ISSN: 0973-7510
E-ISSN: 2581-690X
Carbapenems are the preferred last-line agents for treating multidrug-resistant (MDR) bacterial infections frequently observed in hospital environments. The objective of this study was to estimate the prevalence of different genes linked to carbapenemase production in Enterobacteriaceae, utilizing samples obtained from a tertiary healthcare center located in South India. A retrospective investigation included isolates from blood, tracheal aspirate, bronchoalveolar lavage (BAL), pleural fluid, and mini-BAL from chosen patients, which were inoculated onto blood agar or MacConkey agar and underwent Carba-R testing with the GENE Xpert Carba-R Assay. The study included the various samples sent to the Diagnostic Microbiology Laboratory obtained from all patients between January 2020 to August 2023. The results showed that majority were respiratory samples (31%) followed by blood culture samples (28%) and urine samples (23%). Among the total infections that are culture-positive for Enterobacteriaceae, 10.6% were carbapenem-resistant Enterobacteriaceae. The most common organisms detected was Klebsiella pneumoniae (83%) followed by Escherichia coli (10%), and other various organisms. Among the genotypes, the OXA-48 genotype (31%) was the most prevalent, followed by the NDM genotype (25%) and the KPC genotype (1.3%) as resistance inducers. Additionally, 37% of isolates exhibited a combination of both OXA-48 and NDM genes, while 13.5% of organisms demonstrated the presence of OXA-48, NDM, and KPC. The results helped conclude that OXA-48 followed by NDM genotype were responsible for inducing resistance in more than 50% of the CROs detected. The results elucidate the diverse levels of resistance-acquiring genotypes, which can enhance the formulation of improved patient treatments.
Bacteria, Beta-lactam Antibiotics, Carbapenem-resistant Enterobacteriaceae, Drug-resistance, Genotype
The extensive accessibility of over-the-counter medications is a primary contributor to antibiotic resistance (AMR). Antimicrobial resistance poses substantial challenges to both the healthcare sector and the general populace, with rates increasing fast. A substantial fraction of worldwide morbidity and mortality attributable to bacterial infections is associated with antibiotic resistance. Gram-negative bacteria are the principal contributors to multidrug-resistance (MDR).1
In Gram-negative organisms, phenotypic resistance to carbapenems is typically caused by the acquisition of carbapenemases, or by the synthesis of cephalosporinases in conjunction with mutations that reduce the permeability of the bacterial cell wall to carbapenem entry.2 Depending on the study population, carbapenemase-producing Enterobacterales (CPE) can cause serious infections, lengthen hospital admissions, and increase mortality rates, which can range from 24% to 70%. Although early treatments must be established with efficacy, the limited number of therapeutic choices available restricts the available options.3
The principal carbapenemases identified encompass the following: Class A carbapenemases, particularly those located on plasmids, including GES and KPC. (ii) A recent rise in the prevalence of class B carbapenemases, referred to as metallo-β-lactamases (MBLs), has been observed. These enzymes are generally linked to infections induced by P. aeruginosa, Enterobacterales, and A. baumannii.4,5 The three predominant MBL groups are NDM, IMP, and VIM; class C carbapenemases are quite uncommon and were only recently identified. Although class C β-lactamases do not impart carbapenem resistance, five specific enzymes-ACT-1, DHA-1, CMY-2, CMY-10, and ADC-68-exhibit the notable capability to hydrolyze carbapenems. Furthermore, class D carbapenemases, known as OXAs, were initially discovered many years ago; however, it is only recently that carbapenem-hydrolyzing class D β-lactamases, or CHDLs, have achieved widespread distribution. OXA-48-like is widespread among Enterobacterales, but the carbapenem resistance in A. baumannii mainly arises from the presence of OXA-24/40-like, OXA-23-like, OXA-143-like, OXA-58-like, and OXA-235-like groups.6
From 2000 to 2010, there was a 35% increase in the use of antibiotics across clinical, agricultural, culinary, and aquaculture environments globally; with the BRICS nations being responsible for 79% of this increase.5 Between 2009 and 2015, India exhibited the highest drug-resistance index (range: 71-83), indicating that antibiotic therapy was the least effective.6 While the frequency of multidrug-resistant organisms differs between nations and establishments, an average of 13% of Carbapenem-resistant Organisms (CRO) are found in India.7
Early diagnosis of carbapenemase synthesis or alternative resistance mechanisms is essential to commence antibiotic therapy and manage CRO infections. There is a significant gap in knowledge about the different types of resistance conferred among different CRO infections and their various mechanism of action. There is a deficiency of evidence concerning MDR organisms in ICU environments in South India, particularly regarding the induction of resistance. The objective of this study was to evaluate the prevalence and molecular characteristics of Carbapenem-resistant gram-negative bacilli within a tertiary healthcare facility located in South India. The objective of this study was to estimate the prevalence of different genes linked to carbapenemase production in Enterobacteriaceae, utilizing samples obtained from a tertiary healthcare center located in South India.
This study was performed retrospectively within a clinical microbiology laboratory located at a tertiary healthcare facility in Coimbatore, Tamil Nadu. The research was carried out over a period of three years following the approval from the institutional Human Ethics Committee (IHEC), referenced as (PSG/IHEC/2023/Renew/034). The investigation involved inpatients, outpatients as well as patients admitted to the ICU, with the specimens including blood, tracheal aspirate, pleural fluid, and bronchoalveolar lavage (BAL) sent to the Diagnostic Microbiology laboratory.
Sample selection
Bacteria were identified using conventional biochemical methods. Antibiotic susceptibility testing was performed using the VITEK 2 automated system (bioMerieux, Marcy-l’Toile, France) and the Kirby Bauer method, in accordance with the CLSI guidelines 2021. The Centers for Disease Control and Prevention (CDC) defines CRE as Enterobacterales that test resistant to at least one of the carbapenem antibiotics (i.e., minimum inhibitory concentrations of ≥4 mg/mL for doripenem, meropenem, and imipenem, and ≥2 µg/mL for ertapenem) or Enterobacterales that produce a carbapenemase.8
All Carbapenem-resistant Enterobacteriaceae (CRE) isolated during the study period, for which treating physicians requested the molecular study (Xpert Carba-R) for further patient management, were included in the study. The exclusion criteria included carbapenem-resistant gram-negative bacteria, with the exception of Enterobacteriaceae, and also accounted for repeat samples obtained from the same patient during the same hospitalization period. After applying the inclusion and exclusion criteria, all selected 308 isolates were analysed using Carba-R test.
Biochemical reactions used for identification
The following biochemical tests were conductedto discern between the different bacterial species: Klebsiella pneumoniae [Indole negative, Citrate positive (98%) Urease positive (95%) Methyl red negative, Voges Proskauer positive (98%)], Escherichia coli (Indole positive (100%) Citrate negative, Urease negative, Methyl red positive (99%), Voges Proskauer negative), Enterobacter cloacae [Indole negative, Citrate positive (90%) Urease positive (75%) Methyl red negative Voges Proskauer positive (100%) Lysine positive (95%), Arginine positive (99%), ONPG (o-nitrophenyl-β-D-galactopyranoside positive)] (99%), Proteus mirabilis [Indole negative, Citrate negative, Urease positive (95%) Methyl red positive (97%) Voges Proskauer negative, Phenyl Pyruvic Acid positive (PPA)(98%)], Citrobacter freundii [Indole negative, Citrate positive (95%) Urease positive (70%) Methyl red positive (100%) Voges Proskauer negative, ONPG positive (95%)], Klebsiella oxytoca [Indole positive (99%), Citrate positive (95%) Urease positive (90%) Methyl red negative, Voges Proskauer positive (99%)], Serratia marcescens [Indole negative, Citrate positive (97%) Urease negative, Methyl red negative, Voges Proskauer positive (61%), ONPG 94%], Providencia rettgeri [Indole positive (97%), Citrate positive (71%) Urease positive (98%) Methyl red positive 98%, Voges Proskauer negative, Phenyl Pyruvic Acid positive (99%)].
Carba-R test
Isolates from above mentioned samples from Blood agar or MacConkey agar were subjected to Carba-R testing by GENE Xpert Carba-R Assay. The Xpert Carba-R test was conducted using the GeneXpert platform from Cepheid, based in Sunnyvale, California, USA. It is a qualitative in vitro real-time PCR assay that detects five critical carbapenemase-producing genes: NDM, KPC, OXA-48, IMP, and VIM. The colonies from the culture plate were vortexed at high speed for 10 seconds in an elution reagent tube prior to inserting the Xpert Carba-R test cartridge into the machine, following the manufacturer’s instructions. The analysis of the results necessitated a period of forty-seven minutes. Previously conducted studies have reported that the test exhibits a positive predictive value of 96% and a negative predictive value of 100%, reflecting robust sensitivity and specificity of 100% and 77%, respectively.9,10
Statistical analysis
The gathered data were input into Excel spreadsheets and analyzed using SPSS version 28.0. A descriptive analysis of the data was performed, and the percentile, mean, and standard deviation were computed. The error rates of the two procedures were computed to evaluate the reliability of the direct-testing methodology. Analyses at the patient level and within subgroups were conducted to elucidate the consequences of the direct testing method.
The findings indicate that, among the samples that were collected, blood samples were the most prevalent, followed by urine samples, pus samples, and sputum samples, with a total of 204786 sample requests being made. Most of the samples tested were from males (54.22%), and a large proportion of the samples were acquired from individuals who were between the ages of 20 and 40 years old, closely followed by those who were between the ages of 40 and 60 years old (36.69%) (Table 1 and 2).
Table (1):
Distribution of study samples obtained
| Samples | Year | |||
|---|---|---|---|---|
| 2020 | 2021 | 2022 | 2023 | |
| Ascitic Fluid | 1608 | 1381 | 1606 | 936 |
| Semen | 205 | 260 | 184 | 123 |
| Blood | 26667 | 35344 | 32259 | 19292 |
| Tracheal Aspirate | 428 | 533 | 405 | 634 |
| Broncho Alveolar Lavage (BAL) | 369 | 426 | 459 | 412 |
| Cerebro Spinal Fluid (CSF) | 527 | 571 | 563 | 269 |
| Conjunctival Swab | 5 | 5 | – | – |
| Ear Discharge | 2 | 2 | 7 | – |
| Nasal Swab | 16 | 56 | 48 | 15 |
| Sterile Fluids | 788 | 736 | 701 | 462 |
| Pus | 2916 | 2987 | 3471 | 1770 |
| Sputum | 1552 | 1707 | 1930 | 2145 |
| Tissue | 68 | 424 | 380 | 336 |
| Urine | 11346 | 13892 | 13960 | 9944 |
| Vaginal Swab | 160 | 200 | 570 | 365 |
| Wound Swab | 1413 | 1576 | 1967 | 1403 |
| Total | 48070 | 60100 | 58510 | 38106 |
| Total of all samples | 204786 | |||
Table (2):
Demographic details of patients
| Variable | Sub category | Frequency |
|---|---|---|
| Age | 10-20 Years | 28 |
| 20-40 Years | 108 | |
| 40-60 Years | 113 | |
| 60-80 Years | 59 | |
| Gender | Male | 167 |
| Female | 141 |
There was a total of 204786 requests for culture and sensitivity that were received throughout the course of the study session. Of those, 17002 samples were culture positive, which accounts for 8.30% of the total. Among these, 1911 (11.24%) were determined to be Klebsiella pneumoniae, while 2934 (17.26%) were found to be Escherichia coli. Among the Klebsiella pneumoniae isolates, 258 (13.50%) were resistant to carbapenems. Of the E. coli isolates, 10 (0.34%) were found to be resistant to carbapenems.
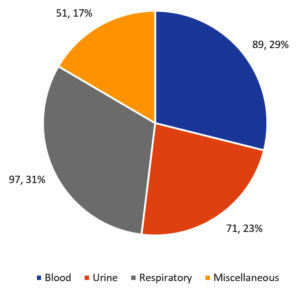
CRE accounted for 10.6% of all the infections that were caused by Enterobacteriaceae and were culture-positive. Among the 1819 CRE samples that were found to be positive, the requests for performing Xpert Carba-R test were 421. However, 113 of these samples were non-fermenters, leaving just 308 samples that were ultimately tested based on the inclusion and exclusion criteria. Of the samples that were analyzed, 31% were respiratory samples (tracheal aspirate and BAL), 29% were blood culture samples, and 23% were urine samples with the remaining 17% samples studied being taken from wound swabs, cerebrospinal fluid, tissue samples, sputum, or other body fluids (Figure 1).
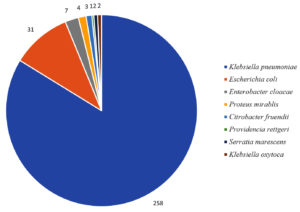
The many organisms that were identified have been listed in Figure 2, and Klebsiella pneumoniae was shown to be the most prevalent organism, accounting for 83% of the total number of samples collected (258). Escherichia coli was found in 31 samples (10%), followed by Enterobacter cloacae in 7 samples (2.2%), Proteus mirabilis in 4 samples (1.2%), Citrobacter freundii in 3 samples (0.9%), Klebsiella oxytoca in 2 samples (0.6%), Serratia marcescens in 2 samples (0.6%), and Providencia rettgeri in a single sample (0.3%).
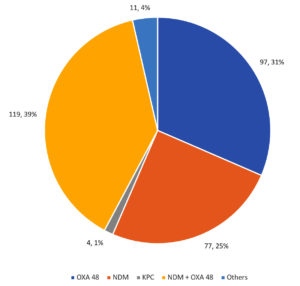
Among the CRE isolates, 97 were isolated with the OXA-48 genotype alone (representing 31% of the total), 77 were isolated with the NDM genotype alone (representing 25% of the total), and 4 were isolated with the KPC genotype (representing 1% of the total) as the resistance inducer with 11 organisms exhibited the presence of NDM, OXA-48, and KPC, which accounts for 4% of the total. A total of 119 isolates were a mix of both OXA-48 and NDM genes, which accounted for 39% of the total (Figure 3).
It was stated in Table 3 that the genotypic distribution of the genes was distributed over a variety of bacterial species, with Klebsiella pneumoniae being the most abundant organism, accounting for 83% of the isolates. There was a co-production of NDM and OXA-48 genes in 48% of the Klebsiella pneumoniae isolates, followed by the production of OXA-48 alone in 34% of the isolates, and then the synthesis of NDM alone in 17% of the isolates. Only a small percentage, 0.3%, of the population carried the KPC gene. Ten percent of the isolates were composed of Escherichia coli, which had a high prevalence of the NDM gene (83%) and a minimal presence of OXA-48 (9.6%) or co-production (6.4%). A significant presence of OXA-48 (66%) and NDM (33%), respectively, was observed in Citrobacter freundii. Isolates of Proteus mirabilis were shown to have the NDM gene equally. It was found that Providencia had a single isolate that produced both NDM and OXA-48 simultaneously. A fifty percent portion of the Serratia isolates were distributed between NDM and KPC. In a vast majority of cases, the NDM gene was carried by Enterobacter cloacae (85%) while the Klebsiella oxytoca isolates were distributed evenly between NDM and OXA-48.
Table (3):
Genotypic distribution of Carbapenem-resistant Enterobacterales
Organisms |
NDM |
OXA-48 |
VIM |
NDM & OXA 48 |
KPC |
|---|---|---|---|---|---|
Klebsiella pneumoniae 83% (n = 258) |
44 (17%) |
89 (34%) |
0 |
124 (48%) |
1 (0.3%) |
Escherichia coli 10% (n = 31) |
26 (83%) |
3 (9.6%) |
0 |
2 (6.4%) |
0 |
Citrobacter freundii 0.9% (n = 3) |
1 (33%) |
2 (66%) |
0 |
0 |
0 |
Proteus mirabilis 1.2% (n = 4) |
4 (100%) |
0 |
0 |
0 |
0 |
Providencia 0.3% (n = 1) |
0 |
0 |
0 |
1 (100%) |
0 |
Serratia 0.6% (n = 2) |
1 (50%) |
0 |
0 |
1 (50%) |
|
Enterobacter Cloacae 2.2% (n =7) |
6 (85%) |
1 (15%) |
0 |
0 |
0 |
Klebsiella oxytoca 0.6% (n = 2) |
1 (50%) |
1 (50%) |
0 |
0 |
0 |
In the last twenty years, the rising prevalence of antimicrobial resistance (AMR) has emerged as a critical global public health issue, endangering healthcare systems. A 2019 study by the Global Research on AMR Project (GRAM) predicted that 4.95 million deaths were associated with bacterial antimicrobial resistance, particularly affecting South Asia. A 2016 analysis on AMR estimated that by 2050, there may be up to 10 million fatalities annually, resulting in trillions in economic losses worldwide.4,11 This study aimed to further understanding of the different genotypes linked to carbapenem-resistant bacterial infections to facilitate efficient antibiotic therapy for affected patients.
Of the samples analyzed in the present study, most of the samples were respiratory samples (tracheal aspiration and BAL) (31%) followed by blood culture samples (29%), urine samples (23%), and the remaining 17% were collected either from other sources. A similar distribution was observed in a study conducted by Zhu et al., in which respiratory specimens were the most prevalent samples (53.77%).12 Another previously conducted study reported that the most common samples were urine (23.2%), followed by swabs (20.1%), and other types of samples which contrasted the results of the present study.13
The demographic distribution indicates that 54.22% of samples were male, with the majority of cases occurring in those aged 20-40, closely followed by those aged 40-60. This pattern mirrors general trends in CRE epidemiology, although geographical and methodological variances are present. The male predominance corresponds with research from China and India, where males represented 56.73% of CRE cases in both instances.12,14 Nonetheless, certain studies indicate elevated CRE resistance rates in females (21.95% vs 15.81% in males),14 implying that although males may be sampled more often, resistance mechanisms or healthcare exposure patterns may vary by gender. A German study indicated that Citrobacter and VIM carbapenemases were markedly more prevalent in females.15 The incidence of instances among younger age groups (20-40 and 40-60 years) differs from research conducted in South Korea and Germany, where CRE prevalence was highest in older persons (>70 years).15,16 Nonetheless, same patterns were observed in China and India, where the 16-45 and 21-40 age cohorts exhibited the highest resistance rates.12,14 This may indicate geographical variations in antibiotic prescribing patterns, healthcare accessibility, or occupational exposures among younger demographics. Demographic patterns may also arise from sampling biases, like elevated hospitalization rates in particular age or gender cohorts. For example, research pertaining to intensive care units (ICUs) frequently documents elderly patients,16 but community-based surveillance may encompass younger populations.12,14 The distribution of carbapenemase genes, such as the predominance of NDM in South Asia compared to KPC in the Americas, may affect population risk profiles.15
Of the various organisms detected in the present study, Klebsiella pneumoniae made up the majority (83%), followed by Escherichia coli (10%), Enterobacter cloacae (2.2%), Proteus mirabilis (1.2%), Citrobacter freundii (0.9%), Klebsiella oxytoca (0.6%), Serratia marcescens (0.6%), and Providencia rettgeri (0.3%). Of the different genotypes assessed, a majority were the OXA-48 genotype alone (31%), which was followed by the NDM genotype alone (25%) and 1.3% were KPC genotype as the inducer of resistance. Among the combined genotypes assessed, 37% of organisms had a mix of both NDM genes and OXA-48 while 13.5% of organisms demonstrated the presence of NDM, OXA-48, and KPC in tandem. A study evaluating global AMR conducted by Lam et al. revealed that 70.2% of identified carbapenem-non susceptible genomes possessed carbapenemases, predominantly NDM-1, KPC-2, OXA-48 and KPC-3.17 According to a recent study by Verma et al., A. baumannii or P. aeruginosa accounted for 82.3% of all discovered CROs, whereas K. pneumoniae or E. coli only made up 17.7% of the same.18 A study by Cai et al. likewise produced contrasting results, reporting that A. baumannii accounted for 45% of the observed CRO and P. aeruginosa for 19%, while Enterobacteriaceae, which was the most often reported species in this study, accounted for only 1%.19
A study by Singh-Moodley et al. examined phenotypic profiles of suspected CPE isolates and established a correlation with carbapenemase production through molecular methods. The results showed that most isolates were Klebsiella pneumoniae (62.9%). Carbapenemases made up more than three fourths of the investigated isolates of which blaOXA-48 was most common (36.5%), which was followed by blaNDM (33.8%), blaVIM (4.0%), blaIMP (1.3%), blaGES (0.9%) and blaKPC (0.7%).13 Both of these results are in complete agreement with those obtained in the present study. Zhu et al. in their study identified different strains of CRE as well as the genotypes reported similar results that the most common CRE strain was K. pneumoniae (70.16%) followed by E. coli (18.99%). However, among the 76 strains that carried the carbapenemase gene, 27 strains of blaNDM (32.5%) and 49 strains of blaKPC (61.25%) were the most common carbapenemase genes detected and this was dissimilar to the present study.12 The differences in outcomes can be linked to the varying sample sizes utilized in the research studies. In a similar vein, Prabhala et al. reported contrasting findings, indicating that among the 2,351 gram-negative bacilli, 26.5% exhibited Carbapenem resistance, with K. pneumoniae identified as the most common isolate from the CRO at 59.9%. The molecular testing performed on the 190 CR isolates revealed that NDM was detected most frequently among the resistant gene, accounting for 33.68%. This was followed closely by a combination of OXA-48 and NDM at 32.63%, while OXA-48 alone made up 14.2%.20 The varied evidence obtained by different studies can be explained by the different types of samples employed as well as the different sample sizes.
In relation to studies that align fully with the findings of the current research, Al-Tawfiq et al. carried out an investigation in a hospital from Saudi Arabia, examining prevalence of different mechanisms of CRE. The findings showed that among the 200 CRE isolates, Klebsiella pneumoniae made up the majority (48%), followed by Escherichia coli (25.5%) and Pseudomonas aeruginosa (22.5%). When carbapenemases were detected, OXA-48 (41.5%), NDM (2.5%), and a combination of OXA-48 and NDM (2.5%) were reported. The findings indicated that OXA-48 emerged as the predominant carbapenemase, with a notable proportion exhibiting no detectable genes.21 Joshi et al. in their investigation, conducted by researchers from a tertiary healthcare facility in South India, estimated the incidence of various genes involved in the generation of carbapenemase in GNB. The 164 detected samples showed the presence of mainly Klebsiella pneumoniae (92.68%) while OXA-48 (84.5%) and NDM (58.6%) were the most prevalent genes responsible, correspondingly. Individually, OXA-48 (40.7%) detection was followed by NDM with 14.8%. There was no presence of IMP or KPC genes.22 Enterobacteriaceae carbapenem resistance is mostly caused by OXA-48-like carbapenemases, which are likely currently underreported.23
The employed genotypic technique possesses specific limitations that require consideration. False negative results may occur due to the failure to identify genes linked to rare carbapenemase types and genetic variants excluded from the screening assay, including IMI, SPM, SIM, and others. This suggests that certain variations and other carbapenemases may have been neglected, indicating that the actual prevalence of CPE in the study population could exceed the stated figures.13 A significant restriction was the utilization of the Xpert Carba-R assay, which is capable of identifying only four of the ten variant genes (blaOXA-163, blaOXA-162, blaOXA-204, and blaOXA-48) associated with OXA-48. The advantages of the present study can be attributed to the vast sample size employed as well as the various types of samples assessed which helps in the generalizability of the obtained results. The precise cause of the disparities in the molecular epidemiology of CR isolates is indeterminate; nevertheless, it is likely that the empirical antibiotics employed in various regions and the length of the investigations influence this variation. Most research in the Indian subcontinent primarily monitors and documents CRE isolates; however, there has also been a rise in carbapenem resistance among non-fermenters. Insufficient data in specific domains may also be attributed to a deficiency in molecular testing resulting from financial limitations.24,25
Since CRE has become more common in medical facilities worldwide in recent years, patient fatality rates are high because of the disease’s laborious diagnostic procedures and constrained treatment options. Thus, clinical treatment and infection prevention depend on the prompt and precise diagnosis of CRE, particularly CPE. In the present study, more than half of all carbapenem-related resistance was caused by the OXA-48 and NDM genotypes and this evidence can help in the formulation of effective treatment regimens to reduce the adverse effects of these MDR organisms among ICU patients.
ACKNOWLEDGMENTS
None.
CONFLICT OF INTEREST
The authors declare that there is no conflict of interest.
AUTHORS’ CONTRIBUTION
BA conceptualized the study and applied methodology. MMR performed visualization and Investigation. SS performed data curation, formal Analysis and validated the data. BA and JVK performed supervision and project administration. SS wrote original draft. SS and MMR wrote, reviewed and edited the manuscript. All authors read and approved the final manuscript for publication.
FUNDING
None.
DATA AVAILABILITY
The datasets generated and/or analysed during the current study are available from the corresponding author on reasonable request.
ETHICS STATEMENT
This study was approved by the Ethical Clearance from the Institutional Human Ethics Committee (IHEC) with reference number (PSG/IHEC/2023/Renew/034).
- Rabaan AA, Eljaaly K, Alhumaid S, et al. An Overview on Phenotypic and Genotypic Characterisation of Carbapenem-Resistant Enterobacterales. Medicina (Kaunas). 2022;58(11):1675.
Crossref - Aurilio C, Sansone P, Barbarisi M, et al. Mechanisms of Action of Carbapenem Resistance. Antibiotics. 2022;11(3):421.
Crossref - Inigo M, Del Pozo JL. Treatment of infections caused by carbapenemase-producing Enterobacterales. Rev Esp Quimioter. 2022;35 Suppl 3 (Suppl 3):46-50.
Crossref - Vazquez-Ucha JC, Arca-Suarez J, Bou G, Beceiro A. New Carbapenemase Inhibitors: Clearing the Way for the b-Lactams. Int J Mol Sci. 2020;21(23):9308.
Crossref - Das S. The crisis of carbapenemase-mediated carbapenem resistance across the human-animal-environmental interface in India. Infect Dis Now. 2023;53(1):104628.
Crossref - Taneja N, Sharma M. Antimicrobial resistance in the environment: the Indian scenario. Indian J Med Res. 2019;149(2):119-128.
Crossref - Garg A, Garg J, Kumar S, Bhattacharya A, Agarwal S, Upadhyay GC. Molecular epidemiology & therapeutic options of carbapenem resistant Gram-negative bacteria. Indian J Med Res. 2019;149(2):285-289.
Crossref - CLSI. Performance Standards for Antimicrobial Susceptibility Testing, M100 32nd Edition. Clinical and Laboratory Standards Institute, Wayne, PA. 2022.
- Li HH, He ZJ, Xie LM, et al. Evaluation of XpertCarba-R assay for the detection of carbapenemase genes in gram-negative bacteria. BioMed Res Int. 2021;2021(1):6614812.
Crossref - Bonomo RA, Burd EM, Conly J, et al. Carbapenemase-producing organisms: a global scourge. Clin Inf Dis. 2018;66(8):1290-1297.
Crossref - Wilson H, Torok ME. Extended-spectrum b-lactamase-producing and carbapenemase-producing Enterobacteriaceae. Microb Genom. 2018;4(7):e000197.
Crossref - Zhu X, Guo C, Xu S, et al. Clinical distribution of carbapenem genotypes and resistance to ceftazidime-avibactam in Enterobacteriaceae bacteria. Front Cell Infect Microbiol. 2024;14:1345935.
Crossref - Singh-Moodley A, Perovic O. Phenotypic and genotypic correlation of carbapenememase-producing Enterobacteriaceae and problems experienced in routine screening. S Afr Med J. 2018;108(6):495-501.
Crossref - Sinha A, Gour M, Seth RJ. Phenotypic detection of carbapenem resistant Enterobacterales in clinical isolates at a tertiary care hospital. J Curr Res Sci Med. 2024;10(1):74-78.
Crossref - Neidhofer C, Buechler C, Neidhofer G, et al. Global distribution patterns of carbapenemase-encoding bacteria in a New Light: Clues on a role for ethnicity. Front Cell Infect Microbiol. 2021;11:659753.
Crossref - Kang M, Jung B, Kim J. Distribution and Risk Factors of Carbapenem-Resistant Enterobacterales in General Hospitals in South Korea. Am J Biochem Biotechnol. 2025;21(1):100-110.
Crossref - Lam MMC, Wick RR, Watts SC, Cerdeira LT, Wyres KL, Holt KE. A genomic surveillance framework and genotyping tool for Klebsiella pneumoniae and its related species complex. Nat Commun. 2021;12(4188):4188.
Crossref - Verma S, Suyasha ST, Bisure K. Prevalence of Carbapenem resistant Enterobacteriaceae-a study in Tertiary Care Hospital in Mumbai. J Evol Med Dent Sci. 2018;7(45):4909-4912.
Crossref - Cai B, Echols R, Magee G, et al. Prevalence of Carbapenem-Resistant Gram-Negative Infections in the United States Predominated by Acinetobacter baumannii and Pseudomonas aeruginosa. Open Forum Infect Dis. 2017;4(3):176.
Crossref - Prabhala S, Sundaresan A, Varaiya A. Prevalence and molecular characterization of carbapenem resistant gram-negative bacilli in a tertiary care hospital in Mumbai. IJMMTD. 2023;9(3):150-154.
Crossref - Al-Tawfiq JA, Rabaan AA, Saunar JV, Bazzi AM. Genotypes and prevalence of carbapenemase-producing Enterobacteriaceae and Pseudomonas aeruginosa in a hospital in Saudi Arabia. Trans R Soc Trop Med Hyg. 2022;116(1):50-53.
Crossref - Joshi DN, Shenoy B, Bhavana MV, Adhikary R, Shamarao S, Mahalingam A. Prevalence of Carbapenem-Resistant Enterobacteriaceae and the Genes Responsible for Carbapenemase Production in a Tertiary Care Hospital in South India. EMJ. 2023.
Crossref - Singh S, Pathak A, Fatima N, Sahu C, Prasad KN. Characterisation of OXA-48-like Carbapenemases in Escherichia Coli and Klebsiella Pneumoniae from North India. Biotech. 2023;13(5):134.
Crossref - Parimala TV. Screening of carbapenem resistant Enterobacteriaceae among nosocomial isolates: a study from south India. Indian J Curr Microb App Sci. 2017;6(4):460-465.
Crossref - Mathur P, Malpied P, Walia M, et al. Health-care-associated bloodstream and urinary tract infections in a network of hospitals in India: a multicenter, hospitalbased, prospective surveillance study. Lancet Global Health. 2022;10(9):e1317-25.
Crossref
© The Author(s) 2025. Open Access. This article is distributed under the terms of the Creative Commons Attribution 4.0 International License which permits unrestricted use, sharing, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.