ISSN: 0973-7510
E-ISSN: 2581-690X
Accurate identification of fungal causes for onychomycosis is essential for proper treatment. Presently available laboratory methods show unreliable sensitivity; so there is a requirement for innovative detection techniques. The aim for this work was to assess the efficiencies of fluorescent staining and internal transcribed spacer (ITS) ribosomal DNA (rDNA) polymerase chain reaction (PCR)-based sequencing in comparison to conventional techniques for diagnosis of onychomycosis. Nail specimens obtained from 100 patients with clinically- diagnosed onychomycosis were analyzed. Nail scrapings or clippings were subjected to direct microscopic examination by KOH mount, culture by using Sabouraud’s dextrose agar and histopathological examination with periodic-acid Schiff (PAS). Collected specimens were subsequently examined by fluorescent staining and PCR-based sequencing (30 specimens only) to compare the feasibility, sensitivity and diagnostic accuracy for these two methods. The most frequently isolated fungi were yeasts (39/76: 51.3%), dermatophytes (24/76; 31.6%) and non-dermatophyte molds (NDMs) (13/76; 17.1%). Mixed mycotic infections were recovered from 6% of the collected nail specimens. The positive detection rates were significantly different between KOH examinations (52%), nail plate histology (55%), fungal culture (70%) and fluorescent staining (80%). Considering fungal culture as the gold standard, the most sensitive technique was PCR (100%) followed by fluorescent staining (89%), PAS staining (69%) while the least sensitive technique was KOH mount (53%). Fluorescence staining can be used as a rapid and high-yield technique for identification of fungi in the specimens. PCR-based sequencing was highly sensitive and faster compared to culture. Whenever possible, it enables species identification with higher adequacy.
Onychomycosis, Direct KOH mount, Fluorescent staining, Fungal culture, PCR-based sequencing
Onychomycosis is a mycotic infection that affects fingers and toenails, leading to nail plate and nail bed distortion. It is considered one of the most common nail diseases globally and constitutes about 50% of whole clinical nail syndromes.1
The etiological causes of onychomycosis may be dermatophytes, non-dermatophyte molds (NDMs) and yeasts. Onychomycosis is categorized into five clinical types: Distolateral subungual onychomycosis; DLSO (discoloration, onycholysis, subungual hyperkeratosis and thickening of the distal and/or lateral nail), White superficial onychomycosis; WSO (white spots and textural changes on or in the nail plate), Proximal subungual onychomycosis; PSO (discoloration and onycholysis affecting the proximal part of the nail), Candidal onychomycosis; CO (marked thickening and roughness of nail plates) and Total dystrophic onychomycosis; TDO (involves the entire nail bed and plate with thickening and roughening or destruction of nail plate.1,2
According to etiologic agents of onychomycosis, dermatophytes are considered the main fungal agents (90% and 40% of toenail and fingernail infections respectively) of which Trichophyton rubrum, T. mentagrophytes and T. tonsurans are the most prevalent species. Onychomycosis of fingernails is usually triggered by yeast infection, mainly Candida spp. The main NDM involved in onychomycosis are Curvularia, Aspergillus, Fusarium, Penicillium and Scytalidium species.2 The disease necessitates long-duration systemic antifungal treatment; hence, accurate mycological identification is crucial for proper treatment consequences. Therefore, fast and sensitive approaches for identification of fungal species are necessary.3
Direct microscopy of KOH mounts requires experienced investigators and may yield 5–15% of false-negative results. Regarding PAS staining; not only the specific etiologic agent is difficult to be established but also, the fungal viability isn’t documented. Also, fungal culture is a slow procedure that requires 3-4 weeks up to 6 weeks to obtain the result and has a higher proportion of false- negatives results (30–50%). Despite all these drawbacks, identification of the causative organism by microbiological techniques is still the cornerstone for disease diagnosis.2,3
Molecular techniques centered on the polymerase chain reaction (PCR) have been introduced that provide a timely and accurate diagnosis of onychomycosis and other mycotic diseases.3 Direct diagnosis of fungal diseases in clinical samples by PCR-based methods would permit early and detailed identification of the causative agents.4
The amplification of internal transcribed spacer (ITS) regions and 28SnrDNA (D1/D2 region) followed by Sanger sequencing have been used for identification of isolated fungi in clinical specimens directly.4,5 A limited number of studies were done to compare between conventional and non-conventional methods for diagnosis of onychomycosis and identification of causative clinical isolates.
The current study aimed to highlight risk factors associated with onychomycosis, to identify the causative fungal pathogens and to compare the diagnostic validity of fungal fluorescent staining and ITS rDNA PCR-based sequencing with conventional methods (KOH direct microscopy, mycological culture and histopathology by PAS stain) for diagnosis of onychomycosis.
Study subjects and data collection
This cross-sectional study was carried out during the period from January 2020 to April 2021 at the Medical Microbiology and Immunology Department in collaboration with the Central Laboratory of Faculty of Medicine, Menoufia University. It involved 100 patients (15 males and 85 females with a mean age of 47.36±15.204 years) clinically- diagnosed to have onychomycosis at the Outpatient Clinic of Dermatology Department.
The Local Ethics Committee of Faculty of Medicine, Menoufia University (11/2018MICR3) approved the study protocol. Informed consents were taken from all participants.
All the patients submitted to full history taking including risk factors of onychomycosis in order to focus on the factors affecting and motivating the occurrence of onychomycosis.
Patients with permanent or semi-permanent discoloration of the nail plate due to topical therapeutics, cosmetics or professional dye exposure, subungual hematoma, nevoid subungual formation or any generalized skin disease associated with nail disorders for example psoriasis, lichen planus and atopic dermatitis were excluded.6
Specimen collection and transport
Nail specimens (60 toenail and 40 fingernail samples) were collected after removing dirt and dust with distilled water from the affected site and cleaning with 70% alcohol. Nail plate clipping, scrapings of nail bed or subungual debris was obtained in a sterile plastic Petri dish plate.7
Processing of nail specimens:
- Each nail specimen was evenly divided into five parts.
- The first portion of specimen was examined directly by 20% KOH mounts. The presence of fungal structures in the form of fungal hyphae, pseudohyphae, arthroconidia, spores, or budding cell was reflected as positive.8
- The second portion of the nail specimen was stained with PAS stain for histopathological examination. Specimens were fixed in paraffin blocks. Approximately 3mm thin slices were taken and mounted on glass slides. PAS staining was then done and examined microscopically for the appearance of any fungal elements. A positive histology was reported on presence of magenta-colored hyphae, arthroconidia, or yeasts.9
- The third portion of the nail specimen was stained with acridine orange (AO) fluorescent dye. One ml of stock solution of AO (A6014; Sigma, Welwyn Garden City, UK) was added to 9ml of a 20% KOH solution. Two drops were placed on a glass slide; then, nail scrapings were added and covered by a cover slip, gently heated over an open flame, and examined using a fluorescence microscope (N.14800, Humascope Flu LED with fluorescence filter; Germany). The fungal fluorescence was identified by tubular or annular elements with a thin rim of bright orange fluorescence outlining the usual shape of the fungal cell wall as described in Mann’s recommendations.10
- The fourth portion of nail specimen was firstly minced and then divided into two parts to be cultured onto two Sabouraud’s dextrose agar plates (one with cycloheximide and chloramphenicol and the other without). These plates were incubated at 25°C and 37°C for 1-4 weeks and examined daily for colony growth. Dermatophytes were identified macroscopically (color and consistency of the surface and reverse, topography, and texture). For suspected yeast and yeast-like growth, microscopic examination of isolates was performed after staining by Gram’s stain to evaluate their response to stain, shapes, their arrangement and yeast budding form.11
- The fifth portion of nail specimen was examined by internal transcribed spacer (ITS) ribosomal DNA (rDNA) PCR-based sequencing. Fungal DNA was extracted using the DNeasy Plant Mini Kit (cat. nos. 69104 and 69106) according to the Manufacturer’s instructions. The fungus-specific universal primers ITS1 (5ʹ- TCCGTAGGTGAACCTGCGG-3ʹ) and ITS4 (5ʹ-TCCTCCGCTTATTGATATGC-3ʹ) were used to amplify the full ITS sequence (product size 700-900 bp).12
PCR was performed for the 100 samples in a thermocycler (Biometra, Germany). PCR products were separated by gel electrophoresis, subsequently purified with the Invitrogen™ PureLink™ PCR Purification Kit-Fisher Scientific. Sequencing of 30 purified PCR products was done using Taq DyeDeoxy™ and ABI Prism™ terminator cycle sequencing kits (Applied Biosystems, Foster City, CA, USA) and the above-mentioned forward and reverse PCR primers. The sequencing results were evaluated using the nucleotide Basic Local Alignment Search Tool (BLAST) to determine the closest relatives on the NCBI website (http: //www. ncbi.nlm.nih.gov).13
Statistical Analysis
SPSS version 26 on IBM compatible computer was utilized to perform statistical analysis. Qualitative data were stated as number and percentage and comparison between the tests of diagnosis was done using Chi-square and Z tests. P-value <0.05 was considered significant. Agreement between the tests was evaluated by the Cohen kappa statistic to evaluate the level of agreement. To determine the efficacy and performance characteristics of each test, fungal culture was selected as the cornerstone. The sensitivity, specificity, accuracy, PPV, and NPV were determined for other methods (Foundation for Statistical Computing, Vienna, Austria, 2015).
One hundred patients with clinically-diagnosed onychomycosis were enrolled in this study. About 59%, 46%, 25% and 20% of the studied patients had chronic hepatic diseases, anemic profile, diabetes and peripheral vascular diseases (PVD), respectively. Notably, 71%, 67% and 54% of the studied patients were excessively exposed to water, chemicals and soil, respectively. The most prevalent clinical type was DLSO (38%) and the mostly affected site was toenails (60%) (Table 1).
Table (1):
Socio-demographic data, risk factors and clinical characteristics of the studied patients.
Characteristics |
No. =100 |
% |
|---|---|---|
Age (mean=47.36±15.204) • 0-20 years • 21-40 years • 40-60 years • >60 years |
5 28 50 17 |
5% 28% 50% 17% |
Sex • Male • Female |
15 85 |
15% 85% |
Diabetes mellitus • Present • Controlled • Uncontrolled • Absent |
25 15 10 75 |
25% 15% 10% 75% |
Peripheral vascular diseases • Present • Absent |
20 80 |
20% 80% |
Chronic hepatic disease (HCV) • Present • Absent |
59 41 |
59% 41% |
Anemia • Present • Absent |
46 54 |
46% 54% |
Excessive exposure to • Water • Chemicals • Soil |
71 67 54 |
71% 67% 54% |
Occlusive footwear • Present • Absent |
8 92 |
8% 92% |
Clinical types • DLSO • TDO • WSO • PO |
38 24 18 20 |
38% 24% 18% 20% |
Affected nails • Toenail • Fingernail • Both |
60 40 0 |
60% 40% 0% |
Severity of onychomycosis • Mild • Moderate • Severe |
15 |
15% 60% 25% |
The phenotypic diagnostic tests; KOH direct microscopy, PAS staining, fungal culture and fluorescent staining were respectively positive in 52%, 55%, 70% and 80% of the processed specimens. There was a statistically- significant difference between positive and negative results regarding fungal culture and fluorescent staining procedures (P<0.001) (Table 2 & Fig. 1).
Table (2):
Diagnosis of onychomycosis by phenotypic tests.
| Diagnostic tests | Positive No. (%) |
Negative No. (%) |
Total (n=100) | Z test | P-value | |
|---|---|---|---|---|---|---|
| Conventional | KOH mount | 52 (52%) | 48 (48%) | 100 | 0.42 | 0.335 |
| PAS staining | 55 (55%) | 45 (45%) | 100 | 1.27 | 0.101 | |
| Mycological culture | 70 (70%) | 30 (30%) | 100 | 5.52 | <0.001** | |
| Non-conventional | Fluorescent staining | 80 (80%) | 20 (20%) | 100 | 8.34 | <0.001** |
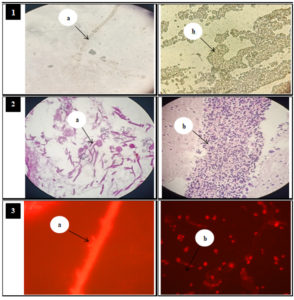
Fig. 1. KOH mount, PAS staining, fluorescent staining results
1: KOH mount showing: (a): Branched septate hyphae (b): Budding yeast cell.
2: PAS stain film showing: (a): Branched hyphae with conidiophores. (b): Budding yeast cell.
3: Fluorescent microscopic examination showing: (a): Branched hyphae with microconidia. (b): Budding yeast cell with pseudohyphae.
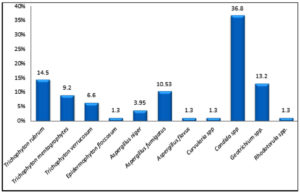
A total of 76 fungal species were isolated from 70 positive fungal cultures. This study showed that the highest frequency was for yeasts (39/76: 51.3%) of which the most frequently isolated species were Candida spp. (28/76: 36.8%). Dermatophytes accounted for 31.6 % (24/76) of the totally-obtained fungal isolates of which Trichophyton rubrum was the most prevalent species (11/76:14.5%). NDMs represented 17.1 % (13/76) where; Aspergillus fumigatus was the predominant species (8/76: 10.5%). Mixed mycotic infections were recovered from 6% of the cultured nail specimens (Aspergillus fumigatus + Candida spp.; 3% and Aspergillus fumigatus+ Trichophyton rubrum; 3%) (Fig. 2 & Fig. 3).
Fig. 2. Mycological profile of the collected nail specimens 76 fungal species were isolated from 70 positive fungal cultures.
Fig. 3. Macroscopic appearance of the obtained fungal isolates on SDA
1: Trichophyton mentagrophytes showing: (a): Fluffy surface appearance. (b): Reddish brown colored reverse.
2: Epidermophyton floccosum showing: (a): Cottony white aerial mycelium surface appearance. (b): Yellowish brown reverse colony.
3: Aspergillus fumigatus showing blue-green granular colony.
4: Curvularia spp. showing fluffy and gray to gray-brown colony.
5: Rhodotorula spp. showing moist, creamy colonies.
6: Candida spp. showing smooth, creamy white colonies
7: Geotrichium spp. showing white to cream-colored yeast like colony.
In this study, 98 out of 100 nail specimens (98%) were positive by PCR assay (Fig. 4).The results of ITS rDNA PCR-based sequencing that was applied for species identification from 30 nail specimens revealed that yeasts, dermatophytes and NDMs accounted for 36.7% (11/30), 33.3% (10/30) and 30% (9/30), respectively. Among yeast fungi, Candida tropicalis was the most frequent isolate (5/30:16.7%). The most dominant isolated dermatophytes were Trichophyton rubrum representing 16.7% (5/30). For NDMs, Aspergillus niger, Aspergillus fumigatus and Curvularia lunata equally represented 10% (3/30) (Fig. 5 & Fig. 6a & b).
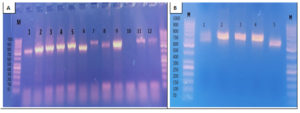
Fig. 4. Agarose gel electrophoresis for the PCR amplified products of ITS gene applied for diagnosis of onychomycosis
A: Before purification
• Lane M: DNA molecular size marker (100-1000 bp).
• Lanes 1, 2, 3, 4, 5, 6, 7, 8, 9, 11 and 12 were positive for ITS gene.
• Lane 10 was negative for ITS gene.
B: After purification
• Lane M: DNA molecular size marker (100-1000 bp).
• Lanes 2, 3 and 4 were purified PCR amplified products.
• Lane 1 and 5 were non-purified PCR amplified products.
Fig. 5. Distribution of the obtained fungal species according to ITS rDNA PCR- based sequencing.
Sequencing was performed for 30 PCR products of nail specimens.
Agreement between the diagnostic tests (KOH, PAS, fluorescent staining and PCR) and fungal culture was evaluated by the Cohen kappa test. For each pair of tests, the kappa coefficient interval was listed in Table 3. The agreement of fungal culture with PAS and fluorescent staining was fair (0.396 and 0.315, respectively) and proved to be higher than that with KOH and PCR (slight agreement; 0.024 and 0.091, respectively).
Table (3):
Agreement and diagnostic accuracy of KOH mount, PAS staining, fluorescent staining and PCR in relation to mycological culture as the gold standard for diagnosis.
| Diagnostic test | Culture (gold standard) | Sensitivity (%) | Specificity (%) | Accuracy (%) | PPV (%) | NPV (%) | Kappa test* | P-value | |
|---|---|---|---|---|---|---|---|---|---|
| Positive (n=70) | Negative (n=30) | ||||||||
| KOH Positive (n=52) | 37(52.9%) | 15 (50%) | 53% | 50% | 52% | 71% | 31% | 0.024 | 0.793 |
| mount Negative (n=48) | 33 (47.1%) | 15 (50%) | |||||||
| PAS Positive (n=55) | 48 (68.6%) | 7 (23.4%) | 69% | 77% | 71% | 87% | 51% | 0.396 | <0.001** |
| Negative (n=45) | 22 (31.4%) | 23(76.6%) | |||||||
| Fluorescent Positive (n=80) | 62(88.6 %) | 8(40%) | 89% | 40% | 74% | 78% | 60% | 0.315 | 0.029*** |
| staining Negative (n=20) | 8(11.4%) | 12(60%) | |||||||
| PCR Positive (n=98) | 70(100%) | 28(93.3%) | 100% | 7% | 72% | 71% | 100% | 0.091 | 0.029*** |
| Negative (n=2) | 0(0%) | 2(6.7%) | |||||||
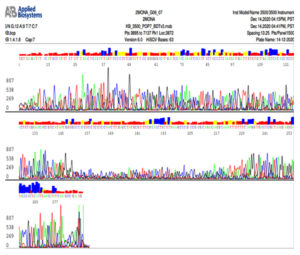
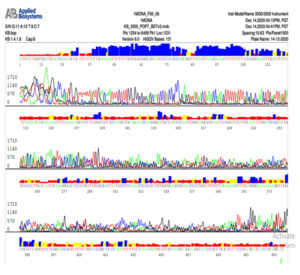
Fig. 6b. Nucleotide sequence of ITS gene of Trichophyton mentagrophytes isolate
Note that C is colored by red, A is colored by green, G is colored by black and T is colored by blue color
Considering fungal culture as the gold standard for diagnosis of onychomycosis, the most sensitive method was PCR (100%) followed by fluorescent staining (89%), PAS staining (69%), and finally KOH mount (53%). The diagnostic accuracy was as follows: KOH mount 52%; PAS staining 71%, PCR 72% and fluorescent staining 74% (Table 3).
Table (4):
The clinical usefulness and performance of each technique used in diagnosis of onychomycosis.
Diagnostic tests |
Time to obtain the result |
Cost (national Egyptian pounds) |
Nail plate penetrance |
Fungal viability |
Species Identification |
|---|---|---|---|---|---|
KOH examination |
15 min |
5 |
No |
No |
No |
PAS |
24 hrs. |
35 |
Yes |
No |
No |
Fluorescent staining |
30 min |
20 |
No |
No |
No |
Fungal culture |
1-3 weeks |
45 |
No |
Yes |
Yes |
PCR-based sequencing |
8 hrs. |
600 |
No |
No |
Yes |
Table 4 shows the clinical usefulness and performance of each technique used in diagnosis of onychomycosis. The most rapid test was KOH direct microscopy (15 min). Strikingly, fluorescent microscopic examination proved to be a rapid and low cost technique while the most expensive was PCR-based sequencing.
Onychomycosis is a fungal nail infection which constitutes approximately half of all nail disorders. The disease continues to spread and persist with both psychosocial and economic effects. Therefore, presence of available and good laboratory techniques is mandatory for quick and accurate identification of causative fungi for proper treatment and prevention.14 The idea of this research was to study the outcomes of fungal fluorescent staining and ITS rDNA PCR-based sequencing in relation to conventional methods (direct microscopy of KOH mount, PAS staining and mycological culture) for diagnosis of onychomycosis.
In this study, onychomycosis was more commonly encountered among elderly patients, a finding which was attributed to the reduced peripheral circulation, poor immune status, diabetes, and frequent nail injury.15,16 This result had been also proven by other researches in Egypt17 and in Iraq1 where onychomycosis had occurred more frequently in patients aged 40-60 years. Additionally, onychomycosis was higher in women (85%) than men (15%) in this study, perhaps because of work habits such as those of housewives, who generally perform household activities, including childcare, laundry, and cooking. Approximately, similar results had been published by other studies conducted in Egypt,17,18 and other countries as in India, Pakistan and Iraq.1,19,20
According to current results, the prevalence of onychomycosis had been shown to be significantly higher in patients with concomitant diseases such as anemia (46%), diabetes mellitus (25%) and PVD (20%) a finding which came in line with that reported by Emam and Abd El-salam18 in Egypt and Dogra et al.21 in India. Also, in this study, toenails (60%) were more affected than fingernails (40%) and the observed patterns of onychomycosis were DLSO (38%), TDO (24%), PO (20%) and WSO (18%), these were in agreement with that published by Mohtady et al.22 in Egypt and Gautam et al.14 in India.
Although, the conventional methods of fungal detection (direct microscopy of KOH mount, fungal culture, and PAS staining) are the most broadly used for diagnosis of onychomycosis, these methods have disadvantages as low sensitivity. Therefore, non-conventional methods like fluorescent staining and molecular biology methods are developed for diagnosis of fungal infections.2
In this study, 52%, 55% and 70% out of the 100 processed nail specimens were fungal positive by KOH direct microscopy, PAS, and mycological culture, respectively. Results obtained by other researchers displayed variable detection rates for these methods. Haghani et al.,23 Tyagi et al.24 and Zhao et al.25 had nearly similar results (74.2%, 70% and 77%, respectively for culture) and their KOH method results were 57.6%, 68.2% and 62.7%, respectively. A higher positivity for KOH direct microscopy reaching up to 85.1% and 85.7% was published by other studies23,26 respectively. For KOH direct microscopy, Lin et al.27 and Patel et al.28 reported positivity rates of 67.3% and 62.12%, respectively.
Regarding the diagnosis of onychomycosis by culture, there was a disparity in the results between different studies. Accordingly, mycological culture yielded positive fungal growth in 60% of the processed samples for Abdo et al.26 and 42.1% for lin et al.27 A lower positivity by culture (29.29%) was reported by Patel et al.28
Some previous studies declared that, however, fungal culture was more precise than the KOH preparation in defining the etiological agents of onychomycosis, false-negative results may occur due to presence of non-living organisms which won’t grow in culture,29 inaccessibility of the organism trapped in nail keratin to the culture growth media,18 previous treatment with anti-fungal drugs that hinder culture growth,29 sampling inaccuracy29 and the slow growth rate of dermatophytes in culture media.29 False-positive results of the culture may also happen due to contamination of the culture medium by the normal fungal flora of skin or laboratory contaminants or saprophytes.23 In the same context, our study demonstrated that of out of 30 culture-negative nail specimens, 93.3% (28/30) were positive by PCR assay. Also, Dhib et al30 showed that culture-negative specimens were 91.3% PCR positive.
In current work, mycological culture proved that yeasts were the main fungi involved in onychomycosis by 51.3% [with Candida spp. as the most isolated spp. (36.8%)] followed by dermatophytes 31.6% [with Trichophyton rubrum as the most frequent spp. (14.5%)] and NDMs represented 17.1% [with Aspergillus fumigatus as the predominant mold (10.5%)]. Some previous studies revealed the same trends, where yeasts accounted for 56%, 65%, 43.3%, 66% and 75.9%, respectively.1,3,10,17,31 On the other hand, Gautam et al.14 reported that dermatophytes were the mostly isolated fungi (60%) followed by NDMs (30%) and finally Candida spp. (10%) in India.
Molecular biology methods for identification of fungi directly from specimens are reliable and reproducible compared to other methods.12 PCR may detect DNA in dead fungal agents and can identify one or more species with specific primers in clinical samples without post-PCR analysis. However, post-PCR procedures such as sequencing raise the sensitivity of detection in dermatological samples and are valuable for identification of causative fungi at the species level from amplicon picked up with pan-fungal primers.32
In this study, PCR was positive in 98% of the processed nail specimens. This finding agreed with Jha et al.19 in India who found that 100% of 231 phenotypically- confirmed onychomycosis cases were genotypically positive by PCR. On the other hand, lower positive results of PCR (67%) were detected by another study.33
The results of ITS rDNA PCR-based sequencing for 30 nail specimens assured that yeasts were the most frequently isolated fungi (36.7%)[with Candida tropicalis as the most prevalent yeast (16.7%)]. Our results were in agreement with Fatahinia et al.34 in Iran who showed that the most frequently isolated fungi were yeasts (57.15%) with C. albicans as the most common species (29.35%). This contrasted with Ahmed et al.35 in Egypt who observed the predominance of NDMs (74.5%) [with Aspergillus spp. as the most prevalent molds (30%)].
The higher percentage of yeasts in our patients may be related to ageing, female predominance, chronic exposure to moisture, wet work in housewives, soil in agricultural activities and chemicals including detergents, soaps, etc. Yeasts may colonize the skin, hair, and nails but may become pathogenic in patients with poor immunity consequent to diabetes mellitus, vascular diseases, and anemia.17,31 Accordingly, 25%, 20% and 46% of our patients complained of diabetes mellitus, vascular diseases, and anemia, respectively.
Considering fungal culture as the cornerstone, this study showed that the most reliable technique for diagnosis of onychomycosis was PCR (100%) followed by fluorescent staining (89%), PAS staining (69%) and then KOH direct smear (53%). Similarly, other studies revealed that PCR had 100% sensitivity.22,2 In this study, the use of fungal fluorescent staining of nail specimens for studied patients enhanced sensitivity of direct examination by 36% compared to KOH (53%). Also, another study by Bao et al.2 showed that the use of fungal fluorescent staining of nail specimens improved the sensitivity of direct examination by 12% and the use of PCR-based sequencing enhanced the sensitivity 6% compared to culture.
Our current results proved that PAS staining microscopy was significantly more sensitive (69%) with diagnostic accuracy 71% which was higher than direct KOH microscopy (52%) in relation to culture. Other studies showed that PAS sensitivity ranged from 84-100%.33 In addition, PAS histopathology staining has other advantages such as detection of fungal structures as spores and hyphae, detection of degree of nail plate involvement, detection of rare hyphae and spores, and the low ratio of false-negative results in comparison with culture.36 However, problems of PAS microscopy include higher cost, a longer turnaround time than direct KOH or PCR and failure to discriminate among fungal species.37,33 KOH microscopy lack sensitivity with false-negative results of 5–15% because of low visibility, dispersed fungal elements in nail scraping, and inexpert examiner. Moreover, the technique cannot distinguish between dermatophytes and other fungal species.2
The time and cost for each technique were evaluated in this research. The fastest test for diagnosis of onychomycosis was KOH direct microscopy (15 min) and fluorescent staining (30 min). PAS stain required experienced dermatopathologists or pathologists to avoid false-positive or false-negative results. PAS approach could not identify the species and fungi, and relatively consumed higher cost than KOH direct microscopy and fluorescent staining. Moreover, fungal culture as well as PCR-based sequencing was able to identify the fungal genus and species. Similar findings were reported by other researchers in China2 and in Taiwan.27
Yeast fungi still were of the commonest etiologic agents for onychomycosis. The most sensitive technique for diagnosis was PCR. Fluorescent staining proved higher sensitivity compared with KOH and PAS approaches. It can be applied as a potential new tool for primary detection and identification of onychomycosis agents, its conjunction with PCR-based sequencing for species identification may help clinicians for more accurate diagnosis and effective treatment of onychomycosis.
ACKNOWLEDGMENTS
The authors would like to thank their colleagues at the Dermatology Department for their great help and assistance to accomplish this work.
CONFLICT OF INTEREST
The authors declare that there is no conflict of interest.
AUTHORS’ CONTRIBUTION
AFM conceptualized and designed the work. MSS collected the samples. SMEH and MSS contributed in the practical section of the work. SMEH performed the molecular study. SAMA and AME Interpreted the clinical and laboratory data. AFM, MMG, SAMA and AME wrote the manuscript.MMG revised and performed the language editing of the manuscript. All authors read and approved the final manuscript for publication.
FUNDING
None.
ETHICS STATEMENT
Not applicable.
AVAILABILITY OF DATA
All datasets generated or analyzed during this study are included in the manuscript.
- Mahmood LJ, Qurtas DS. Mycological Study of Onychomycosis in Erbil City. Annals of RSCB. 2021;25(4):20470-20479.
- Bao F, Fan Y, Sun L, et al. Comparison of fungal fluorescent staining and ITS rDNA PCR-based sequencing with conventional methods for the diagnosis of onychomycosis. J Eur Acad Dermatol Venereol. 2018;32(6):1017-1021.
Crossref - Verrier J, Monod M. Diagnosis of dermatophytosis using molecular biology. Mycopathologia. 2017;182(1-2):193-202.
Crossref - Tartor YH, Abo Hashem ME, Enany S. Towards a rapid identification and a novel proteomic analysis for dermatophytes from human and animal dermatophytosis. Mycoses. 2019;62(12):1116-1126.
Crossref - Tsang C, Teng J, Lau S, Woo P. Rapid Genomic Diagnosis of Fungal Infections in the Age of Next-Generation Sequencing. J Fungi. 2021;7(8):636.
Crossref - Nada EA, El Taieb MA, El-Feky MA, et al. Diagnosis of onychomycosis clinically by nail dermoscopy versus microbiological diagnosis. Arch Dermatol Res. 2020;312:207-212.
Crossref - El-Hamd MA, Elhameed MA, Fattouh M, Shalaby M, Saleh R. In vitro antifungal susceptibility testing of fungi in patients with onychomycosis. Dermatologic Therapy. 2020;33(3):e13429.
Crossref - Arora S, Pal R, Suhag DV, Dabas R, Arora G, Chand S. KOH Wet Mount versus Cellophane Strip without Mounting Medium for Rapid Diagnosis in Superficial Mycoses. Indian Journal of Dermatopathology and Diagnostic Dermatology. 2021;8(1):1-5.
Crossref - Hayette M, Seidel L, Adjetey C, et al. Clinical evaluation of the DermaGeniusR Nail real-time PCR assay for the detection of dermatophytes and Candida albicans in nails. Medical Mycology. 2019;57(3):277-283.
Crossref - Taha M, Saleh HM, Fathy G, Hamed MA. Evaluation of fluorescence microscopy using acridine orange stain versus hematoxylin and eosin stain in the diagnosis of onychomycosis. Journal of Egyptian Women Dermatologic Society. 2017;14(1):66-70.
Crossref - Banik A, Durairaj E, Lyngdoh WV, Khyriem AB, Sabhapandit D. Clinico-aetiologic profile of Onychomycoses in a tertiary care centre in northeast India. Trop Doct. 2018;48(2):136-142.
Crossref - Tartor YH, El Damaty HM, Mahmmod YS. Diagnostic performance of molecular and conventional methods for identification of dermatophyte species from clinically infected Arabian horses in Egypt. Vet Dermatol. 2016;27(5):401-e102.
Crossref - Arammehr A, Dehghan P, Chadeganipour M, Katoueezadeh M, Shadzi S. Detection of Dermatophytes from Dermatophytosis-Suspected Cases in Iran, Evaluation of Polymerase Chain Reaction-Sequencing Method. Adv Biomed Res. 2020;9:56.
Crossref - Gautam M, Shah N, Bhattar P, Nadkarni N, Patil S. Comparative Evaluation of Potassium Hydroxide Mount, Fungal Culture, and Histopathology of Nail Clipping with Periodic Acid- Schiff Stain in the Diagnosis of Onychomycosis. Indian Journal of Dermatopathology and Diagnostic Dermatology. 2021;8(1):6-12.
Crossref - de Carvalho Ribeiro CS, Zaitz C, de Souza Framil VM, de Carvalho Ottoboni TS, de Carvalho Tonoli MS, Ribeiro RP. Descriptive study of onychomycosis in a hospital in Sao Paulo. Braz J Microbiol. 2015;46(2):485-492.
Crossref - Gupta AK, Nakrieko KA. Onychomycosis infections do polymerase chain reaction and culture reports agree? J Am Podiatr Med Assoc. 2017;107(4):280-286.
Crossref - Bedaiwy MY, Metwally MA, El Zawawy NA, Saba HE. Epidemiology, Causative Agents and Clinical Features of Onychomycosis in El-Gharbia Governorate, J.Bot. The 7th Inter. Conf.” Plant & Microbial Biotech. & their Role in the Development of the Society”, 2017:187-196.
Crossref - Emam SM, Abd El-salam OH. Real-time PCR: A rapid and sensitive method for diagnosis of dermatophyte induced onychomycosis, a comparative study.Alexandria Journal of Medicine. 2016;52(1):83-90.
Crossref - Jha BK, Mahadevamurthy S, Sudisha J, Bora A. Isolation, Identification and Antifungal Susceptibility Test of Dermatophytes from the Patients with Onychomycosis in Central Nepal. American Journal of Dermatology and Venereology. 2015;4(3):30-36.
- Rana M, Altaf F, Bashir B, Rani Z. Frequency of associated factors of onychomycosis. Journal of Pakistan Association of Dermatologists. 2017;27(3):226-31.
- Dogra S, Kumar B, Bhansali A, Chakrabarty A. Epidemiology of onychomycosis in patients with diabetes mellitus in India. Int J Dermatol. 2002;41(10):647-651.
Crossref - Mohtady H, Nasar A, Elbadawy N E, Abdelhafiz M. The role of PCR in the diagnosis of dermatophytes in onychomycosis. The International Arabic Journal of Antimicrobial Agents. 2019;9(3):4.
Crossref - Haghani I, Shokohi T, Hajheidari Z, Khalilian A, Aghili SR. Comparison of Diagnostic Methods in the Evaluation of Onychomycosis. Mycopathologia. 2013;175(3-4):315-321.
Crossref - Tyagi S, Kaur R, Rawat D. Mycological Profile and Prevalence of Superficial Mycoses Agents: A Study from North India. International Journal of Medical and Dental Sciences. 2021;10(1):1925-1931.
Crossref - ZhaoY, Yu Z, Yue X. Evaluating the accuracy and diagnostic value of CFW and a new fluorescent reagents, fluorescent brightener 85, for the diagnosis of vulvovaginal candidiasis. J Clin Lab Anal. 2021;35(8):1-7.
Crossref - Abdo HM, AbdElrahem SI. Clinicomycological study of coexisting toenail onychomycosis and tineapedis. Gulf J Dermatol Venereol. 2016;23(2):8-19.
- Lin YC, Sun PL, Hsiao PF, Sun FJ, Wu YH Methods for diagnosing onychomycosis: A comparative study of 459 cases. Dermatol Sin. 2019;37(2):63-66.
Crossref - Patel M, Ndlovu N, Owen C, Veale R. Properties of a new mouthrinse for patients receiving radiation therapy. SADJ. 2010;65(9):410-412. PMID: 21180287
- Gordon AK, McIver C, Kim M, Murrell DF, Taylor P. Clinical application of a molecular assay for the detection of dermatophytosis and a novel non-invasive sampling technique. Pathology. 2016;48(7):720-726.
Crossref - Dhib I, Fathallah A, Yaacoub A, Slama FH, Said MB, Zemni R. Multiplex PCR assay for the detection of common dermatophyte nail infections. Mycoses. 2014;57(1):19-26.
Crossref - Hamdinoa M, Elsayed S, Tahac M, El kadyd E. Evaluating the role of non dermatophyte fungi as a causative agent of tineapedis and its relation to diabetes. The Scientific Journal of Al-Azhar Medical Faculty, Girls. 2020;4(2):256-261.
Crossref - Sylla K, Tine RT, Sow D, et al. Epidemiological and Mycological Aspects of Onychomycosis in Dakar (Senegal). J. Fungi. 2019;5(2):35.
Crossref - Gustafson E, Bakotic W, Bennett L, Page L, McCarthy L. DNA-based detection for onychomycosis correlates better to histopathology than does fungal culture. Dermatology Online Journal. 2019;25(7):3.
Crossref - Fatahinia M, Jafarpour S, Rafiei A, Taghipour S, Makimura K, Rezaei-Matehkolaei A. Mycological aspects of onychomycosis in Khuzestan Province, Iran: A shift from dermatophytes towards yeasts. Curr Med Mycol. 2017;3(4):26-31.
Crossref - Ahmed F, Elkholy I, Mehalawy A, Abdou D. Prevalence of onychomycosis due to non-dermatophyte yeasts and molds in Cairo, Egypt. Egypt J Exp Biol (Bot.). 2020;16(1):1-10.
Crossref - Idriss M, Khalil A, Elston D. The diagnostic value of fungal fluorescence in onychomycosis. J CutanPathol. 2013;40:385-390.
Crossref - Blake N, Zhu J, Hernandez G, Juliano P. Retrospective Review of Diagnostic Testing for Onychomycosis of the Foot. J Am Podiatr Med Assoc. 2015;105(6):503-508.
Crossref
© The Author(s) 2022. Open Access. This article is distributed under the terms of the Creative Commons Attribution 4.0 International License which permits unrestricted use, sharing, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.