniques. With respect to resistance against erythromycin, the elective treatment to penicillin unfavorably susceptible patients, results got in this investigation far surpassed the rates recently detected in the equivalent geological area. Despite the fact that there is no accessible information on macrolide utilization in other area in Saudi Arabia, it is well recognized that, for related species, protection from this antibiotic is specifically connected to its utilization25. Generally, it well known that the phenotypes of macrolide resistance isolates are well associated with genotypes, excluding for that one isolate displayed a cMLSB phenotype and did not have any of the related genes. Similar results for recognizin germ and mef genes in S. agalactiae group B phenotype have been reported somewhere else8. Also, several strains that have ermA+ ermB genes and displayed an iMLSB phenotype has been reported in different areas, including Saudi Arabia15,19. But, combination of ermA+ mefA/E genes in an iMLSB phenotype had never been detected in erythromycin-resistant isolates that found in Saudi Arabia.
In the present study, it was found that the GBS isolates are resistance to Tetracycline due to the existence of tet Oand tetM genes. Also most of the isolates were resistance to the antibiotic Erthromycine, that is because all isolates are carrying ErmB1, ErmB2, and Erm(A|TR)genes. Therefore, according to the results found in this study, the antibiotics Meropenem, Cefotaxime, Cefepime, Amoxicillin, Pencillin G, Daptomycin, Chloramphenicol, Lienezolid and Levofloxacin can be reliable replacements of erythromycin in curing or avoiding GBS infection in women who suffer from GBS colonization.
This study was funded by the Deanship of Scientific Research, Taif University, KSA (research project number 1-349-6050).
Authors have declared that no competing interests exist.
- Romanik, M., K.Nowosielski, R.Poreba, U.Sioma-Markowska, G.Martiroisian, and J.Groborz, Streptococcus group B serotype distribution in anovaginal isolates of women in term pregnancy. Neuro Endocrinol. Lett., 2014; 301–305.
- Al, S.R., S.Amor, G. Hery-Arnaud, B. Spellerberg, P. Lanotte, L. Mereghetti, et al. Enhanced expression of lmb gene encoding laminin-binding protein in Streptococcus agalactiae strains harboring IS1548 in scpB-lmb intergenic region. PLoS ONE 2010; 5: E10794.
- Dutra, V.G., V.M. Alves, A.N. Olendzki, C.A. Dias, A.F. de Bastos, G.O. Santos, et al., Streptococcus agalactiae in Brazil: serotype distribution, virulence deter-minants and antimicrobial susceptibility. BMC Infect. Dis., 2014; 14: 323.
- Sendi P., L. Johansson, andA. Norrby-Teglund. Invasive group B strep-tococcal disease in non-pregnant adults: a review with emphasis on skin and soft-tissue infections. Infection, 2008.; 6: 100-111.
- Phares, C.R., R. Lynfield, M.M., Farley, J.Mohle-Boetani, L.H.,Harrison, S. Petit,et al. Epidemiology of invasive group B streptococcal disease in the United States, 1999-2005. JAMA, 2008; 299: 2056-2065.
- Duarte, R.S., B.C.Bellei, O.P.Miranda, M.A.Brito, L.M.Teixeira. Distribution of antimicrobial resistance and virulence-related genes among Brazilian group B streptococci recovered from bovine and human sources. Antimicrob. Agent. Chemother., 2005; 49: 97-103.
- Tazi, A., T.Gueudet, E.Varon, L.Gilly, P.Trieu-Cuot, C.Poyart. Fluoroquinolone-resistant group B streptococci in acute exacerbation of chronic bronchitis. Emerg. Infect. Dis., 2008; 14: 349-350.
- Pinheiro, S., H.Radhouani, C.Coelho, A.Gonחalves, E. Carvalho, J.A. Carvalho, F.Ruiz-Larrea, C.Torres, G.Igrejas, P.Poeta. Prevalence and mechanisms of erythromycin resistance in Streptococcus agalactiae from healthy pregnant women. Microb. Drug. Resist., 2009; 15: 121-124.
- de Azavedo J.C., M.McGavin, C.Duncan, D.E.Low, A.McGeer. Prevalence and mechanisms of macrolide resistance in invasive and non-invasive group B streptococcus isolates from Ontario, Canada. Antimicrob. Agent. Chemother., 2001; 45: 3504-3508.
- Kawamura, Y., H.Fujiwara,N.Mishima, Y.Tanaka, A.Tanimoto, S.Ikawa, Y.Itoh, T.Ezaki. First Streptococcus agalactiae isolates highly resistant to quinolones with point mutations in gyrA and parC. Antimicrob. Agent. Chemother., 2003; 47:3605-3609.
- Hassan, M.M. and E.B. Belal. Antibiotic resistance and virulence genes in enterococcus strains isolated from different hospitals in Saudi Arabia. Biotechnol. Biotechnol. Equip., 2016; 30: 726-732.
- Almuneef, M., S.Alalola, S.Ahmed, Z.Memish, M.Y. Khan,and M.Alshaalam. The changing spectrum of Group B Streptococcal (GBS) infection in infants of Saudi Arabia. Chemother., 2000; 12: 48-52.
- Bahar, A.M., N.Bilal,and M.A.Eskander. High vaginal swab cultures in normal and preterm labor. Inter. J. Gynecol. Obstet., 2004; 87: 145- 146.
- Al-sunaidi, M., S.Abong, M.Al-sharani, et al. Prevalence of Group B Streptococcus Colonization in Mothers and Babies at Abha General Hospital, Kingdom of Saudi Arabia. Med. J. Cairo Univ., 2011; 79(2):163-165.
- Musleh, J., and N.Al-Qahtani. Group B Streptococcus colonization among Saudi women during labor. Saudi J. Med. Sci., 2018; 6: 18-22.
- CLSI. Performance Standards for Antimicrobial Susceptibility Testing, Twenty-Fourth Infor-mational Supplement, M100-S24. Wayne, MI: CLSI, 2014.
- Poyart C., J. Laurence, Q. Gilles,B. Patrick, and T. Patrick. Genetic Basis of Antibiotic Resistance in Streptococcus agalactiae Strains Isolated in a French Hospital. Antimicrob. Agent. Chemother, 2003; 47: 794–797.
- Gharailoo, Z., S.F.Mousavi, N.Halvani, M.M.Feizabadi. Antimicrobial Resistant Pattern and Capsular Typing of Streptococcus Pneumoniae Isolated from Children in Sistan -Baluchestan. Maedica., 2016; 11(3): 203-207.
- Uduman, S.A., T.K.Chatterjee, M.I.Al-Mouzan, S.Al-Suleiman. Group B streptococci colonization among Saudi women in labor and neonatal acquisition. Inter. J.Gynaecol. Obstet., 1985; 23(1):21-24.
- Arain, F.R., .N.A. Al-Bezraha, K.Y. Al-Aalib. Prevalence of Maternal Genital Tract Colonization by Group B Streptococcus from Western Province, Taif, Saudi Arabia. J. Clin. Gynecol. Obstet., 2015; 4(3): 258-264.
- Dzowela, T., O.O.Komolafe,and A.Lgbigbia. Prevalence of group B Streptococcus colonization in antenatal women at the Queen Elizabeth Central Hospital Blantyre a preliminary study. Malawi Med. J., 2005; 17: 97-99.
- Elbaradie, S.M., M.Mahmoud, M.Farid. Maternal and neonatal screening for Group B streptococci by SCP B gene based PCR: a preliminary study. Indian J. Med.Microbiol., 2009; 27(1):17-21.
- Mavenyengwa, R.T., J.E.Afset, B.Schei, S.Berg, T.Caspersen, H.Bergseng, S.R. Moyo. Group B Streptococcus colonization during pregnancy and maternal-fetal trans-mission in Zimbabwe. Acta Obstet. Gynecol. Scand., 2010; 89(2): 250-255.
- Benchetrit, L.C., S.E Fracalanzza, H. Peregrino, A.A.Camelo, and LA.Sanches. Carriage of Streptococcus agalactiae in women and neonates and distribution of serological types: a study in Brazil. J. Clin.Microbiol., 1982; 15(5): 787-790.
- Muller-Vranjes, A., D.Puntaric, D.Curzik, S.Sijanovic, Z.Topolovec, Z.Kasac, M. Miskulin. Prevalence and signi-ficance of vaginal group B strep-tococcus colonization in pregnant women from Osijek, Croatia. Coll.Antropol., 2011; 35(1):21-26.
- Emaneini, M. A.Mirsalehian, R.Beigvierdi, A.A.Fooladi, F.Asadi, F.Jabalameli, M. Taherikalani. High Incidence of Macrolide and Tetracycline Resistance among Streptococcus agalactiae Strains Isolated from Clinical Samples in Tehran, Iran. Maedica, 2014; 9(2):157-61.
- Wang, P., J. Tong, X.Ma, F. Song, L.Fan, C.Guo, et al., Serotypes, Antibiotic Susce-ptibilities, and Multi-Locus Sequence Type Profiles of Streptococcus agalactiae Isolates Circulating in Beijing, China. PLoS ONE. 2015; 10(3): e0120035. doi:10.1371/journal.pone. 0120035.
© The Author(s) 2019. Open Access. This article is distributed under the terms of the Creative Commons Attribution 4.0 International License which permits unrestricted use, sharing, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.


