ISSN: 0973-7510
E-ISSN: 2581-690X
In an in vitro study of antagonistic mangrove actinobacteria for its antifungal compounds to eliminate Fusarium oxysporum f. sp. lycopersici, the extracellular metabolites from the actinobacterium Kutzneria sp. strain TSII was documented. A total of 24 useful ligands were identified when profiled with Gas Chromatography-Mass Spectrometry after purification through silica gel column chromatography. Molecular modeling of Short-chain Dehydrogenases / Reductases (SDRs) was performed in the prime module of Schrodinger 2018-4 software using SDR of Bacillus anthracis as a template (PDB: 3I3OA). In silico studies utilizing the molecular docking of the 24 ligands with SDR responsible for pathogenicity, distributed on the membrane of the Fusarium oxysporum were carried out in Schrodinger 2018-4. Results were indicated as 1,4-Benzenedicarboxylic acid, Bis(2-ethylhexyl) ester, Spiro[Cyclopentane-1,2’(1’H)-quinoxaline], Decanedioic acid, Didecyl ester with docking scores (Kcal/mol) of – 8.151, -7.231, – 6.031 and low binding free energy (ΔGb) values (Kcal/mol) of – 68.11, -42.23, -79.41 respectively, relating to their potential use as antifungal agents in Fusarium wilt infections. As SDRs are involved in melanin biosynthesis, binding of these ligands may interfere with melanin biosynthetic pathways conferring pathogenicity to Fusarium oxysporum f. sp. lycopersici. Hence, the present research study uncovers in vitro antagonistic potential of marine actinobacterial metabolites towards the most devastating vascular wilt pathogen Fusarium oxysporum in tomatoes and identifies the probable involvement of SDR as the potential target in an in silico molecular docking approach.
Short-chain Dehydrogenase/Reductases, Antifungals, Marine actinobacteria, Melanin biosynthesis, Fusarium oxysporum.
Tomato forms a vital component of daily food intake as both raw and processed vegetables1. It is one of the heavily consumed horticulture crops worldwide and stands next to the potatoes. However, a profitable yield is often threatened by several fungal diseases. Among the potentially threatening fungal diseases, vascular wilt caused by the fungus Fusarium oxysporum f. sp. lycopersici (Sacc.) W.C. Snyder and H.N. Hansen (FOL) is a very serious fungal disease affecting the overall yield globally2. Disease incidence is favored by warmer temperatures (27°C to 28°C), acidic pH of soils (pH-5.0 to pH-5.6) and dry weather. Percentages of worldwide yield losses due to wilt were found within the range of 10% to 90%, alarming the need for intensive research to sustain the tomato yield3. This fungus can be isolated from both aerial and underground parts of the infected tomato plants. Infection to the newer hosts can be furnished by spores and hypha components carried over by dead plant materials, infested soils, and wind4. Tomato roots are the gateways for FOL. Following the entry, fungus proliferates internally along the cortex and publicize to different parts of the host through vascular bundles affecting the transportation of water to the aerial parts5. It produces characteristic obstruction of xylem vessels and secretes toxins such as Dehydrofusaric acids, Fusaric acids, and Lycomarasmins, responsible for the symptomatic wilt disease6,7. Though several fungicides have been chemically synthesized and routinely employed in tomato cultivations as effective against Fusarium oxysporum, it is quite challenging to neglect their negative impacts on the released environment. Continual application of these expensive and harmful chemical antifungals into the soil imparts fungicide resistance in Fusarium spp. and also may invariably eliminate non-target and beneficial native microbial communities of the cultivable lands8. In other words, soil fertility is affected by chemical pesticide implications.
An in vivo study performed by Singha et al., (2011)2, Piper betle leaf extracts were observed to be the source of antifungals for vascular wilt in tomatoes. Though the search for antifungals from plant resources may be cost-effective, plants are supposed to be the non-renewable resources. It is presumed that mangrove ecosystem because of its versatility of nutrients shall persist as an ecological niche for a diverse group of microorganisms especially hosts a diverse group of actinobacteria with potentials to be explored9. Hence a search for antagonistic compounds among the natural resources especially from the pristine mangrove actinobacterial metabolites is an attracting choice of the study to fulfill the needs for novel antifungals to control Fusarium wilt of tomatoes. In the recent research set-up, numerous literatures narrate the production of antifungal compounds with pyrrole ring structures by a marine10 and rhizosphere soil11 actinobacteria belonging to the genus Streptomyces spp. Short-chain dehydrogenases/reducatses (SDRs) are the essential enzymes of melanin biosynthetic pathways12. SDRs are responsible for the development of key virulent factors and contribute to the pathogenic behaviors such as appressorium formation, conidial germination, penetration, and propagation inside the host tissues13. Chemical fungicides such as Oxathiapiprolin and Famoxadone were assessed for their in vitro antagonism and in silico docking with SDRs of FOL. Antibacterial metabolites produced by Nocardiopsis dassonvillei, marine actinobacterium were docked with bacterial DNA gyrases and observed to have interfering potentials in silico14. At this juncture, studies caring for the exploration of antifungals to FOL from marine actinobacterial sources with the involvement of computational tools revealing SDRs as the novel antifungal targets have not been established yet. Hence, we conducted an investigation to test the antagonistic behavior associated with mangrove sediment inhabiting actinobacterial majority followed by identification of the participating antifungal metabolites using GCMS analysis and an in silico molecular docking tactics to carefully identify the effective metabolite capable of inhibiting Short-chain Dehydrogenase / Reductase (SDR) in FOL.
Sample collection and processing
Mangrove soil sediments (50g) were collected from the Muthupet mangrove ecosystem situated at the Southeast Coast of India (10° 20` N, 79° 35`E), Tamilnadu, India, at 5 cm depth using sterile sample collection containers and transferred immediately to the laboratory at the Department of Microbiology, Alagappa University, Karaikudi, Tamilnadu, India. The mangrove sediments were air-dried for 34 h at 37°C to isolate the associated actinobacteria9. Vascular wilt affected tomato plants were collected in sterile polythene bags from agricultural fields located at the southern region of Tamilnadu, India and brought to the laboratory for the isolation of wilt fungus Fusarium oxysporum f. sp. lycopersici (FOL).
Isolation of marine actinobacteria
Pure and Distinct actinobacterial colonies were obtained using standard serial dilution and spread plating on Yeast Malt Agar (ISP2 – International Streptomyces Project 2 medium) plates supplemented with Nalidixic acid (100 mg/l) (Himedia) and Nystatin (100U/L) (Himedia) to suppress the fungal and gram-negative bacterial growth. Plates were incubated at 28°C for 7 days to attain optimum growth of actinobacterial colonies9. Purified actinobacterial cultures were stored as 20% v/v glycerol stock cultures. Early log-phase cells of potential strain (TSII) were harvested from ISP2 medium and genomic DNA was extracted by adopting the protocol described in the Molecular Cloning: A Laboratory Manual19. 16S rRNA gene was amplified specifically using the universal primers such as 27F and 1492R and sequenced in DNA sequencer (GeneAmp PCR System 9700, Applied Biosystems) at Xcelris, Ahmadabad, India.
Isolation of Fusarium oxysporum f. sp. lycopersici
Reference pathogenic fungal strain of Fusarium oxysporum f. sp. lycopersici MTCC 10270 was procured from Microbial Type Culture Collection (MTCC), India. Test pathogenic strains of Fusarium oxysporum f. sp. lycopersici were isolated from infected tomato plant tissues on Potato dextrose agar (PDA) and the virulence of the test strains were assayed by challenging each fungal strain in 30 days old healthy tomato seedlings (from Indian PKM1 variety seeds) followed by detection of wilt symptoms and re-isolation of the same pathogen proving Koch postulates15. Purified spores and mycelium of virulent strains of FOL were stored as 20% v/v glycerol stock cultures.
Assaying the antagonism to FOL
Dual culture method
The actinobacterial isolates were screened for their ability to inhibit the radial growth of FOL by employing dual culture method16 with slight modification utilizing mixture of 50% PDA + 50% ISP2 agar plates. A loopful of 7 days old actinobacterial cultures grown in ISP2 broth were line streaked on one side leaving 1 cm from the margin, and 6mm disc of fungal growth was placed at the center and incubated at 25±2°C for 7 days. Plates without antagonistic bacterial inoculum served as control. Antifungal activity was observed by the inhibition of mycelial growth of the fungal colony in the direction of actively growing actinobacteria. The level of inhibition was calculated by deducting the distance (mm) of fungal growth in the direction opposite to the antagonistic actinobacterial growth from the control fungal colony radius. The percentage of inhibition was calculated using the following formula:
% of growth inhibition = (R – r) / R ×100
Where ‘r’ is the radial growth of the fungal colony opposite the bacterial colony in mm and, R is the radial growth of the fungal pathogen in the control plate (without actinobacterial inoculum) in mm.
Agar well diffusion assay
Reference and test pathogenic fungal cultures were spread evenly onto PDA plates using sterile cotton swabs. Agar wells were punched (about 6mm in diameter) on the inoculated plates and loaded with 100μl ethyl acetate (EtOAc) extracts of cell free supernatants (CFS) from actinobacterial isolates observed to exhibit growth inhibitory effects in dual culture method along with 100μl pure ethyl acetate serving as negative control loaded into an adjacent well and incubated at 28°C ± 1°C for 2 days. After incubation, the diameter of the zone of fungal growth inhibition was measured and expressed in millimeters (mm)17.
Determination of Minimum Inhibitory Concentration (MIC) of antifungals
In vitro Minimum Inhibitory Concentration (MIC) of ethyl acetate extracts inhibiting Fusarium oxysporum f. sp. lycopersici were determined in microtiter plate dilution technique18. Briefly, the spore suspension of FOL was adjusted to 1 × 104 cells / mL by hemocytometric count and plate count on PDA. In a 96-well microtiter plate, 100µL of the extract was diluted to two folds in Potato Dextrose Broth (PDB) and 100µL of fungal spore suspension was added to each well followed by incubation at 27˚C for 3 days. Optimal fungal growth and media background were determined by growing FOL without inhibitors (control) and without fungal spores (blank), respectively. Percentage of growth inhibition at MIC can be expressed using the formula:
% growth inhibition at MIC (% GI) = [Control OD630 – Test OD630]/Control OD630 ×100
Purification and Characterization of the antifungals
Bioassay guided fractionation
Ethyl acetate extracts of actinobacteria exhibiting excellent antifungal activity against FOL in agar well diffusion assay was fractionated through thin layer chromatography (TLC) silica gel GF254 using Benzene: Dichloromethane: Methanol (6:2:2) as a solvent system and UV/iodine vapor as detection system and the active spots were identified by persistent antifungal activity in an autobiography assay on PDA. Briefly, the developed TLC plate was kept onto PDA plate surface already spread with test fungal isolates in such a way that the silica layer along with separated spots come in contact with the inoculated agar surface and the plates were incubated at 28°C for 48 h. After the detection of fungal growth inhibition in the respective compound spots of autobiography assay, the same solvent ration was used to collect the active spot as a separate fraction in silica gel column chromatography. Antifungal fractions were collected using a silica gel column (mesh size: 200 to 400) with a linear gradient (% v/v) of Benzene and Dichloromethane. The active fractions were pooled and again subjected to silica gel column with linear gradients of chloroform and ethyl acetate. Purified fraction (eluted in Chloroform: ethyl acetate at 9:1) was further separated into its isomeric forms in HPLC equipped with ODS/C18 column with linear gradient of chloroform and ethyl acetate (95% / 5%, V/V) and a flow rate of 0.5mL/min where Kirby Bauer disc diffusion assay was used to guide the antifungal activity of the elutes.
Gas Chromatography-Mass Spectral analysis
Active antifungal compounds fractionated by HPLC (Shimadzu UFLC) were subjected to GC-MS analysis to profile the list of volatile compounds whose m/z ratio were matched with library (WILEY 8 LIB) of compounds in conjunction with a literature search reporting antifungal properties previously11. In GC-MS, Fractions were injected into Gas chromatographic column (0.25µm thickness; 30 m length; 0.25mm internal diameter; capillary type column SH-Rxi-5Sil MS) with injector port temperature of 250°C, temperature of the column oven at 50°C with constant increment in the temperature of 6°C per min, hold time of 2 min and temperature of detector port at 280°C. The entire procedure lasted for 40.33 minutes with development of Gas chromatogram and respective mass spectra using Quadrupole Mass Spectrometer (Shimadzu GCMS/QP 2020).
Homology modeling and Molecular docking
Homology modeling is a reasonable theoretical method, useful to predict the unknown 3D structure by relying on the experimentally solved known 3D structure of the homologous protein. A Sequence of Short-chain Dehydrogenases/Reductase (SDR) (FOXG_00472) in Fusarium oxysporum was retrieved from NCBI and modeled using dehydrogenase of Bacillus anthracis used as a template (PDB: 3I3OA)12. The model of SDR in Fusarium oxysporum was constructed by homology modeling through the prime module of Schrodinger 2018-4. The constructed SDR model was docked with 24 ligands profiled in GCMS analysis as antifungals using Schrodinger 2018-4.
Marine actinobacterial isolates were recovered from mangrove sediments collected at Muthupet mangrove ecosystem, India. All the isolates were purified by continuous subculture on ISP2 medium and stored in 20% v/v glycerols. Isolation of mangrove sediment-associated actinobacteria was well documented in numerous reports after the completion of International Streptomyces Project20,21,9. The potential actinobacterial strain producing antifungals (TSII) was identified by 16S rRNA gene sequence homology using bioinformatics tool (NBLAST) as Kutzneria sp. and the sequence was submitted in GenBank at NCBI database with accession number MN565961.
Several strains of Fusarium oxysporum were isolated from wilt infected tomato plant tissues associated with roots, root hairs, stem, twigs, leaves, and petioles. Virulence of the test strains was investigated in planta assays with 30 days old tomato seedlings and fulfilled the Koch postulates. Virulent fungal strains were identified by microscopic and macroscopic key morphological features of the fungal elements of Fusarium oxysporum resembling the reference strain of FOL MTCC 10270.
Cell free supernatant Kutzneria sp. strain TSII exhibited in vitro antagonism to the isolated test virulent strains of Fusarium oxysporum f. sp. lycopersici (FOL) and the reference strain of FOL MTCC 10270 in Dual culture and Agar well diffusion assays indicates the involvement of extracellular antifungal metabolites (Fig. 1). A prominent antifungal activity was observed with ethyl acetate extract of TSII with percentages of growth inhibition to test and reference FOL at the minimum inhibitory concentration (MIC) (0.78µL/100µL of culture broth) were 98.88± 0.01, 98.02± 0.02, and 97.44± 0.01 respectively, indicating the solubility of antifungal metabolites (Table 1). The investigation of Harikrishnan and Shanmugaiah, (2012)17, strongly support our findings in a way the ethyl acetate extract of culture filtrate from Streptomyces sp., an actinobacterium showed a marked broad-spectrum antifungal activity towards Fusarium udum, Fusarium oxysporum, and Alternaria alternata and Macrophomina phaseolina, with inhibition of conidial and mycelial growth along with the germination of sclerotium.
Table (1):
Representation of antagonistic activity of marine actinobacteria to virulent strains of FOL.
| Isolate ID | Antagonism to Fusarium oxysporum f. sp. lycopersici(FOL) | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| Strain MTCC10270 | Strain MFOL | Strain NFOL | |||||||
| DZFGI (mm) | DCM (% GI) | MIC (% GI) | DZFGI (mm) | DCM(% GI) | MIC (%GI) | DZFGI (mm) | DCM (%GI) | MIC (%GI) | |
| TSII | 37 | 44.44 ± 0.02 | 97.40 ± 0.02 | 39 | 96.22±0.02 | 98.85 ±0.01 | 40 | 44.44 ± 0.05 | 96.01 ± 0.01 |
Note: DZFGI-Diameters of the zone of fungal growth inhibition (DZFGI) measured in agar well diffusion assay; DCM-Dual Culture Method; MIC- Minimum Inhibitory Concentration; % GI-Percentage of fungal growth inhibition. Observed values for percentage of growth inhibitions are significantly different by LSD test, values are means ± SD (n = 6–10).
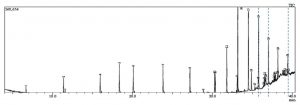
Fig. 1. GC-MS chromatogram represents the elution of Decanedioic acid, Didecyl ester at Rt (Retention time) of 36.65 min, 1,4-Benzenedicarboxylic acid, Bis(2-ethyl hexyl) ester at Rt of 37.47 min and Spiro [Cyclopentane-1,2′(1’H)-quinoxaline] at Rt of 39.06 min. Dashed lines drawn across specific elution peaks represent Decanedioic acid, Didecyl ester, 1,4-Benzenedicarboxylic acid, Bis(2-ethyl hexyl) ester and spiro [Cyclopentane-1,2′(1’H)-quinoxaline] at respective retention time.
The extract was fractionated through TLC silica gel GF254 using Benzene: Dichloromethane: Methanol (6:2:2) as a solvent system and UV/iodine vapor as detection system and the active spot (Rf value: 0.83) was identified by persistent antifungal activity in an autobiography assay on PDA. Antifungal fractions collected using silica gel column with a linear gradient of Benzene and Dichloromethane at 6:4 were pooled and further eluted in Chloroform: ethyl acetate at 9:1. This partially purified fraction was further separated into its isomeric forms in HPLC equipped with ODS/C18 column with linear gradients of chloroform and ethyl acetate where Kirby Bauer disc diffusion assay was used to guide the antifungal activity of the elutes. GCMS profiling of HPLC purified active fractions from TSII was performed in Shimadzu GCMS/QP2020 and resulted in 24 bioactive metabolites whose m/z ratio, retention time and molecular formula were matched with compounds available with Wiley Library (Version 8.0) (Fig. 1). Of these, eight compounds were found to be antimicrobials reported in earlier studies as follows and might be responsible for the in vitro antagonism to FOL (Table 2.).
Table (2):
Gas Chromatography Mass Spectrometry predicted major antimicrobial compounds from marine actinobacteria Kutzneria sp. strain TS-II.
Name of the Compound (Ligands) |
Molecular Formula |
Peak area (%) |
Structures of ligands |
Functions reported previously |
|---|---|---|---|---|
Eicosane |
C20H42 |
8.43 |
Antifungals |
|
1,2-Benzene Dicarboxylic acid (Phthalic acid) |
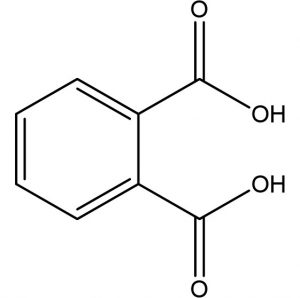
C8H6O4 |
3.42 |
Antimicrobials |
|
1,4-Benzenedicarboxylic acid, Bis(2-ethylhexyl) ester |
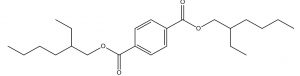
C24H38O4 |
1.82 |
Antifungals |
|
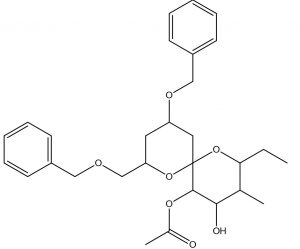
1,7-Dioxaspiro[5.5]undecane-4,5-diol, 2-ethyl-3-methyl-10-(phenylmethoxy)-8-[(phenylmethoxy)methyl]-, 5-acetate |
C29H38O7 |
0.4 |
Antimicrobials |
|
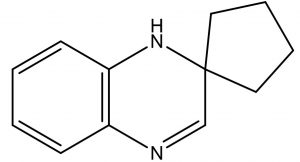
Spiro[Cyclopentane-1,2′(1’H)-quinoxaline] |
C12H14N2 |
2.4 |
Antifungal,
Anticandidal, |
|
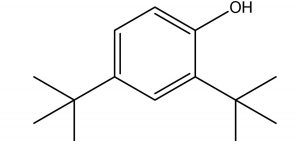
Phenol, 2,4-Bis(1,1-Dimethylethyl)- |
C14H22O |
4.32 |
Antifungals |
|
Tritriacontane |
C33H68 |
3.36 |
No activity reported |
|
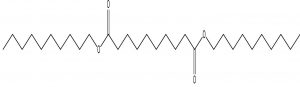
Decanedioic acid, Didecyl ester |
C30H58O4 |
2.23 |
Antimicrobials |
The previous report of Velikorodov et al., (2012)22, evaluated in vitro antifungal activity of carbamate-functionalized spiro compounds against dermatophytic fungi such as Microsporum canis and Trichophyton rubrum. However, the functionalized Spiro compounds were found to exhibit fungistatic properties. Glucosamine-6-phosphate synthases and Methionyl tRNA synthetases are the key enzymes in the pathway of cell wall chitin synthesis and aminoacylation of tRNA synthesis in fungi. These enzymes were the antifungal targets in an in silico virtual screening by Sapnakumari et al., (2017)23, who identified spirooxindoles for their ability to inhibit methionyl tRNA synthetase (PDB ID: 1PFV) and glucosamine-6-phosphate synthase (PDB ID: 1JXA) enzymes which was supported by in vitro antagonism by chemically synthesised spirooxindoles. Similarly, Wu et al., (2015)24, have also contributed synthetic spirooxindole derivatives for antifungal activity against phytopathogenic fungi including Fusarium graminearum. Synthetic spiro [cyclopropane-1, 4′-pyrazol-3-one] derivatives were found to have inhibitory effects on Candida albicans and shall serve as leads for anticandidal drugs25. Spiro pyrrolidines were synthesized and assayed for antibacterial and antifungal activities26. Though the enlisted reports describe the antifungal potential of synthetic compounds with a spiro ring system, we report for the first time a similar group of spiro compound, SPIRO[Cyclopentane-1,2’(1’H)-Quinoxaline], from marine Kutzneria sp. strain TSII sp. with antifungal activity against FOL.
Ren et al., (2019)27, have also identified the antifungal activity of Phenol, 2, 4-Bis (1,1-Dimethylethyl)- from Pseudomonas fluorescens towards several phytopathogenic fungi including Fusarium oxysporum. In parallel, Antifungal activity of Phenol, 2,4-Bis(1,1-Dimethylethyl)-, pyrrolizidines and fatty acid esters produced by Streptomyces sp. from the rhizosphere of Opuntia stricta11 correlates the antifungal potential of Phenol, 2,4-Bis(1,1-Dimethylethyl)-, and fatty acid methyl esters observed in our findings. Anticandidal and antibacterial activity was observed in the ink of squid Loligo duvauceli due to the presence of bis(2-ethylhexyl) phthalate (BEHP) in a study carried out by Girija et al., (2014)28 relating the possible antimicrobial efficacy of Phthalates documented in our GCMS data.
Wang et al., (2012)29, observed the production of Benzeneacetic acid, 2-propenyl ester in addition to 3-phenyllactic acid by the lactic acid bacterium Lactobacillus plantarum from Koumiss with broad-spectrum antifungal activity against F. oxysporum, Penicillium citrinum and P. drechsleri. Fatty acid methyl esters such as Octadecanoic acid, methyl esters and hexadecanols extracted from marine macroalgae were the antifungals to Fusarium oxysporum and Fusarium solani30. Thus the earlier reports stand as added evidence indicating the sensitivity of FOL to the fatty acids present in the extract of Kutzneria sp. TSII.
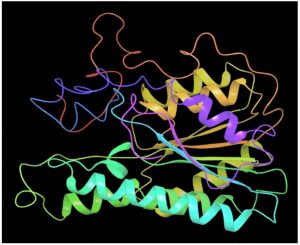
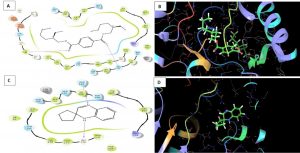
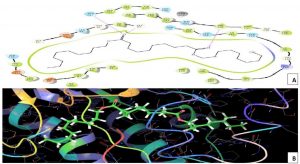
Melanin biosynthesis is essential to contribute pathogenicity to many plant pathogenic fungi. Several commercial fungicides target enzymes of the melanin biosynthetic pathway especially the class of Short-chain Dehydrogenases / Reductases (SDRs)13. Molecular docking studies are recently taking birth to investigate the probable interaction of FOL SDR with commercial fungicides like Famoxadone 12. However, our study finds novelty through molecular docking of actinobacterially synthesized antifungal compounds against the modeled SDR (FOXG_00472) of FOL. Sequence of Short-chain dehydrogenases/reductase (SDR) (FOXG_00472) in Fusarium oxysporum was retrieved from NCBI and modeled using SDR of Bacillus anthracis as a template (PDB: 3I3OA) since the template exhibited highest similarity index of 52.02% and query coverage of 97%. A total of 24 useful ligands have been detected in GCMS analysis and were docked with the modeled protein SDR of FOL (FOXG_00472). Among the 24 compounds docked, 1, 4-Benzenedicarboxylic acid, Bis(2-ethylhexyl) ester – FOXG_00472 complex, Spiro [Cyclopentane-1,2’(1’H)-quinoxaline] – FOXG_00472 and Decanedioic acid, Didecyl ester – FOXG_00472 have the docking scores (Kcal/mol) of – 8.151, -7.231, – 6.031 and the lowest binding free energy (ΔGbind) value (Kcal/mol) of – 68.11, – 42.33 and -79.41 respectively (Table 3). 2D and 3D structures of modeled FOXG_00472 alone and ligand-FOXG_00472 complexes were represented in Fig 2A; 2B; 2C. Changes occurred at active site region of modeled SDR protein (FOXG_00472) during the interaction of the 1, 4-Benzenedicarboxylic acid, Bis(2-ethylhexyl) ester especially in the residues such as Ile66, Ser192. Similarly, changes were also common upon the interaction of Spiro [Cyclopentane-1, 2’(1’H)-quinoxaline] with FOXG_00472 in the residues of Gly236, Gln145. In case of interaction between Decanedioic acid, Didecyl ester and FOXG_00472, changes can be detectable in the residues such as Asn141, Gly67, Val238. Hence, the interaction of three docked ligands [1, 4-Benzenedicarboxylic acid, Bis(2-ethylhexyl) ester, Spiro [Cyclopentane-1,2’(1’H)-quinoxaline] and Decanedioic acid, Didecyl ester] with the modeled SDR of FOL may synchronously inhibit melanin biosynthesis relating to the antifungal action of the ligands explored from marine actinobacterium Kutzneria sp. Further studies encompassing the development of antifungal formulations and application in field trials may explore novel alternatives for chemical fungicide use in tomato cultivations.
Table (3):
Docking parameters analysed for the three antifungal lead compounds in Schrodinger 2018-4.
Compound Name |
Docking score (Kcal/mol) |
Glide emodel (Kcal/mol) |
Glide Energy (Kcal/mol) |
Interacting aminoacid residues |
ΔGbind(Kcal/mol) |
|---|---|---|---|---|---|
1,4-Benzenedicarboxylic acid, Bis(2-ethylhexyl) ester |
-8.151 |
-79.193 |
-54.932 |
Ile66, Ser192 |
-68.11 |
Spiro[Cyclopentane-1,2′(1’H)-quinoxaline] |
-7.231 |
-43.211 |
-31.096 |
Gly236, Gln145 |
-42.23 |
Decanedioic acid, Didecyl ester |
-6.031 |
-80.819 |
-57.471 |
Asn141, Gly67, Val238 |
-79.41 |
Fig. 2A. Modeled 3D structure of SDR protein (FOXG_00472) in Fusarium oxysporum f. sp. lycopersici (FOL) using prime module of Schrodinger 2018-4
Fig. 2B. Molecular Docking of actinobacterially synthesized ligands with modeled SDR protein FOXG_00472 in Fusarium oxysporum f. sp. lycopersici (FOL). (A) 2D structure of FOXG_00472-1,4-Benzenedicarboxylic acid, Bis ( 2-ethyhexyl) ester complex showing the interacting aminoacid residues. (B) 3D structure of FOXG_00472- ligand complex showing the interaction of 1, 4-Benzenedicarboxylic acid, Bis(2-ethylhexyl) ester at the active site of modeled SDR protein FOXG_00472. (C) 2D structure of FOXG_00472 – Spiro [Cyclopentane-1,2’(1’H)-quinoxaline] complex showing the interacting aminoacid residues. (D) 3D structure of FOXG_00472 – ligand complex showing the interaction of Spiro [Cyclopentane-1,2’(1’H)-quinoxaline] at the active site of modeled SDR protein FOXG_00472
Fig. 2C. Molecular Docking of actinobacterially synthesized ligands with modeled SDR protein FOXG_00472 in Fusarium oxysporum f.sp. lycopersici (FOL). (A) 2D structure of FOXG_00472- Decanedioic acid, Didecyl ester complex showing the interacting aminoacid residues. (B) 3D structure of FOXG_00472 – ligands complex showing the interaction of Decanedioic acid, Didecyl ester at the active site of modeled SDR protein FOXG_00472
The present investigation deals with the effective in vitro antagonistic potential of marine actinobacterium Kutzneria sp. strain TSII through the secretion of antifungal compounds targeting Short-chain Dehydrogenases/Reductases of fungal melanin biosynthetic pathway and exerting inhibitory effects in an in silico study. Furthermore, this study uncovers potential metabolome of marine actinobacteria having antifungal products which shall be applied as biocontrol formulates to control fungal pests like Fusarium oxysporum in agriculture. On the birth of this investigation, we conclude that these bioactive compounds may replace the usage of chemically synthesized fungicides for eco-friendly and sustainable crop protection.
ACKNOWLEDGMENTS
All the authors are very much thankful for the research grant provided by DST-SERB-ECRA project sanctioned vide Letter No. DST/SERB/ECRA/000446/2017, under Government of India and RUSA Phase 2.0 scheme grant sanctioned vide Letter No. F. 24-51/2014-U, Policy (TNMulti-Gen), Department of Education, Govt. of India, dated 09.10.2018. All the authors gratefully acknowledge the lab support provided by the Department of Bioinformatics, Alagappa University, Karaikudi, India, for the bioinformatics and molecular docking part of the manuscript.
CONFLICT OF INTEREST
All the authors declare no conflict of interest.
AUTHORS’ CONTRIBUTION
TK designed, supervised the research, prepared and prompted the manuscript. SS performed in silico experiments and RMV directed in silico studies. KKS and TSD assisted in vitro studies. KV performed in vitro experiments, analysed the data and prepared the manuscript. All the authors have read and approved the manuscript.
FUNDING
DST-SERB, India [Grant no. DST/SERB/ECRA/000446/2017 and RUSA phase 2.0 scheme [F. 24-51/2014-U, Policy (TNMulti-Gen), Department of Education, Govt. of India, dated 09.10.2018.
ETHICS STATEMENT
Not applicable.
AVAILABILITY OF DATA
All datasets analysed in this study are included in the manuscript.
- KL Brookie, GI Best and T S Conner. Intake of raw fruits and vegetables is associated with better mental health than intake of processed fruits and vegetables. Front Psychol, 2018; 9: 487.
- IM Singha, Y Kakoty, BG Unni, MC Kalita, J Das, A Naglot, ….. and L Singh. Control of Fusarium wilt of tomato caused by Fusarium oxysporum f. sp. lycopersici using leaf extract of Piper betle L.: a preliminary study. World J Microb Biot, 2011; 27(11): 2583.
Crossref - AK Singh and S Kamal. Chemical control of wilt in tomato (Lycopersicon esculentum L.). Int J Hortic, 2012; 2.
- BA Summeral, B Salleh, JF Leslie. A utilitarian approach to Fusarium identification. Plant Dis, 2003; 87: 117–128.
Crossref - BE Kane and GK Link. Production of Toxic Substances in vitro by Fusarium lycopersici. Proc Soc Exp Biol Med, 1927; 24(6):578-580.
Crossref - VK Singh, HB Singh and RS Upadhyay. Role of fusaric acid in the development of ‘Fusarium wilt’ symptoms in tomato: Physiological, biochemical and proteomic perspectives. Plant Physiol Biochem, 2017; 118: 320-332.
Crossref - C Srinivas, DN Devi, KN Murthy and CD Mohan, TR Lakshmeesha, B Singh, ….. and B Tabassum. Fusarium oxysporum f. sp. lycopersici causal agent of vascular wilt disease of tomato: Biology to diversity–A review. Saudi J Biol Sci, 2019.
Crossref - JA Lucas, NJ Hawkins and BA Fraaije. The evolution of fungicide resistance. In Advances in Applied Microbiology, 2015; 90: 29-92, Academic Press.
Crossref - ZA Zainal Abidin, N Abdul Malek, Z Zainuddin and AJK Chowdhury. Selective isolation and antagonistic activity of actinomycetes from mangrove forest of Pahang, Malaysia. Front Life Sci, 2016; 9(1): 24-31.
Crossref - K Kannabiran. Bioactivity of Pyrrolo [1, 2-a] pyrazine-1, 4-dione, hexahydro-3-(phenylmethyl)- Extracted from Streptomyces sp. VITPK9 Isolated from the Salt Spring Habitat of Manipur, India. Asian J Pharm, 2016; 10(4).
- D Qi, L Zou, D Zhou, Y Chen, Z Gao and R Feng, M Zhang, K Li, J Xie, and W Wang. Taxonomy and Broad-Spectrum Antifungal Activity of Streptomyces sp. SCA3-4 Isolated From Rhizosphere Soil of Opuntia stricta. Front Microbiol, 2019; 10.
Crossref - M Aamir, VK Singh, MK Dubey, M Meena, SP Kashyap, SK Katari, RS Upadhyay, A Umamaheswari and S Singh. In silico Prediction, Characterization, Molecular Docking and Dynamic Studies on Fungal SDRs as Novel Targets for Searching Potential Fungicides against Fusarium Wilt in Tomato. Front Pharmacol, 2018; 9: 1038.
Crossref - MJ Kwon, KS Kim and YH Lee. A short-chain dehydrogenase/reductase gene is required for infection-related development and pathogenicity in Magnaporthe oryzae. Plant Pathol J, 2010; 26(1): 8-16.
Crossref - S Priyanka, M Jayashree, R Shivani, S Anwesha and KB Rao. Characterisation and identification of antibacterial compound from marine actinobacteria: In vitro and in silico analysis. J Infect Public Heal, 2019; 12(1): 83-89.
Crossref - K Inami, T Kashiwa, M Kawabe, A Onokubo-Okabe, N Ishikawa, ER Perez, Hozumi T, Caballero LA, de Baldarrago FC, MJ Roco, KA Madadi, TL Peever, T Teraoka, M Kodama and T Arie. The tomato wilt fungus Fusarium oxysporum f. sp. lycopersici shares common ancestors with nonpathogenic F. oxysporum isolated from wild tomatoes in the Peruvian Andes. Microbes Environ, 2014; ME13184.
Crossref - F Mela, K Fritsche, W De Boer, JA Van Veen, LH De Graaff, M Van Den Berg and JH Leveau. Dual transcriptional profiling of a bacterial/fungal confrontation: Collimonas fungivorans versus Aspergillus niger. ISME J, 2011; 5(9): 1494.
Crossref - H Harikrishnan and V Shanmugaiah. Streptomyces sp. VSMGT1014-mediated antifungal activity against fungal plant pathogens. In Prospects in Bioscience: Addressing the Issues, 2012; 335-341.
Crossref - J Nam, MY Jung, PI Kim, HB Lee, SW Kim and CW Lee. Structural characterization and temperature-dependent production of C 17-fengycin B derived from Bacillus amyloliquefaciens subsp. plantarum BC32-1. Biotechnol Bioprocess Eng, 2015; 20(4): 708-713.
Crossref - MR Green and J Sambrook. Molecular cloning. A Laboratory Manual 4th ed., 2012.
- B Gong, S Chen, W Lan, Y Huang and X Zhu. Antibacterial and antitumor potential of actinomycetes isolated from mangrove soil in the Maowei Sea of the southern coast of China. Iran J Pharm Res, 2018; 17(4): 1339.
- E Rosmine and S A Varghese. Isolation of actinomycetes from mangrove and estuarine sediments of Cochin and screening for antimicrobial activity. J Coast Life Med, 2016; 4(3): 207-210.
Crossref - AV Velikorodov, VA Ionova, OV Degtyarev and LT Sukhenko. Synthesis and antimicrobial and antifungal activity of carbamate-functionized spiro compounds. Pharm Chem J, 2013; 46(12): 715-719.
Crossref - M Sapnakumari, B Narayana, KS Shashidhara and BK Sarojini. Multicomponent synthesis, biological evaluation and molecular docking of new spiro-oxindole derivatives. J Taibah Univ Sci, 2017; 11(6): 1008-1018.
Crossref - JS Wu, X Zhang, YL Zhang and JW Xie. Synthesis and antifungal activities of novel polyheterocyclic spirooxindole derivatives. Org & Biomol Chem, 2015; 13(17): 4967-4975.
Crossref - H Maruoka, N Kashige, T Eishima, F Okabe, T Fujioka, F Miake, … and R Tanaka. Synthesis and antifungal activity of spiro [cyclopropane-1, 4′-pyrazol-3-one] derivatives. J Heterocycl Chem, 2008; 45(6):1883-1887.
Crossref - AA Raj, R Raghunathan, MR Sridevi Kumari and N Raman. Synthesis, antimicrobial and antifungal activity of a new class of spiro pyrrolidines. Bioorg Med Chem, 2003; 11(3): 407-419.
Crossref - J Ren, J Wang, S Karthikeyan, H Liu and J Cai. Natural anti-phytopathogenic fungi compound phenol, 2, 4-bis (1, 1-dimethylethyl) from Pseudomonas fluorescens TL-1. Indian J Biochem Bio, 2019; 56(2): 162-168.
- S Girija, V Duraipandiyan, PS Kuppusamy, H Gajendran, and R Rajagopal. Chromatographic characterization and GC-MS evaluation of the bioactive constituents with antimicrobial potential from the pigmented ink of Loligo duvauceli. Int Sch Res Notices, 2014.
Crossref - H Wang, Y Yan, J Wang, H Zhang and W Qi. Production and characterization of antifungal compounds produced by Lactobacillus plantarum IMAU10014. PloS one, 2012; 7(1): e29452.
Crossref - AH Shobier, SAA Ghani and KM Barakat. GC/MS spectroscopic approach and antifungal potential of bioactive extracts produced by marine macroalgae. Egypt J Aquat Res, 2016; 42(3): 289-299.
Crossref
© The Author(s) 2020. Open Access. This article is distributed under the terms of the Creative Commons Attribution 4.0 International License which permits unrestricted use, sharing, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.