ISSN: 0973-7510
E-ISSN: 2581-690X
In the present study twenty five human urine samples of hospital patients were screened for Staphylococcus aureus. Twenty percent of the samples were positive for Staphylococcus aureus as yellow colonies were observed aerobically on mannitol salt agar, out of the five isolates two were catalase positive. On the basis of resistance against methicillin, out of the three catalase positive isolates, two were MSSA and one was MRSA. One MRSA and one MSSA strain was examined using PCR technique, for gene identification. The mecA and nuc gene were amplified using PCR technique. The presence of mecA gene and nuc gene in MRSA and MSSA respectively, confirmed the strains. Comparative gene analysis of mecA gene and nuc gene was performed using BLAST and CLUSTALW tools. Thereafter rooted phylogenetic tree was obtained for both mecA and nuc gene sequences explaining the genetic evolutionary lineage. Present study analyzed the resistance mechanism of MRSA and to identify the gene responsible in order to control the re-emerging methicillin resistant Staphylococcus aureus infection.
MRSA, Antibacterial activity, mecA gene, nuc gene, PCR.
Staphylococcus aureus is one of the most important opportunistic pathogen among Staphylococci belonging to Micrococaceae family causing significant infections under appropriate conditions (Narmeen and Jubrael 2009). Methicillin-resistant S. aureus, abbreviated MRSA is one of a number of greatly feared strains of S. aureus which have become resistant to most â-lactam antibiotics. MRSA was first isolated in 1960 as a nosocomial pathogen, now known as hospital-acquired MRSA (HA-MRSA). Another class of MRSA, designated community-acquired MRSA (CA-MRSA), emerged in the community from 1997 to 1999, posing a novel threat worldwide (Goldstein et al. 2012). Recently strains of multiple drug resistant Staphylococcus aureus have appeared and proven very difficult to treat. During the late 1950s and early 1960s, Staphylococcus aureus caused considerable morbidity and mortality as a nosocomial or hospital acquired pathogen and has become the head leading cause of nosocomial infection during the last two decades. Unfortunately, MRSA (methicillin resistant Staphylococcus aureus) strains isolated are resistant to multiple non-ß-lactam containing antimicrobial drugs (Abraham et al. 2009). Resistance is conferred by the mecA gene, which codes for an altered penicillin-binding protein (PBP2a or PBP2′) that can function as transpeptidase (Lopez et al. 2012). In the absence of ß-lactam antibiotics the Staphylococci utilize usual PBPs to synthesize the cell wall which is composed of peptidoglycan. In contrast, an additional low affinity penicillin binding protein PBP designated PBP 2a is encoded by a unique MRSA-PBP gene and is the main factor responsible for the expression of the ß-lactam resistance of MRSA which is an intrinsic resistance of the cells to all ß-lactam antibiotics is mediated by the methicillin resistance determinant (Haroun and Hameed 2011). Much of the research aimed at identifying novel therapeutic targets for the treatment of Staphylococcus aureus infections has been directed at identification of specific virulence factors and regulatory circuits that are relevant to the disease process. Some organisms rapidly acquire resistance e.g. coliforms and Staphylococcus aureus. The organisms exhibit remarkable versatility in their behaviour towards antibiotics, with some strains having overcome most commonly used drugs (Uwaezuoke and Aririatu 2004).
Comparative genomics is an attempt to take advantage of the information provided by the signatures of selection to understand the function and evolutionary processes that act on genomes (Chandrakanth et al. 2010). A typical alignment procedure involves the application of a program such as CLUSTAL W, followed by a tree building program. For further identification at genus level, bacterial isolates were identified through homology search with BLAST and FASTA using mecA and nuc gene sequences. Keeping in mind the increasing mortality rate due to MRSA infections in the last two decades, in the present study the sequence of the mecA gene which accounts for the resistance of Staphylococcus aureus towards â-lactam antibiotic methicillin and the sequence of nuc gene, which is found in all Staphylococcus aureus strains were subjected to phylogenetic analysis. This study suggests that further research must continue on analyzing the resistance mechanism of the genes responsible for imparting resistance in Staphylococcus aureus strains, in order to control the reemerging MRSA infections.
Isolation and identification of Staphylococcus aureus Bacteria
Twenty five human mixed stream urine samples were collected from the pathology department, Jeevan Jyoti Hospital, Allahabad, INDIA. The urine samples were inoculated on mannitol salt agar (MSA) plates. The plates were incubated aerobically at 37p C for 24 h to obtain colonies (Abraham et al. 2009) that were identified on the basis of cultural, morphological and biochemical characteristics as described in the Bergey’s Manual of Determinative Bacteriology (Holt et al. 1984).
Antibiotic Susceptibility Pattern of Staphylococcus aureus against selected antibiotics
The test organism was tested for its sensitivity towards the given antibiotics using the Disk Diffusion Method (Bauer et al., 1966). Overnight broth cultures were spread on the surface of Nutrient Agar Media plates with the help of sterile swabs. The antibiotic discs (Hi Media) were placed on the agar surface and plates were incubated aerobically at 37p C for 24 h. Zone of inhibition was measured in ‘mm’ and the result interpreted on the basis of CLSI standards (Wayne, 2003). Antibiotic discs of various concentrations i.e. Amoxicillin (AMX) 10µg, Ceftriaxon (CTX) 30µg, Cefuroxime (CFX) 30µg, Ciprofloxacin (CIP) 5µg, Gentamicin (GEN) 10µg, Methicillin (MET) 5µg, Norfloxacin (NOR) 10µg, Penicillin-G (P) 1µg, Tetracycline (TE) 30µg, Vancomycin (VA) 30µg were used in the present study.
Gene Amplification and Comparative Gene Analysis
DNA was isolated followed by PCR to amplify the gene sequences of mecA gene present in MRSA strain and nuc gene in MSSA strain and then gene sequencing was done. The gene sequences of mecA and nuc gene were analyzed using BLAST (basic local alignment search tool) to search the sequences available on the data base. Four sequences were selected having high similarity with nuc gene and six sequences were selected having high similarity with mecA gene. ClustalW tool was used for multiple sequence alignment of the nuc gene sequence along with the four related sequences. Then multiple sequence alignment of the mecA gene sequence along with the six related sequences was performed. The data obtained as a result of multiple sequence alignment, was represented in the form of rooted UPGMA (unweighted pair group method of arithmetic mean) phylogenetic tree. The phylogenetic tree was interpreted in order to analyze the genetic evolutionary lineage.
Culture Characterization
Out of twenty five samples, five samples (20%) were positive for Staphylococcal spp as yellow colonies were observed aerobically on mannitol salt agar, this yellow pigmentation was due to the mannitol fermentation by the organism. One colony was picked from each of the five plates which were positive for Staphylococcal spp and noticed that these five isolates were gram-positive cocci, arranged in clusters, when screened for morphological characteristics.
On the basis of biochemical characterization the five isolates were found to be non-motile, also gave positive result for nitrate reduction test and methyl red test, while negative result was obtained for indole test. Tube Coagulase test was performed, which is the most commonly used method to identify S. aureus and the five isolates were coagulase positive. Another study showed comparable results in which thirty six out of hundred isolates (36%) were gram-positive cocci in cluster form, catalase positive and eighteen out of thirty six isolates fermented mannitol. Six out of eighteen isolates were tube coagulase positive, which was presumptive as MRSA (Amini et al. 2012). The carbohydrate fermentation test revealed that Staphylococcus aureus isolates demonstrated fermentation of glucose, galactose, sucrose, fructose, mannitol, whereas could not ferment raffinose (Table 1).
Table (1):
Cultural, morphological and biochemical identification of Staphylococcus aureus isolates.
| Characteristics | Staphylococcus aureus | ||
|---|---|---|---|
| MRSA | MSSA | ||
| Cultural characteristics | Colour of colony | Golden Yellow | Golden Yellow |
| Shape of colony | Small, spherical | Small, spherical | |
| Elevation | Convex | Convex | |
| Margin | Regular | Regular | |
| Morphological characteristics | Gram’s reaction | Gram positive | Gram positive |
| Arrangement of cells | Clusters | Clusters | |
| Spore formation | Non –sporing | Non –sporing | |
| Biochemical characteristics | Indole test | Negative | Negative |
| Methyl red test | Positive | Positive | |
| Catalase test | Positive | Positive | |
| Motility test | Negative | Negative | |
| Nitrate reduction test | Positive | Positive | |
| Coagulase test | Positive | Positive | |
| Carbohydrate fermentation | D-Glucose | A+G– | A+G– |
| D- Galactose | A+G– | A+G– | |
| Sucrose | A+G– | A+G– | |
| Maltose | A+G– | A+G– | |
| Lactose | A+G– | A+G– | |
| D-Mannitol | A+G– | A+G– | |
| Raffinose | A–G– | A–G– | |
A+: Acid Positive; A– : Acid negative; G+: Gas positive; G–: gas negative; WA+: Weakly Positive.
Antibiotic Susceptibity pattern of Staphylococcus aureus
On the basis of resistance against methicillin, out of the three catalase positive isolates, two were MSSA and one was MRSA. The MRSA strain was resistant towards the seven out of eleven antibiotics used, whereas MSSA was resistant towards three out of eleven antibiotics used. Among the eleven antibiotics, gentamicin possessed maximum inhibitory potential whereas penicillin showed least inhibitory effect against MRSA and MMSA strains. MRSA strain was multi-drug resistant as it was resistant to methicillin, penicillin, cefuroxime, ceftazidim, ceftriaxone, ciprofloxacin, tetracycline (Table 2).
Table (2):
Antibiotic susceptibility profile of Staphylococcus aureus strains (MRSA, MSA).
| S. aureus strains | Antibiotics | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| MET (5mg) | P (10mg) | VA (30mg) | GEN (10mg) | NOR (10mg) | AMX (10mg) | CXM (30mg) | CAZ (30mg) | CTR (30mg) | CIP (5mg) | TE (30mg) | |
| MRSA | – | – | ++ | +++ | ++ | ++ | – | – | – | – | – |
| MSSA | ++ | – | +++ | +++ | ++ | ++ | ++ | ++ | ++ | – | – |
(+++) Sensitive, (++) Intermediate, (-) Resistant, Methicillin (MET), Amoxicillin (AMX), Penicillin (P), Cefuroxime (CXM), Vancomycin (Va), Ceftazidim (CEZ), Gentamicin (GEN), Ceftriaxone (CTR), Norfloxacin (NX), Ciprofloxacin (CIP), Tetracycline (TE)
In the present study the antibiotic susceptibility of the MSSA and MRSA isolates was tested. Results revealed that both MRSA and MSSA strains were sensitive to amoxicillin, gentamicin and this was comparable with the reports of other workers (Nwankwo et al. 2010; Khadri and Alzohairy 2010). In another study the isolates were most sensitive to gentamicin (91.7%), cloxacillin (85.4%) and most resistant to penicillin (95.8%) and ampicillin (89.6%). The capacity of Staphylococcus aureus to produce human diseases has not diminished with the introduction of antibiotics. The organisms exhibit remarkable versatility in their behaviour towards antibiotics, with some strains having overcome most commonly used drugs. In this study, a high sensitivity percentage to gentamicin (91.7%), cloxacillin (85.4%) was recorded. Also most of the strains of Staphylococcus aureus were sensitive to erythromycin (66.7%) and streptomycin (66.7%). This finding shows that staphylococcal infections could be treated with gentamicin, cloxacillin, erythromycin (Uwaezuoke and Aririatu 2004).
Amplification and comparative gene analysis of mecA gene and nuc gene sequences
Specific gene identification of MRSA and MSSA strains were performed. PCR was performed to amplify the mecA and nuc gene sequences. After gene amplification gel electrophoresis was performed for both MRSA and MSSA genome sequences for mecA, nuc gene sequences respectively (Fig. 1). mecA is the gene responsible for resistance towards methicillin while nuc gene is responsible in detecting the presence of S. aureus, thus this sample can be considered as MRSA. Those samples which lack nuc gene were not S. aureus. In the previous study five isolates of Staphylococcus aureus were positive for the presence of mecA gene and only one of the samples was positive to nuc gene (Amini et al. 2012).
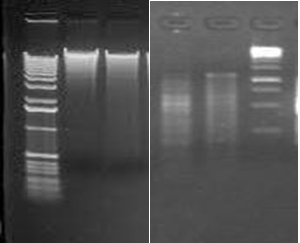 Fig. 1. Gel electrophoresis picture of (a) Genomic DNA isolation, (b) PCR amplification
Fig. 1. Gel electrophoresis picture of (a) Genomic DNA isolation, (b) PCR amplificationFollowing sequences were obtained for nuc gene in methicillin susceptible and mecA gene sequence in methicillin resistant Staphylococcus aureus respectively.
nuc gene sequence in methicillin susceptible Staphylococcus aureus:
>giAAAGGTGAAGATAAAGTGCATGTTCAAA GAGTCGTCGAT GGAGATACTTTCATAGCGAA TCAAAACGGCAAGGAAATCAAGGTTAGA CTCATTGGAGTCGATACGCCTGAAACA GTAAAACCGAATACGCCTGTAC AGCCATTTGGTAAGGAAG CATCAAATTATAGTAAGAA GACTTTAACGAATCAAGACGTTTATTTA GAATACGATAAAGAAAAGCAAGACCGCTATG GTAGAACATTGGCGTATGTA
mecA gene sequence in methicillin resistant Staphylococcus aureus:
>giATGAAAAAGATAAAAATTGTTCCACT TATTTTAATAGTTGTAGTTGTCGGGTTT GGTATATATTTTTATGCTTCAAAAGATA AAGAAATTAATAATACTAT TGATGCAATTGAAGATAAAAATTT AAACAAGTTTATAAAGATAGC AGTTATATTCTAAAAGCGATAA TGGTGAAGTAGAAATGACTGA ACGTCCGATAAAAATATTAATA GTTTAGGCGTTAAAGATATAA ACATTCAGGATCGTAAAATAAA AAAAGTATCTAAAAATAAAAAA CGAGTAGATGCTCAATATAAAATTA AAACAAACTACGGTAACATTGA TCGCAACGTTCAATTTAATTTT GTTAAAGAAGATGGTATGTGGAA GTTAGATTGGGATCATAGCG TCATTATTCCAGGAATGCAGAAA GACCAAAGCATACATATTGAA AATTTAAAATCAGAACGTGGTAA AATTTTAGACCGAAACAATGT GGAATTGGCCAATACAGGAACA GCATATGAGATAGGCATCGTT CCAAAGAATGTATCTAAAAAAGATTATA AAGCAATCGCTAAAGAACT AAGTATTTCTGAAGACTATATCA AACAACAAATGGATCAAAAT TGGGTACAAGATGATACCTT CGTTCCACTTAAAACCGTTAA AAAAATGGATGAATATTTAA GTGATTTCGCAAAAAAATTTCATCTTA CAACTAATGAAACAGAAAGTCGT
On submitting the mecA and nuc gene sequences to nucleotide BLAST (basic local alignment search tool), information about the two test sequences were retrieved (Nuc gene sequence in MSSA: Query ID lcl|1011Database Name: nr; Program: BLASTN 2.2.28; Query Length: 250) and (mecA gene sequence in MSSA: Query ID lcl|17517Database Name: nr; Program: BLASTN 2.2.28; Query Length: 712). As a result of BLAST the sequences available on the database showing high sequence similarity with the mecA and nuc gene sequences respectively, were retrieved. Four sequences were selected having high similarity with nuc gene and six sequences were selected having high similarity with mecA gene. As a result of multiple sequence alignment using ClustalW tool, the nuc gene sequence along with the four related sequences and the mecA gene sequence along with the six related sequences were aligned on the basis of sequence identity (Higgins et al. 1996) and then rooted UPGMA phylogenetic tree was obtained for both mecA and nuc gene sequences explaining the genetic evolutionary lineage (Fig. 2 a,b).
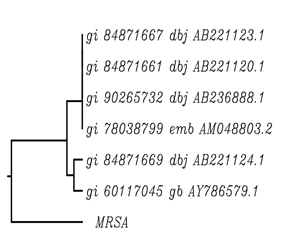
 Fig. 2. Phylogenetic Tree for mecA gene sequence in (a) MRSA (b) MSSA
Fig. 2. Phylogenetic Tree for mecA gene sequence in (a) MRSA (b) MSSAAs a result of the phylogenetic tree retrieved for the mecA gene sequence in MRSA (Fig. 2a) it was concluded that the mecA gene sequence showed sequence similarity with two clusters of sequences viz gi 84b71667 dbj AB221123.1; gi 84b71661 dbj AB221120.1; gi 90265732 dbj AB236888.1; gi 78038799 dbj AM048803.2 and gi 84871669 dbj AB221124.1; gi 60117045 dbj AY786579.1. The three sequences gi 84871667 dbj AB221123.1, gi 84871661 dbj AB221120.1 and gi 90265732 dbj AB236888.1 belong to one clade, thus these genes include the most recent common ancestor of all of its members and all of the descendants of that most recent common ancestor. The two sequences gi 84871669 dbj AB221124.1 and gi 60117045 gb AY786579.1 are called a clade. Gene transfer is observed to occur from mecA gene sequence in MRSA to gi 84871669 dbj AB221124.1 and gi 60117045 gb AY786579 gene sequences (Felsenstein 1985).
While the phylogenetic tree retrieved for the nuc gene sequence in MSSA (Fig. 2b) revealed that the nuc gene sequence showed sequence similarity with three clusters of sequences viz gi 440658993 dbj JX499025.1; gi 440658995 dbj JX499026.1 and gi 440658991 dbj JX499024.1 and gi 300078420 dbj GQ37047.1. The three sequences gi 440658993 gb JX499025.1, gi 440658995 gb JX499026.1 and gi 440658991 gb JX499024.1 were one clade, these gene sequences include the most recent common ancestor of all of its members and all of the descendants of that most recent common ancestor. Gene transfer is observed to occur from the nuc gene sequence in MSSA to gi 440658993 dbj JX499025.1 (Saitou, 1996).
According to the MRSA genome information it was revealed that a circular chromosome of 2809422 bp length and pT811 plasmid of 4440 bp length was present.
Results from present study showed that among the eleven antibiotics used, gentamicin possessed maximum inhibitory potential whereas penicillin showed least inhibitory effect against MRSA and MMSA strains. MRSA strain was multi-drug resistant as it was resistant to methicillin, penicillin, cefuroxime, ceftazidim, ceftriaxone, ciprofloxacin, tetracycline. mecA and nuc gene sequences were retrieved using PCR from MRSA and MSSA strains respectively, followed by BLAST. Evolutionary lineage and genetic diversity of the bacterial strains isolated from different ecological niche, also giving information regarding the horizontal gene transfer was obtained from from phylogenetic tree. This study suggests that comparative genomic analysis can be used to identify the molecular genetic basis of 99.8% of the antimicrobial resistance phenotypes of the isolates, highlighting the potential of pathogen genome sequencing as a diagnostic tool and also screen the genetic changes associated with adaptation to the hospital environment and with increasing drug resistance over time, and how MRSA evolution likely has been influenced by country-specific drug use regimens.
ACKNOWLEDGMENTS
Authors are thankful to Head, Department of Molecular and Cellular Engineering, SHUATS for providing all laboratory facility to conduct this study.
- Abraham M, De N, Sudi IY, Ma’ori L, Isolation of methicillin resistant Staphylococcus aureus (MRSA) from AIDS patients attending state specialist hospital, Yola and Federal Medical Centre, Yola, Adamawa State, Nigeria. Reports and Opinion, 2009; 1:103-107
- Amini R, Abdulamir AS, Jahanshiri F, Shan LC, Ali H, Amini Y, Sekawi Z, Jalilian FA, Isolation and identification of methicillin-resistant Staphylococcus aureus from students’ coins. Afr J Biotechnol, 2012; 11:11143-11149.
- Bauer AW, Kirby WMM, Sherris JC, Truck M, Antibiotic susceptibility testing by a standardized single disk diffusion method. Amer J Clin Pathol, 1966; 45:365-378.
- Chandrakanth K,Virupakshaiah DBM, Gavimath CC, Morabad U Kangralkar VA, Comparative genomics of staphylococcus aureus coagulase gene. J Adv Bioinfo Appl Res, 2010; 1:31-36.
- Felsenstein J, Conûdence intervals on phylogenies: An approach using the bootstrap. Evolution, 1985; 39: 783–791.
- Goldstein RER, Micallef SA, Gibbs SG, Davis JA, He X, George A, Kleinfelter LM, Schreiber NA, Mukherjee S, Sapkota A, Joseph SW, Sapkota AR, Methicillin-resistant Staphylococcus aureus (MRSA) detected at four U.S. waste water treatment plants. Environ Health Perspect, 2012; 120:1551-1558.
- Haroun BO Hameed EAB, Molecular identification of methicillin resistant Staphylococcus aureus isolated from patients with wound infections in Khartoum teaching hospital, Sudan. J Sci Tech, 2011; 12:51-57.
- Higgins DG, Thompson JD Gibson TJ, Using ClustalW for multiple sequence alignments. Meth Enzymol, 1996; 266: 383–402.
- Holt JG, Bergey DH, Kreig NR, Bergey’s Manual of Systematic Bacteriology (2nd). Williiams and Wikins, Baltimore, USA, 1984; pp-1234.
- Khadri H, Alzohairy M, Prevalence and antibiotic susceptibility pattern of methicillin-resistant and coagulase-negative Staphylococcus aureus in a tertiary care hospital in India. Int J Med Sci, 2010; 2:116-120.
- López MCV, Moreno JE, Rueda VR, Chirivi JS, Garzon A, Arévalo SA, Martínez MF, Gardeazábal PA, Baquero C, Methicillin-resistant Staphylococcus aureus (MRSA) isolated from Colombian foods. Bio, 2012; 2: 61-67.
- Narmeen SM, Jubrael JMS, Isolation and identification of Staphylococcus aureus using classical and molecular methods. J Duhok Uni, 2009; 12: 10-16.
- Nwankwo E, Abdulhadi S, Magagi A, Ihesiulor G, Methicillin resistant S. aureus (MRSA) and their antibiotic sensitivity pattern in Kano, Nigeria Afr J Cli Microbiol, 2010; 11:129-136.
- Saitou N, Reconstruction of gene trees from sequence data. Meth Enzymol, 1996; 266: 427–449.
- Uwaezuoke JC, Aririatu LE, A survey of antibiotic resistant Staphylococcus aureus strains from clinical sources in Owerri. J App Sci and Environ Mgmt, 2004; 8:67-69.
- Wayne PA, Performance standards for antimicrobial susceptibility testing, fifteenth informational supplement. CLSI doc, 2003; 26:1-3.
© The Author(s) 2017. Open Access. This article is distributed under the terms of the Creative Commons Attribution 4.0 International License which permits unrestricted use, sharing, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.


