Popularity of Polyglutamate (PGA) in industrial applications has motivated an extensive research in the area of bioprocess engineering for its production, strain improvement, advancements in analytical methods and applications. Some facets of PGA still remain a challenge and offer the researchers with time consuming methods to seek deeper insights into the behaviour of molecules. A few simple analytical approaches have been explained which can be used to address those challenges. This review focuses on understanding each step involved in manufacturing, analysis and applications of PGA. An attempt is made towards understanding the chemistry of PGA and its interaction with various other compounds. Occurrence of PGA in various conformational states under the influence of different environmental conditions is discussed. The properties of the biopolymer largely depend on its chain length which in turn arises due to diverse conditions in which it is biosynthesized by the microorganisms. Applications of PGA in various fields of food, agriculture, medical, cosmetics etc. are also dependent on the molecular weight and conformations of PGA under the given set of conditions. A few major applications of PGA which make this biopolymer an industrially influential molecule have been described.
Polyglutamic acid, properties, conformations, applications.
Biodegradability of the synthetic polymers has always been a most sought after question and has now raised an alarm amongst the environmentalists. The need for the replacement of non-degradable synthetic polymers with biodegradable biopolymers has become indispensable to protect the sustainability of the environment. Biopolymers are the biodegradable polymers that are produced by living organisms. They have been categorised under polypeptides, nucleotides and polysaccharides or sugars. Applications of starch, cellulose, chitosan, polyhyroxy-3-butyrate etc. have been exploited to a large extent at industrial scale. Apart from the existing biopolymers, there are few polyamides that are produced naturally such as Polyglutamic acid (PGA), Poly-e-lysine (PL) and Cyanophycin1. Polyamino acids or polyamides are completely different form proteins. Protein formation is DNA directed where the amino acids are placed in sequential manner to form a molecule whereas polyamino acids contain only one type of amino acid. One such polyamide encompassing an extraordinary and multifaceted feature is PGA which is a biopolymer of glutamic acid and this makes it one of the most attractive biopolymers of today’s generation. PGA is reported to be the main constituent in the sticky mucilaginous Japanese dish ‘Natto’ which is prepared by fermenting the steamed small soybeans with starter culture of Bacillus subtilis (natto)1.
The various properties of PGA such as its biodegradability, non-immunogenicity2, water-solubility of the salt forms, super absorbency3, anionic and homo-polymeric nature offer a stimulus for it to be used in various applications. In foods, PGA can find its place as a bitterness relieving agent4 and cryoprotectant for probiotics5. In medical sector it can be used for sustained release of drug as it is non-toxic to the host6,7. For water purification it is a proven entity to be used as biopolymer flocculent8 and can act as absorber for heavy metals and impurities9. PGA is being now used by cosmetic industries also due to its great moisturising capacity and is comparable to hyaluronic acid10. Hydrogel forming ability of cross-linked PGA can also be used in agriculture, biodegradable plastic and in health care in sanitary napkins.
Structure and conformation of PGA: an influence on its properties
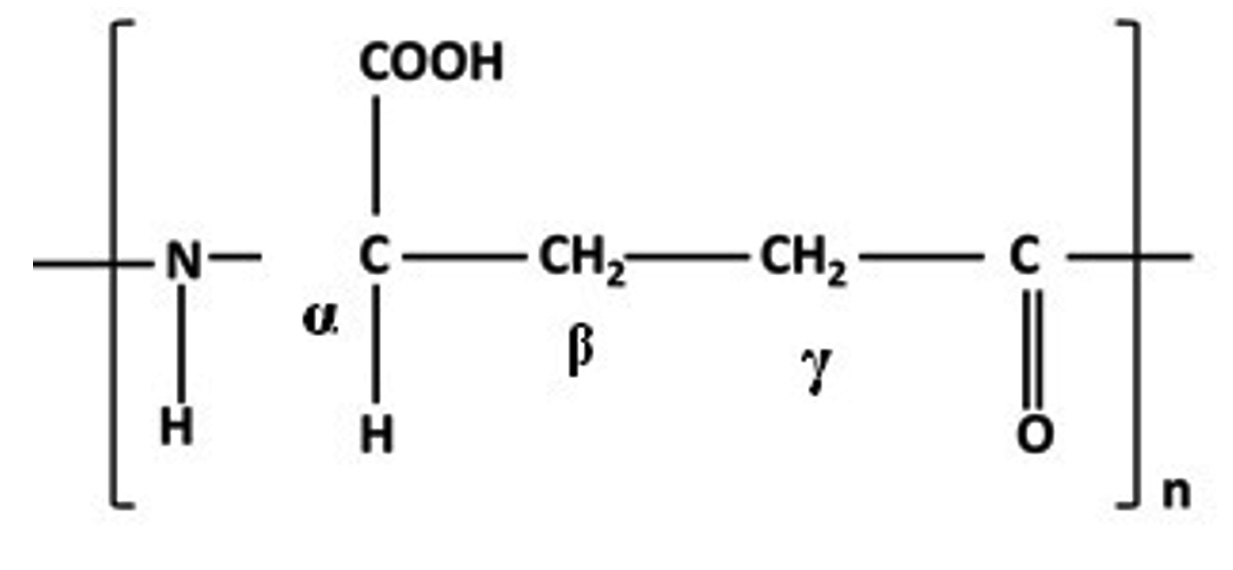
PGA is formed with the chain of its monomeric entity that contains two units of carboxylic acid of which a-carboxylic acid is linked with an amino acid through gamma linkage as shown in Fig 1. This g-linkage makes the biopolymer resistant to proteases as the normal proteases are known to cleave g-amino linkages11.
a-PGA is a result of chemical synthesis where nucleophile-initiated polymerization of N-carboxyanhydride (g-protected) of L-glutamic acid takes place. However, g-PGA is synthesised microbially12,13. Polymerization process of glutamate inside the cell and ribosome independent synthesis of PGA makes it a completely different entity from other proteins3. The organization of D and/or L forms glutamic acid in the chain length of PGA provides it stereo specificity3. The chain length in the biopolymer manoeuvres the molecular weight of the biopolymer that may range between 100 kDa to 2,500 kDa (Kilo Daltons)14. Though the molecular weight of PGA is comparable to the proteins but the arrangement of amino acids makes its structure much simpler. Higher the molecular weight, greater is the viscosity. PGA exists in various conformational states depending on the environmental conditions. Lowering of pH leads to increase in the number of COO– groups in the side chains of PGA and it acquires a-helical conformation15 that exists between pH 2.5 to 5.5. On further increasing the pH of the solution a transition from a-helical structure to random coil has been reported16,17. Secondary structure of PGA is sensitive to temperature variation as well, at higher temperatures it has been reported to aggregate and forms amyloid fibrils like structure18,19. The environmental conditions thus impact the structure of PGA to a large extent that may or may not impact the properties of the biopolymer in return.
Microscopic manufacturers of PGA
Bacillus anthracis is a gram positive, capsulated, spore forming bacterium that was initially discovered to produce PGA by Ivanovic and his co-workers in 1973. They observed the release of PGA during steam sterilization in autoclave, autolysis or ageing of the cells4. Later, several researchers reported the PGA production by other species of Bacillus such as B. licheniformis, B. subtilis, B. amyloliquefaciens, B. megaterium etc. Apart from Bacillus sp., various other microbes like halophilic archaebacteria, some gram-negative bacteria, few eukaryotes can also bio-synthesise this polymer but not to a significant extent due to various reasons20,21,22. PGA production has been successful at industrial scale mostly with B. subtilis and B. licheniformis which has made them the most studied microorganisms amongst the class Bacilli. PGA is present in the capsule of the microbes which is covalently joined with the peptidoglycan layer23 and may help the bacteria survive during the period of starvation as it has been reported to break down in the late stationary phase24.
Types of PGA
Researchers have identified three different types of PGA on the basis of alignment of PGA structure in Levorotatory or Dextrorotatory form. The L and D form of PGA is either produced as the homopolymer of either D or L or as a random copolymer with the mix of L and D forms. B. anthracis is known to be the possibly only bacteria to produce D-form of PGA25. L-PGA has been reported to be produced by an extremely halophilic bacteria, Natrialba aegyptiaca, B. megaterium, B. halodurans and Natronococcus occultus21,26,27 biosynthesise PGA of less than 20kDa molecular weight. PGA production containing both the forms of D and L-PGA has been reported by many strains of B. subtilis and B. licheniformis and few belonging from Staphylococcus epidermidis28,29. Bacillus sp. produces this exo-polymer with the molecular weight ranging from 250- 5000 kDa30.
Why do living organisms produce PGA?
There are many contributing functions of PGA that benefit the PGA producing living beings, out of which few of them are listed below:
The highly pathogenic microbes namely B. anthracis and S. epidermidis biosynthesize PGA to escape phagocytosis, aids them with virulence factor and therefore helps them evade immune evasion31,32.
Biofilms are formed by the matrix of biopolymer and helps the microbial community stick together. This further benefits the microorganisms to undergo accelerated conjugation. Since the microbes remain glued to each other in the biofilm, it protects them from the penetration of chemicals and drugs33. PGA is one such biopolymer that is produced by few Bacillus species and helps the bacteria to survive in adverse conditions by sticking to each other34.
Amongst eukaryotes, PGA is also found in the stinging cells of Hydra that helps them catch prey and it is also produced during their own defence35.
Microbiological Biosynthesis of PGA
Polymer production and its quality is highly dependent on even the small differences in the environmental factors such as media components and its ionic strength, temperature, pH, agitation, aeration, culture time etc. Thus, understanding the biosynthetic pathways and enzymes involved in the PGA production becomes very important. Biosynthesis of PGA is a ribosomal independent process. Microbes contain a multienzyme complex that drives the synthesis either through the thiotemplate mechanism or through the membrane associated amide ligation process. Troy in 1973, while carrying out research with B. licheniformis had proposed the probable existence of enzyme bound sulfhydryl group that may act as a primary acceptor of a glutamyl residue and may provide a platform for polymerization of the other glutamyl moieties within the multienzyme complex36. This work led to a conclusion that PGA biosynthesis was a result of RNA template independent process and contains a protein complex that is membrane associated. Furthermore, the membrane fraction was also reported to produce PGA even in the absence of glutamate which indicated that the biosynthetic mechanisms could be glutamic acid dependent as well.
Ashiuchi, an active and intense researcher in the biopolymer area later mentioned the involvement of amide ligation process in B. subtilis subsp. chungkookjang along with his co-workers and ruled out the thiotemplate mechanism as previously suggested. It was found and substantiated that the PGA synthetic enzyme complex is membrane bound and has DL-PGA synthetic activity in the above mentioned strain. Amide ligases are responsible for amide ligation which is carried out by either of the two types of catalysis. First of which are Rossman type enzymes that activate C-terminal carboxyl moiety of the biopolymer for eg. Murein-biosynthetic enzymes37,38. Second type of catalysis takes place by the ATP grasp type enzymes that phosphorylate the carboxyl group39. Any lack in the stereo specificity ends up in the mix of DL copolymer production. Following steps elucidate the DL PGA synthesis:
Step 1 – Phosphoryl group of ATP gets transferred to the C terminal end of carboxyl group during polymerization followed by discharge of ADP from the active site of enzyme.
L- Glutamic acid + ATP® g-L- Glutamyl-AMP + Ppi
Step 2 – Amide link formation takes place through nucleophilic attack on the phosphorylated carboxyl group by the amino group of glutamyl moiety. Thus, D and/or L glutamic acid units are joined together.
Step 3 – Liberation of DL- PGA takes place after numerous repetitions of step 1 and step 2.
PGA is an exopolymer which always gets released from the membrane bound enzyme.
Genetic configuration of PGA producing units
Few Bacillus sp. have been extensively studied with respect to the genes involved in PGA production. B. subtilis 168, B. subtilis (natto), B. licheniformis and B. anthracis are amongst the selected species on which several reports are now available. S. epidermidis also has been shown to possess PGA producing gene15. The genetic machinery of the bacteria contains various set of genes and their names have been given on the basis of fate of the final product. In B. anthracis PGA forms a capsule and therefore the genes are named as cap (capsule) genes whereas in B. licheniformis and B. subtilis PGA is surface associated due to which their genes have been named as pgs (polyglutamate synthase) genes. Therefore, the nomenclature of the genes is based on either the subsistence or deliverance of PGA. The four set of genes required for PGA synthesis in the most studied bacteria is termed as cap or pgs B, C, A and E and are arranged in the operon as shown in the fig 2. B. anthracis contains four genes – capB, capC, capA and capE on its plasmid which are categorised as necessary for polyglutamate synthesis40. However, synthesis of PGA can also take place even in the absence of these four genes in B. anthracis. In B. subtilis the similar three genes (pgsB, pgsC and pgsAA) were found to be responsible for PGA synthesis41,42. Therefore, the presence of pgs locus was considered to be adequate for PGA synthesis43. Later, pgsE/capE was also found to be present on the pgs locus. Tiwari et al. reported an indispensability of ywsC and ywtA genes in their studies based on microarray44.
To confirm the involvement of pgsBCA system in B. subtilis, pgs null mutants were created which failed to produce PGA. Urushibata et al. have also mentioned the importance of similar genes for PGA production in B. subtilis. They used the nomenclature yws C (pgsB), ywt A (pgsC) and ywt B (pgsA) in their work43.
Proposed Pathway of PGA biosynthesis
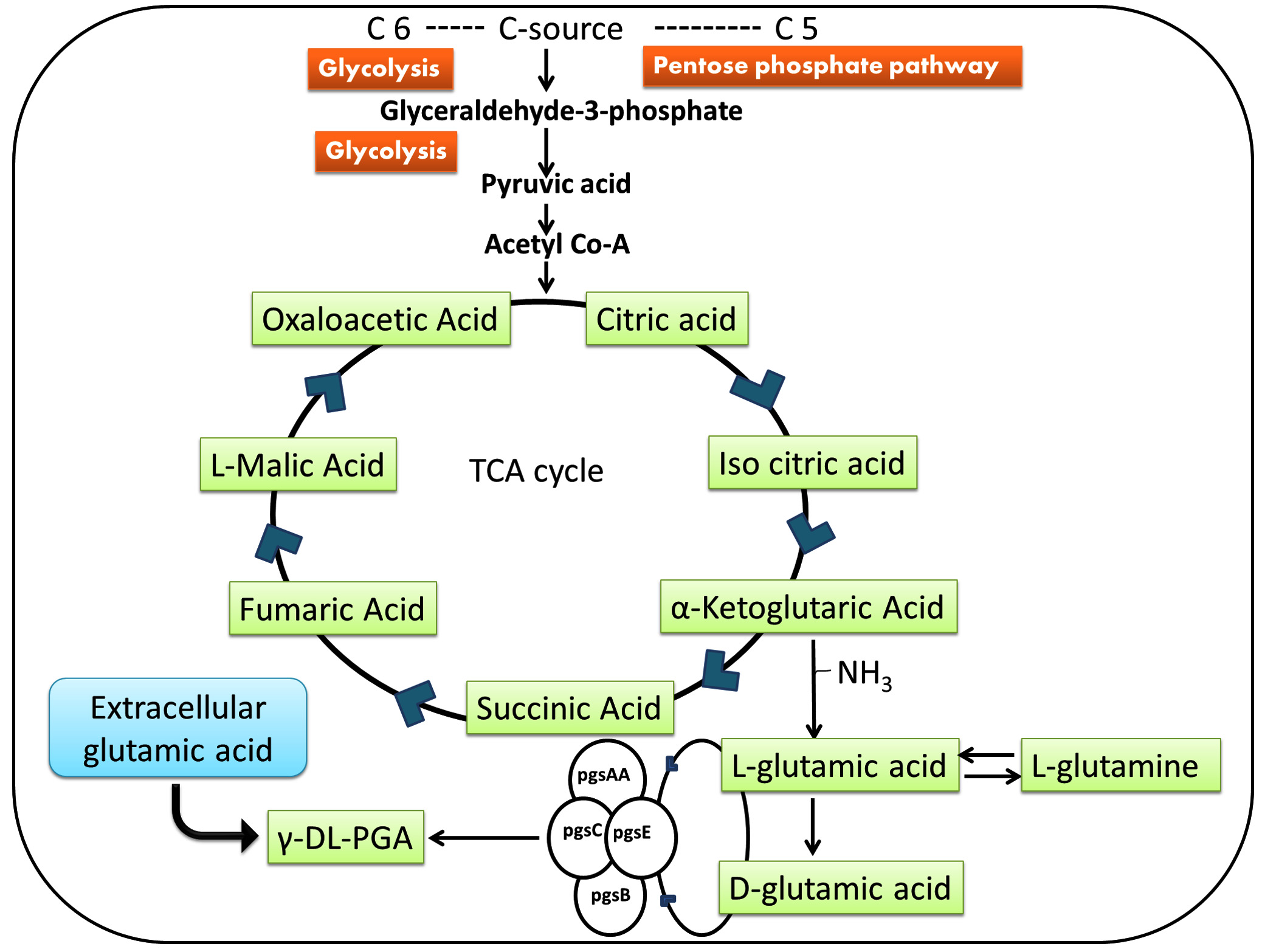
PGA biosynthesis is an intracellular process. Monomeric unit of PGA which is a glutamic acid can either be produced endogenously by the cells or provided exogenously. For endogenous production of glutamic acid, the cell has to utilize the carbon source via acetyl-CoA and TCA cycle. Exogenous supply of glutamic acid leads to its enzymatic conversion to glutamine.
Biosynthesis of PGA has been well explained in four distinct stages; racemization, poly-merization, regulation, and degradation.
Racemization
PGA can exist in both the L and D enantiomeric forms depending on the isomers of intracellular/exogenously supplied glutamic acid. Racemization is the step of conversion of L-glutamate (exo or endogenous) to D-glutamate by glutamate racemase (glr) in B. subtilis42. Other than the above mentioned enzyme racE/glr and yrpC are two more homologs of the glutamate racemase gene. racE/glr and yrpC are highly selective for L-glutamate and are present in cytosol of the cell but none of them are involved in PGA synthesis25,58. racE has been reported to be different from yrpC with respect to its activity in rich media as compared to the minimal media which is preferred by yrpC. Kimura et. al had reported in his studies that both the above mentioned genes were indispensable for the metabolism of D-glutamate24
Polymerization
As shown in fig 2, polyglutamate synthase genes are responsible for PGA biosynthesis which is encoded by pgsB, C, A, and E. pgsBCA has been reported to be involved in polymerising the monomeric units by the synthase enzyme complex (PgsBCA) through ATP-dependent process3. The key segment of catalytic site of the enzyme include PgsB and PgsC and PgsA remains accountable for removal of the polymerized chain from the active site so that the upcoming monomer can be appended. pgsA also transports the PGA through the cell membrane13. Researchers have described that if pgsB, C and A are present in high concentration then PGA production takes place even in absence of pgsE41. The presence of pgsE becomes important if Zn2+ is present in the environment in B. subtilis 101.
Regulation
Regulation of PGA biosynthetic mechanisms encompasses couple of signal transduction systems in B. subtilis where the first one is the ComP-ComA regulator, and the second one includes a two-part DegS-DegU, DegQ, and SwrA system47. DegSU, DegQ and ComPA get activated in response to environmental conditions such as prevalence of quorum sensing, osmolarity or phase variation48. degQ has been shown to be effective in down regulating the production of degradation enzymes49. Osera et al., reported that SwrA and phosphorylated DegU (DegU-P) successfully activate the pgs operon, but their effect on g-PGA production was observed to be negligible50. On the contrary, Ohsawa et. al. later revealed that DegU-P was responsible for the activation of pgs genes for PGA production instead of swrA51.
Degradation
g-Glutamyl-transpeptidases (ggt) are the well-known enzymes that inhibit the trans-peptidation reaction52. The exohydrolase activity of the enzyme causes the release of glutamic acid cleaving the PGA and the monomer is used by the bacterium as a source of carbon and nitrogen25. pgdS (also pgsS) encodes g-glutamyl-hydrolase (PgsS) that causes degradation of PGA. It is located downstream of the pgsBCA operon53. As mentioned in the previous section, the fate of PGA either lies in its attachment to the bacterial surface or freely released where CapD attaches the PGA to peptidoglycan layer and PgsS catalyses the release of PGA11. The strains having active pgsBCA, may fail to produce PGA due to high activity of deploymerase enzyme43 Yao et al. studied B. subtilis NX-2 for the presence and activity of PGA depolymerase enzyme, responsible for the degradation of PGA in batch culture54. They found the enzyme to be extracellular with endo-hydrolase activity. The responsible gene for encoding the enzyme was ywtD (pgsS). The enzyme was observed to be active extracellularly even during the late stationary phase. The study demonstrated that the molecular weight of PGA can get affected by activation/deactivation of the ggt enzyme and can be a better alternative to various methods of physical and chemical methods of degradation.
Screening of PGA producing bacteria
Primary screening is one of the most crucial steps in isolating the desirable microorganism which later decides the fate of the final product. It must be carried out carefully after the collection of the source sample as purification and isolation of the right microbe starts from this stage. Assay methods can either be direct or indirect. Advances in using the chromogenic substrate for enzyme detection leading to morphological change around the colony or use of analytical and sophisticated instruments has been of great help in isolation of desirable PGA producers. Thus, for a good quality of PGA production with high concentration, screening becomes an integral part of the whole process from sample collection to production till the recovery of the final product. However, the screening methods that are employed widely are mainly dependent on morphological changes of the culture colonies. Mucoid colonies that are picked up are subjected to liquid assay where the analysis of PGA production is carried out using high performance liquid chromatography (HPLC)24,55,56. Such methods did bring up the efficient PGA producers but the whole process is very inefficient, cumbersome and time consuming and may also lead to losing out the good PGA producers due to various reasons. Firstly, the colony morphology could be deceiving, secondly in shake flask one has to wait for a good number of hours or days for PGA production to happen and thirdly it involves use of HPLC methods which is a sophisticated analytical tool and screening of broth with numerous cultures is very laborious and time consuming and prolonged process. Thus, the screening of PGA producers becomes important either in the solid agar plate or in the liquid. Nishikawa et. al and Zeng et.al. came up with the exploration of dyes and their use for identifying the PGA producers in agar plate. Zeng et al. focussed on high throughput screening for handling multiple flora at one time57,84. Use of dyes thus could make the identification of PGA producers easy and efficient.
Bacillus species especially B. subtilis and B. licheniformis are few of the established PGA producers that have been exploited at industrial scale. Few native strains have been reported to produce more than 20 g/L of PGA also during fermentation processes.
Due to the highest capability recorded so far by Bacilli, researchers have categorised the PGA producers into two groups. Group I includes all the PGA producing Bacillus species; Group II includes the bacteria belonging to other genera. B. subtilis having GRAS (Generally Regarded As Safe) is a Gram-positive, spore-forming, rod-shaped bacteria and is also used for production of various enzymes such as alpha amylase and proteases that find their use in the sector of food and medicine. B. subtilis strains with exceptional PGA production abilities have been exploited to large extent through simple enrichment techniques. B. licheniformis is also a Gram-positive, endospore-forming bacterium that resembles with B. subtilis in various aspects. Other species such as B. anthracis and B. thuringiensis also produce PGA but the biopolymer remains attached to the peptidoglycan layer and doesn’t get secreted out which makes the recovery of the final product very difficult59.
Bacterial Fermentation of PGA
PGA production has been tried out either in the solid state fermentation (SSF) or submerged fermentation. Reports on submerged fermentation of PGA outnumber the research papers published on SSF. Very handful number of bacteria has been explored for SSF that include B. subtilis and B. licheniformis generally60. The data from the SSF either mentions the production of not very high molecular weight or it does not include much details on the molecular weight of PGA with approximately ranging between 130 to 200 kDa of molecular weight61,62. More than 500 kDa molecular weight of PGA has been reported in submerged fermentation.
Synthetic a- PGA faces the limitation of lower molecular weight with less than 10 kDa limiting its application, but microbially produced g – PGA overcomes this limitation with the production of high molecular weight entity ranging from 10 – >1000 kDa31,63,64. High cost of PGA limits its use in various applications. Therefore the focus of the researchers has been on reducing the cost of its production as much as it is possible. Cutting down the cost of raw material could solve the problem to a certain extent but this should not be a hindering factor in the high quality PGA production. This makes the media of the PGA fermentation a very important factor on which the microbe relies on for the chain length and yield. For example, it has been observed that NaCl plays a very important role in deciding the molecular mass of the final product. PGA fermentation with B. licheniformis ATCC 9945a showed an increase in the molecular weight on increasing the dose of NaCl in the media65.
Bajaj et al. have used statistical approach while using B. licheniformis NCIM 2324 in SSF as well as submerged fermentation. The bacteria was reported to produce 98.64 mg (g dry solids)-1 in solid state and 35.75 g/L PGA in liquid broth under optimized conditions64. Soliman et al. reported 33.5 g/L of PGA production with glutamate independent producer B. licheniformis SAB-26 using Plackett Burman design of experiments. B. licheniformis A13 was observed to produce PGA yield of 28.2 g/l in an optimised medium66.
Bacillus sp. have been divided into two major categories for their capability to produce PGA in presence and absence of glutamic acid. Generally, the glutamic acid independent producers remain more preferable than the glutamic acid dependent producers. The glutamic acid dependent production includes expensive raw materials which raises the cost of production80. Glutamic acid independent process is though economy friendly with respect to raw materials but the yield of PGA remains very low. Attempts have been made to genetically improve the bacteria for increasing the production. An exhaustive list of the Bacillus sp. with the details of the raw materials, production, yield etc. has been detailed by Sirisansaneeyakul et al.67 in their review from the year 1996 to 2017. In addition, in an article published by Zhang et.al 2018, the researchers have promoted the use of amino acids to increase the yield of PGA68.
B. subtilis was first shown by Bovarnick et al. to have produced PGA freely in the fermentation medium69. Extensive research with various strains has been carried out since then for PGA production using B. subtilis. As a result of which B. subtilis has become the most researched microorganism than B. licheniformis for the biopolymer production due to its high capacity of production. Huang et al. reported the use of B. subtilis ZJU-7 (B. subtilis CGMCC1250) using fed batch process which gave the PGA concentration of 101.1 g/L70,70. B. subtilis C1 was used by Shih and co-workers which was dependent on both citric acid and glycerol in the medium. Absence of either of the two above mentioned ingredient was observed to impact the PGA production. The molecular weight of PGA produced by B. subtilis C1 was recorded to be higher (1 x 107 Da) than the studies carried out by Park et al.31 due to the presence of glycerol.
g-PGA production by glutamic acid independent Bacillus subtilis C10 was evaluated with five different organic acids i.e. succinic acid, citric acid, fumaric acid, oxalic acid and acetic acid glutamic Zhang et al.71. Citric acid or oxalic acid was observed to regulate the bioactivity of key enzymes and thus an increase in the productivity of PGA could be achieved. Maximum concentration of PGA reached up to 27.7 g/L by adding 70 and 20 g/L citric acid into the fermentation medium.
B subtilis has also been co-cultured with Corynebacterium glutamicum using glucose and sucrose in the media, where the L-glutamic acid produced by C. glutamicum was used by B. subtilis. Thus, this may avoid the exogenous supply of L-glutamic acid.
B. anthracis was the first microbe to be identified as PGA producer, but it cannot be used for industrial production firstly due to its toxicity and secondly it does not secrete PGA extracellularly making the downstream processing difficult. B. thuringiensis also cannot be used due to similar reasons.
Downstream Processing of PGA
In the submerged fermentation process, the extracellular production of PGA leads to a straightforward recovery from the broth. Fermentation broth is either filtered or centrifuged to remove the cells first after which PGA is precipitated from broth13,29. The three most widely used recovery methods include; use of organic solvents such as ethanol for precipitation, use of cations such as Cu2+, Al3+, Cr3+, and Fe3+ for complex formation with PGA and filtration can be used in the recovery of g-PGA4,13,29. Process of PGA recovery may include co-precipitation of undesired extracellular polysaccharides along with the biopolymer. The disadvantages or inefficiency of PGA purification methods triggered Manocha and Margaritis, 2010 to come up with “copper-sulphate induced precipitation of g-PGA”72. They reported a highly efficient method where the precipitation of impurities was significantly reduced using copper sulphate as compared to ethanol precipitation. Precipitation of 48% of protiens using ethanol was reduced to 3% using the copper sulphate method. Characterization of PGA was followed after the downstream processing.
Characterization of PGA
The macromolecule tends to exhibit different behaviours under diverse set of environments and this property broadens the scope of its applications.
Ultraviolet-Visible Spectroscopy
In absorption spectroscopy the analytical entity absorbs radiations in UV and visible regions. Non-bonding electrons get into excited state to higher anti bonding molecular orbital after absorption of energy. Therefore it works on the basis of excitation of electrons from lower to high energy level. The application of concentration measurement of sample is based on Beer and Lambert law which says that absorbance is proportional to the concentration and path length of the sample73. The range of ultraviolet region falls between 190-380 nm, that of visible region falls between 380-750 nm. Accordingly the chromophore groups absorb visible light in a certain region and other organic entities such as DNA, proteins etc. absorb light basis the aromatic groups of the entities. Zeng et al. have elucidated the absorption of UV light by PGA with local maxima of 216 nm wavelength84. Glutamic acid shows absorption maxima at 215 nm. Therefore the need of a dye to separate the two above mentioned entities becomes indispensable.
Fourier Transform Infra-Red (FTIR) spectroscopy
Due to the diversity in the structure and conformation of PGA, it becomes essential to deduce the properties and characterize the PGA produced by the microorganisms. Use of analytical tool like Fourier transform infra-red (FT-IR) spectroscopy technique can be helpful in differentiation of the biopolymer either in the solid or liquid form. In FTIR, amide bond absorption can be predicted at approximately 1620-1655 cm-1, carbonyl C=O absorption at 1394-1454 cm-1, hydroxyl OH absorption at about 3400-3450 cm-1 and C-N group absorption in the range from 1085 to 1165 cm-1. N-H stretching shows up between 2900 cm-1 and 2800 cm-1, amide groups are exhibited at around 1600-1660 cm-1 and C=O groups between 1390-1450 cm-1 74.
Nuclear Magnetic Resonance (NMR) Spectroscopy
Structure of salt form of PGA can be deduced using NMR as it is one of the best spectroscopic technique capable of providing the detailed dynamics of the biological or organic compounds75. NMR spectroscopy uses radio-frequencies (usually ranging from 10 to 800 MHz). Magnetic properties of compounds are studied to determine the structure of a compound. In general Proton (1H) and carbon (13C) – NMR spectroscopies are implemented to determine the homogeneity and degree of esterification of g-PGA67,76. Shifts in the NMR of the samples are compared with that of the known standard.
Molecular weight determination of PGA
Molecular weight of PGA is the deciding factor for the type of application it would go into. The molecular weight (Mw), molecular number and (Mn) and dispersity index (׀) are determined using gel permeation chromatography (GPC). The Polydispersity (P) represents the wider range of the molecular weight. GPC or Size Exclusion Column (SEC) separates the desired analytes on the basis of their sizes for which Size Exclusion column packed with porous beads are used. Bigger sized analytes are eluted first from the column as they do not get into the pores whereas smaller sized analytes are eluted much later due to their entrapment inside the pores of the column, thus increasing their retention times.
Colouring the PGA with dyes to understand it a bit more
The availability of copious amount of literature indicates the perusal of conventional research techniques for screening of PGA producing bacteria. Normal practise of screening and isolation involves differentiating the microorganisms on the basis of colony morphology on the agar plates followed by analysis of PGA production in liquid media using chromatographic methods56,77,78,79,80,81,82. Very few researchers have focussed on streamlining the screening protocols to selectively isolate the PGA producing microorganisms from the mixed flora 83,84,85. Most discussions for screening depend on use of dyes in the agar media to obtain clear demarcations in the plates. Use of Neutral red and Methylene blue have been shown to display colour change in the solid as well as liquid media34,35. Zeng et. al used neutral red for high through put screening assay to isolate promising and potent PGA producing microbes whereas Chatterjee et. al demonstrated the application of methylene blue in solid as well as liquid media to isolate PGA producers. PGA was also observed to get stained with methylene blue only instead of coomasie blue during SDS PAGE analysis86. Interaction between PGA and methylene blue or neutral red is a result of various weak interactions33. PGA has been studied to adsorb methylene blue from the waste water effluents, widening the scope of its applications. Thus, use of colouring agents to screen and isolate potent PGA producers has significantly reduced the time taken for screening of the bacteria. Apart from the screening applications, dye-polymer systems have been studied to explore the changes occurring in both the systems87. Myhr et.al. used PGA with acridine orange to demonstrate the changes occurring in both the molecules due to their complexation87.
Applications of PGA
PGA is a well appreciated polymer because of its various attractive properties such as its solubility in water, biodegradability, non-toxicity to human beings, edibility etc. These unique properties of PGA make it one of most sought after molecule that offers numerous industrial applications across diverse fields.
PGA in Agriculture
Yu et. al had suggested the application of PGA as a fertilizer synergist88 as it has the capacity to delay the release of the nutrients leading to availability of nutrients for the longer period of time in soil. Zhang et al. had carried out few lab scale trials using PGA and investigated its effect on carbon leaching and nitrogen uptake which they further validated later in the fields89. They mentioned in their report about an increase in the plant biomass as a consequence of enhanced uptake of macronutrients by the roots due to PGA application. Improvement in the uptake of nutrients was stated to be exclusively due to presence of PGA which made the soil nutrients more available to the plant. Yu et al. reported in their studies about the role of PGA in colonization of bacteria in the rhizosphere of the plant roots90. Effect of PGA bound urea fertilizer application was observed to enhance the growth of tomato plants by reducing the loss of applied nitrogen and increased nitrogen content in soil91. Apart from using PGA directly the combinations of PGA along with other components such as chitosan have been proven to enhance the germination of plants like Phaseolus vulgaris92. Xu et al. demonstrated an increase in the yield of wheat grains when the PGA was used as the fertilizer synergist93. They revealed an increase in the immobilised nitrogen content in the soil which led to the gradual release during the late growth phase of wheat97.
Waste water treatment
Unavailability of clean and potable water is now slowly becoming a global concern due to establishment of various small and big scale industries. Attempts are being made to come up with the good flocculants that can purify the waste water and make it available for regular use. Apart from the chemical agents such as potash alum, PGA has been reported to be one of the best flocculants by many researchers94,95. Various chemically derived flocculants still remain a challenge for water purification owing to the impact on health. PGA is an edible biopolymer that serves the purpose. Taguchi et. al have demonstrated the purification of water from the ponds and rivers using crosslinked PGA99. PGA has been reported to flocculate vinasse which is the waste water obtained after the distillation of ethanol in Tequila industries. Vinasse contains lot of organic and inorganic impurities which when was treated with PGA at the pH of 2.5 to 3.5 exhibited removal of around 70% of turbidity in the water96.
Apart from flocculating the organic and inorganic matter from water, PGA has also been explored to chelate the heavy metals in the unsafe contaminated water. Copper ions have been studied well to bind with PGA molecules leading to precipitation97. Inbaraj et al. reported the recovery of 9.8% mercury on using PGA at pH 2. Presence of even the weaker ions in solution hinders with the PGA-metal binding capacity. Campos et al. had proved the high effectiveness of PGA-based coagulants over the other chemically derived entities. It can be inferred from all the researches reporting the use of PGA in the applications of waste water purification that PGA outshines as biodegradable flocculent in the water purification applications without compromising the health of individuals negatively.
Food
PGA is a part of various food items out of which a Japanese dish natto, is one the most popular fermented dish made from soybeans. Indian version of natto is eaten in the east side of India in the name of Kinema. Being a component of food, PGA has been shown to play multiple roles in the food sector solely. PGA has been reported to be used as one of the most effective and efficient cryoprotectant for maintaining the viability of probiotics98. Hydration property of PGA offers its use in preventing the batters, powders etc. from drying up. Lim et al., confirmed the lesser intake of oil by the doughnuts in their preparation due to presence of PGA. Therefore it can be used as an oil reducing agent in deep fried foods99. Monomeric form of PGA which is glutamic acid carries intense taste properties but when the chains of glutamic acid form PGA, the biopolymer becomes tasteless. PGA has also been shown to act as a bitterness relieving agent. Sato et al. have shown the masking of bitterness of potassium chloride with the use of PGA as an ingredient100. Additionally, PGA has also been reported to improve the shelf life and texture of starch based food products. Tanimoto et al. have shown the mineral absorption in the intestines due to presence of PGA. Similar effect was seen in the animals as well where PGA had promoted the mineral absorption and decreased animal fat101.
PGA with the molecular weight below 20 kDa offers an excellent choice as an antifreeze agent102. PGA itself imparts very week taste as compared to the commonly used antifreeze agents such as saccharide, amino acids and inorganic salts; therefore its addition in larger quantities is expected to keep the food’s taste intact without causing any change in it. Antifreeze activity of PGA has also been investigated by Bhat et al. for maintaining the viability of probiotic bacteria5.
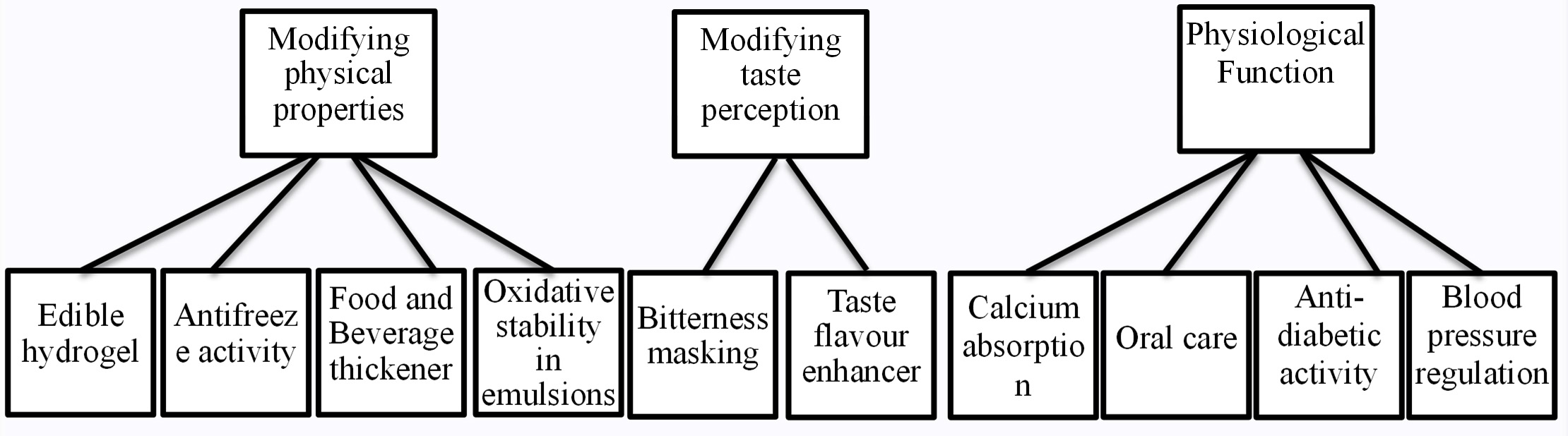
Tanimoto et al. elucidated the physiological role of PGA with respect to absorption of calcium in vitro and in vivo103,104. Phytic acid and oxalic acid are the part of food components which tend to chelate the minerals making them unavailable to the individual in intestines. However, presence of PGA has been proven to enhance the calcium absorption capability in the intestines of rats thereby increasing the Ca content in their bones. Furthermore, the retention of Ca was also assessed to impact positively in the bones of chicks105. Fig 5 represents the overall area of scope of using PGA in food applications.
Medicine
PGA is non-immunogenic harmless and biocompatible biopolymer due to which it finds application in Medical and pharmaceutical sector. Molecular weight of PGA is a crucial parameter basis which its use in medicines is decided. PGA has been widely tested as a carrier for chemotherapeutics. Slow drug release from PGA inside the body makes it one of the most reliable biopolymer. Anticancer drug like paclitaxel has been studied in conjugation with the biopolymer106. PGA- paclitaxel combination was found to have enhanced half-life in plasma as compared to the drug alone. Singer et al. designed a conjugate drug with the combination of PGA and paclitaxel and called it paclitaxel poliglumex which they claimed to extend the tumour exposure duration107. PGA-L-phenyla-lanine nanoparticles were reported to be highly effective against the retinal diseases by regulating the inflammation causing phagocytic cells. PGA-L-phenylalanine nanoparticles were also stated to be used for drug delivery to the affected part of retina108. PGA in combination with chitosan making a polyelectrolyte complex was reported to be used for wound dressings. PGA was used to maintain the moisture content. Development of keratin assimilation was also observed to be more in the wounds treated with the complex as compared to chitosan alone109.
Cosmetics
PGA is a hydrophilic molecule which when used in skin care products has been shown to stimulate the production of natural moisturising factors. It has also been shown to be more effective than collagen and hyaluronic acid in enhancing the elasticity of the skin110. PGA with the molecular weight ranging from 15×106 to 2×108 Da has been prepared as hydrogel for its application in cosmetic products111.
Thermoplastics
Food packaging and pharmaceutical industries are in constant search of biocompatible, antimicrobial and biodegradable plastics. PGA shares its similar kind of structure with polyacrylates but differs majorly in its properties. Hygroscopic nature of PGA restricts its applications in making plastics, fibres etc. Ashiuchi et al. identified a compound, hexadecylpyridinium cation (HDP+) in toothpaste that suppresses the hydrophilicity of PGA on binding with it. They named it as PGAIC. Nanofibers were also made with their combinations by electro spinning them. Furthermore, PGAIC was also found to be antimicrobial against variety of microbes. Thus, it was concluded that potential of PGAIC as bioplastics in food and hygiene industries could be explored112.
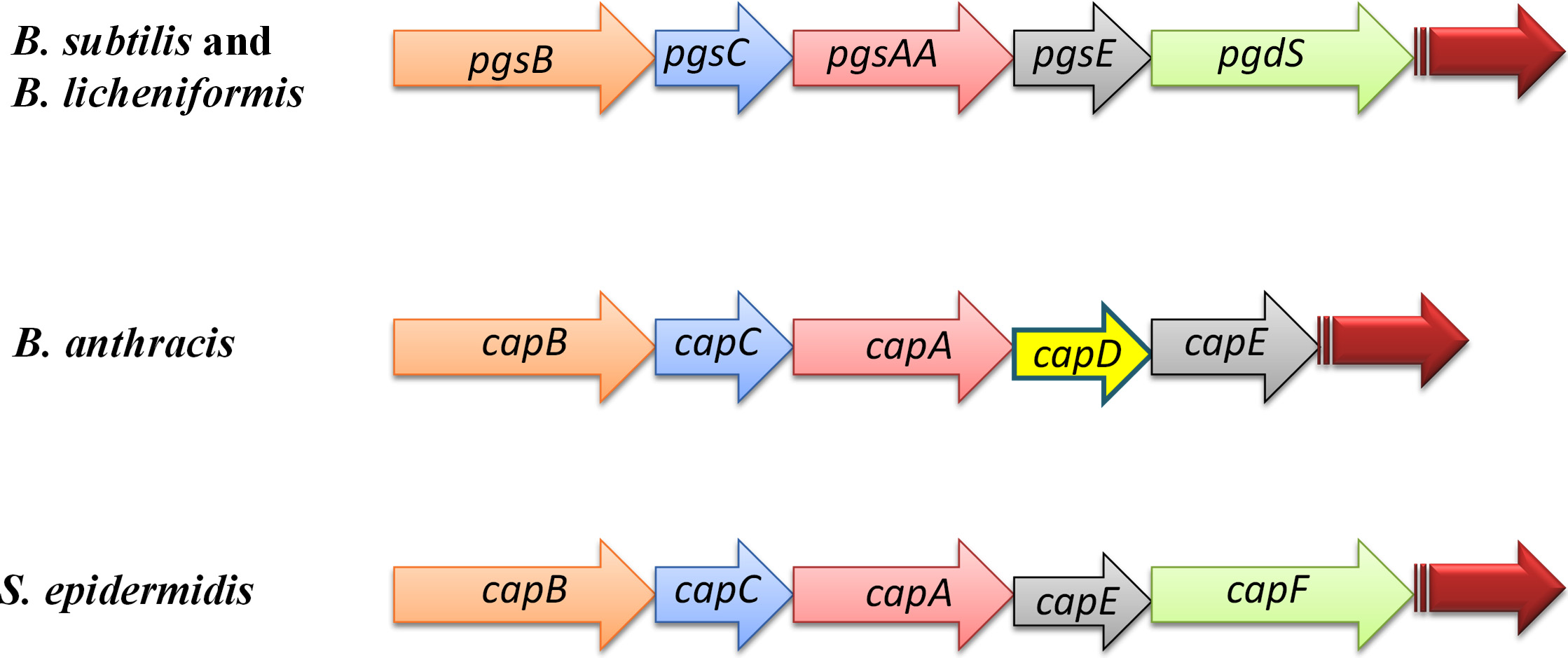 Fig. 2.Genetic assembly of B. subtilis, B. lichenisformis, B. anthracis and S. epidermidis required for PGA biosynthesis. Structures of PgsB (CapB) and PgsC (CapC) are similar to the cytosolic enzyme. PgsA (CapA) is analogous to the serine/threonine phosphatases. PgsE (CapE) stimulates PGA production. CapF of S. epidermidis is similar to B. anthracis enzyme. PgdS in B subtilis and B. licheniformis is an amidase enzyme.
Fig. 2.Genetic assembly of B. subtilis, B. lichenisformis, B. anthracis and S. epidermidis required for PGA biosynthesis. Structures of PgsB (CapB) and PgsC (CapC) are similar to the cytosolic enzyme. PgsA (CapA) is analogous to the serine/threonine phosphatases. PgsE (CapE) stimulates PGA production. CapF of S. epidermidis is similar to B. anthracis enzyme. PgdS in B subtilis and B. licheniformis is an amidase enzyme.Properties of PGA make this biopolymer worth exploring for wide variety of applications. Its biocompatibility with the host, bio-degradability, safety with respect to consumption and usage mark it as a unique biopolymer. A lot of insights have been gathered by the researchers and pioneers in the area of screening the microbes, fermentation, genetic engineering, analytical and application based studies. Hopefully, this review will contribute in expanding the knowledge further in making the biopolymer a successful and useful entity.
Aknowledgements
None
Conflict Of Interest
The authors declare that there is no conflict of interest.
Authors’ Contribution
All authors have made substantial, direct and intellectual contribution to the work and approved it for publication.
Funding
None
Data Availability
All datasets generated or analyzed during this study are included in the manuscript.
Ethics Statement
This article does not contain any studies with human participants or animals performed by any of the authors.
- Oppermann-Sanio F.B., Steinb chel A. Occurrence, functions and biosynthesis of polyamides in microorganisms and bio-technological production. Naturwissenschaften., 2002; 89: 11-22.
- Yoon S.H., Do J.H., Lee S.Y., Chang H.N. Production of poly-g-glutamic acid by fed-batch culture of Bacillus licheniformis. Biotechnol. Lett., 2000; 22: 585-8.
- Sung M.H., Park C., Kim C.J., Poo H., Soda K., Ashiuchi M. Natural and edible biopolymer poly g-glutamic acid: synthesis, production, and applications. Chem. Rec., 2005; 5: 352-66.
- Shih L, Van YT. The production of poly-(g-glutamic acid) from microorganisms and its various applications. Bioresour. Technol., 2001; 79: 207-25.
- Bhat A.R., Irorere V.U., Bartlett T., Hill D., Kedia G., Morris M.R., Charalampopoulos D., Radecka I. Bacillus subtilis natto: a non-toxic source of poly-g-glutamic acid that could be used as a cryoprotectant for probiotic bacteria. AMB Express., 2013; 3: 36.
- Li C., Yu D.F., Newman R.A., Cabral F., Stephens L.C., Hunter N., Milas L., Wallace S. Complete regression of well-established tumors using a novel water-soluble poly (L-glutamic acid)-paclitaxel conjugate. Cancer res., 1998; 58: 2404-9.
- Auzenne E., Donato N.J., Li C., Leroux E., Price R.E., Farquhar D., Klostergaard J. Superior therapeutic profile of poly-L-glutamic acid-paclitaxel copolymer compared with taxol in xenogeneic compartmental models of human ovarian carcinoma. Clin. Cancer Res., 2002; 8: 573-81.
- Taniguchi M., Kato K., Shimauchi A., Ping X., Nakayama H., Fujita K.I., Tanaka T., Tarui Y., Hirasawa E. Proposals for wastewater treatment by applying flocculating activity of cross-linked poly-g-glutamic acid. J. Biosci. Bioeng., 2005; 99: 245-51.
- Yao J., Xu H., Wang J., Jiang M., Ouyang P. Removal of Cr (III), Ni (II) and Cu (II) by poly (g-glutamic acid) from Bacillus subtilis NX-2. J Biomater. Sci. Polym. Ed., 2007; 18: 193-204.
- Moon H.S., Chung P., Chul J.K., Haryoung P., Kenji S. and Makoto A: “Natural and Edible Biopolymer Poly-g-glutamic Acid: Synthesis, production, and applications,” Chem. Rec., 2005; 5: 352-366.
- Candela T., Fouet A. Poly gamma glutamate in bacteria. Mol. Microbiol., 2006; 60: 1091-8.
- Ogunleye A., Bhat A., Irorere V.U., Hill D., Williams C., Radecka I. Poly-g-glutamic acid: production, properties and applications. Microbiology., 2015; 161: 1-7.
- Buescher J.M., Margaritis A. Microbial bio-synthesis of polyglutamic acid biopolymer and applications in the biopharmaceutical, bio-medical and food industries. Crit. Rev. Bio-technol., 2007; 27: 1-9.
- Najar I.N., Das S. Poly-glutamic acid (PGA)—structure, synthesis, genomic organization and its application: a review. Int J Pharm Sci Res., 2015; 6: 2258-80.
- Wang L.L., Chen J.T., Wang L.F., Wu S., Zhang GZ, Yu H.Q., Ye X.D., Shi Q.S. Conformations and molecular interactions of poly-g-glutamic acid as a soluble microbial product in aqueous solutions. Sci. Rep., 2017; 7: 12787.
16 Gooding EA, Sharma S, Petty SA, Fouts EA, Palmer CJ, Nolan BE, Volk M. pH-dependent helix folding dynamics of poly-glutamic acid. Chem. Phys.,2013; 422: 115-23.
- Stanley C.B., Strey H.H. Osmotically induced helix-coil transition in poly (glutamic acid). Biophys. J., 2008; 94: 4427-34.
- Tobias F., Keiderling T.A. Role of side chains in ג-sheet self-assembly into peptide fibrils. IR and VCD spectroscopic studies of glutamic acid-containing peptides. Langmuir., 2016; 32: 4653-61.
- Colaco M., Park J., Blanch H. The kinetics of aggregation of poly-glutamic acid based polypeptides. Biophys. Chem. 2008; 136: 74-86.
- Niemetz, R., Kהrcher, U., Kandler, O., Tindall, BJ., Kצnig, H. The cell wall polymer of the extremely halophilic archaeon Natronococcus occultus. Eur. J. Biochem., 1997; 249: 905-911.
- Tanaka, T., Hiruta, O., Futamura, T., Uotani, K., Satoh, A. Purification and characterization of poly (g-glutamic acid) hydrolase from a filamentous fungus, Myrothecium sp. TM-4222. Biosci. Biotechnol. Biochem., 1993; 57: 2148-2153.
- Hezayen F.F., Rehm B.H., Tindall B.J., Steinb chel A. Transfer of Natrialba asiatica B1T to Natrialba taiwanensis sp. nov. and description of Natrialba aegyptiaca sp. nov., a novel extremely halophilic, aerobic, non-pigmented member of the Archaea from Egypt that produces extracellular poly (glutamic acid). Int. J. Syst. Evol. Microbiol., 2001; 51: 1133-42.
- Soliman N.A., Berekaa M.M., Abdel-Fattah Y.R. Polyglutamic acid (PGA) production by Bacillus sp. SAB-26: application of Plackett–Burman experimental design to evaluate culture requirements. Appl. Microbiol. Biotechnol., 2005; 69: 259.
- Kimura K., Tran L.S., Uchida I., Itoh Y. Characterization of Bacillus subtilis g-glutamyl-transferase and its involvement in the degradation of capsule poly-g-glutamate. Microbiology., 2004; 150: 4115-23.
- Ashiuchi M., Misono H. Biochemistry and molecular genetics of poly-g-glutamate synthesis. Appl. Microbiol. Biotechnol., 2002; 59: 9-14.
- Shimizu K., Nakamura H., Ashiuchi M. Salt-inducible bionylon polymer from Bacillus megaterium. Appl. Environ. Microbiol., 2007; 73: 2378-9.
- Aono R. Characterization of structural component of cell walls of alkalophilic strain of Bacillus sp. C-125. Preparation of poly (g-l-glutamate) from cell wall component. Biochem. J., 1987; 245: 467-72.
- Kunioka M., Goto A. Biosynthesis of poly (g-glutamic acid) from L-glutamic acid, citric acid, and ammonium sulfate in Bacillus subtilis IFO3335. Appl. Microbiol. Biotechnol., 1994; 40: 867-72.
- Ogawa Y., Yamaguchi F., Yuasa K., Tahara Y. Efficient production of g-polyglutamic acid by Bacillus subtilis (natto) in jar fermenters. Biosci. Biotechnol. Biochem., 1997; 61: 1684-7.
- Park C., Choi J.C., Choi Y.H., Nakamura H., Shimanouchi K., Horiuchi T., Misono H., Sewaki T., Soda K., Ashiuchi M., Sung M.H. Synthesis of super-high-molecular-weight poly-g-glutamic acid by Bacillus subtilis subsp. chungkookjang J. Mol. Catal., B Enzym., 2005; 35: 128-33.
- Kocianova S., Vuong C., Yao Y., Voyich J.M., Fischer E.R., DeLeo F.R., Otto M. Key role of poly-g-DL-glutamic acid in immune evasion and virulence of Staphylococcus epidermidis. J. Clin. Invest., 2005; 115: 688-94.
- Ezzell Jr J.W., Welkos S.L. The capsule of Bacillus anthracis, a review. J. Appl. Microbiol. Biochem., 1999; 87: 250.
- Watnick P., Kolter R. Biofilm, city of microbes. J. Bacteriol.. 2000; 182: 2675-9.
- Morikawa M., Kagihiro S., Haruki M., Takano K., Branda S., Kolter R., Kanaya S. Biofilm formation by a Bacillus subtilis strain that produces g-polyglutamate. Microbiology. 2006; 152: 2801-7.
- Holstein T., Tardent P. An ultrahigh-speed analysis of exocytosis: nematocyst discharge. Science, 1984; 223: 830-3.
- Troy F.A. Chemistry and biosynthesis of the poly (g-D-glutamyl) capsule in Bacillus licheniformis I. Properties of the membrane-mediated bio-synthetic reaction. J. Biol. Chem., 1973; 248: 305-15.
- Eveland S.S., Pompliano D.L., Anderson M.S. Conditionally lethal Escherichia coli murein mutants contain point defects that map to regions conserved among murein and folyl poly-g-glutamate ligases: identification of a ligase superfamily. Biochemistry., 1997; 36: 6223-9.
- Sheng Y., Sun X., Shen Y., Bognar A.L., Baker E.N., Smith C.A. Structural and functional similarities in the ADP-forming amide bond ligase superfamily: implications for a substrate-induced conformational change in folylpolyglutamate synthetase1. Journal of molecular biology, 2000; 302(2): 425-38.
- Kino K., Kotanaka Y., Arai T., Yagasaki M. A novel L-amino acid ligase from Bacillus subtilis NBRC3134, a microorganism producing peptide-antibiotic rhizocticin. Biosci. Biotechnol. Biochem., 2009; 73: 901-7.
- Candela T., Mock M., Fouet A. CapE, a 47-amino-acid peptide, is necessary for Bacillus anthracis polyglutamate capsule synthesis. J. Bacteriol., 2005; 187: 7765-72.
- Ashiuchi M., Soda K., Misono H. A poly-g-glutamate synthetic system of Bacillus subtilis IFO 3336: gene cloning and biochemical analysis of poly-g-glutamate produced by Escherichia coli clone cells. Biochem. Biophys. Res. Commun., 1999; 16263: 6-12.
- Urushibata Y., Tokuyama S., Tahara Y.. Characterization of the Bacillus subtilis ywsC gene, involved in g-polyglutamic acid production. J. Bacteriol., 2002; 184: 337-43.
- Ashiuchi M., Nawa C., Kamei T., Song J.J., Hong S.P., Sung M.H., Soda K., Yagi T., Misono H. Physiological and biochemical characteristics of poly g-glutamate synthetase complex of Bacillus subtilis. Eur. J. Biochem., 2001; 268: 5321-8.
- Tiwari D.P., Chatterjee P.M., Rotti H., Chand B., Raval R., Dubey A.K. Expression dynamics of the poly-g-glutamic acid biosynthesis genes of Bacillus subtilis in response to glucose and glutamic acid—a pilot study. FEMS microbiology letters. 2018; 365(22): fny248.
- Ho G.H., Ho T.I., Hsieh K.H., Su Y.C., Lin P.Y., Yang J., Yang K.H., Yang S.C. g-Polyglutamic Acid Produced by Bacillus Subtilis (Natto): Structural Characteristics, Chemical Properties and Biological Functionalities. J. Chin. Chem. Soc., 2006; 53: 1363-84.
- Luo Z., Guo Y., Liu J., Qiu H., Zhao M., Zou W., Li S. Microbial synthesis of poly-g-glutamic acid: current progress, challenges, and future perspectives. Biotechnol. Biofuels., 2016; 9: 134.
- Tran L.S.P., Nagai T., Itoh Y. Divergent structure of the ComQXPA quorumsensing components: molecular basis of strain-specific communication mechanism in Bacillus subtilis. Mol. Microbiol., 2000; 37: 1159–71.
- Stanley, N.R. & Lazazzera, B.A. Defining the genetic differences between wild and domestic strains of Bacillus subtilis that affect poly-c-DL-glutamic acid production and biofilm formation. Mol. Microbiol., 2005; 57: 1143–1158.
- Do T.H., et al. Mutations suppressing the loss of DegQ function in Bacillus subtilis (natto) poly-g-glutamate synthesis. Appl. Environ. Microbiol., 2011; 77: 8249–58.
- Osera C., Amati G., Calvio C., Galizzi A. SwrAA activates poly-gamma-glutamate synthesis in addition to swarming in Bacillus subtilis. Microbiology, 2009; 155: 2282–7
- Ohsawa T., Tsukahara K., Ogura M. Bacillus subtilis response regulator DegU is a direct activator of pgsB transcription involved in gamma-poly-glutamic acid synthesis. Biosci. Biotechnol. Biochem., 2009; 73: 2096–102
- Morelli C. F., Calvio C., Biagiotti M. & Speranza G. pH dependent hydrolase, glutaminase, trans-peptidase and autotranspeptidase activities of Bacillus subtilis c-glutamyltransferase, FEBS J., 2014; 281: 232–245.
- Ashiuchi M., Kamei T. & Misono H. Poly-gamma-glutamate synthetase of Bacillus subtilis. J. Mol. Catal B Enzym., 2003; 23: 101–106.
- Yao J., Jing J., Xu H., Liang J., Wu Q., Feng X. & Ouyang P. Investigation on enzymatic degradation of c-polyglutamic acid from Bacillus subtilis NX-2. J. Mol. Catal B Enzym., 2009; 56: 158–164.
- Bajaj I.B., Singhal R.S. Sequential Optimization Approach for Enhanced Production of Poly (g-Glutamic Acid) from Newly Isolated Bacillus subtilis. Food Technol. Biotechnol., 2009; 47.
- Ju W.T., Song Y.S., Jung W.J., Park R.D. Enhanced production of poly-g-glutamic acid by a newly-isolated Bacillus subtilis. Biotechnol. Lett., 2014; 36: 2319-24.
- Nishikawa M., Ogawa K.I. Distribution of microbes producing antimicrobial e-poly-L-lysine polymers in soil microflora determined by a novel method. Appl. Environ. Microbiol., 2002; 68: 3575-81.
- Ashiuchi M., Kuwana E., Komatsu K., Soda K., Misono H. Differences in effects on DNA gyrase activity between two glutamate racemases of Bacillus subtilis, the poly-g-glutamate synthesis-linking Glr enzyme and the YrpC (MurI) isozyme. FEMS Microbiol Lett., 2003; 223: 221-5.
- Cachat E., Barker M., Read T.D., Priest F.G. A Bacillus thuringiensis strain producing a polyglutamate capsule resembling that of Bacillus anthracis. FEMS Microbiol. Lett. 2008; 285: 220–6.
- Bajaj I., Singhal R. Poly (glutamic acid)–an emerging biopolymer of commercial interest. Bioresour. Technol., 2011; 102: 5551-61.
- Bajaj I., Lele S., Singhal R. A statistical approach to optimization of fermentative production of poly (g-glutamic acid) from Bacillus licheniformis NCIM 2324. Bioresour. Technol., 2009; 100: 826-32.
- Yong X., Raza W., Yu G., Ran W., Shen Q., Yang X. Optimization of the production of poly-g-glutamic acid by Bacillus amyloliquefaciens C1 in solid-state fermentation using dairy manure compost and monosodium glutamate production residues as basic substrates. Bioresour. Technol., 2011; 102: 7548-54.
- Bajaj I.B., Singhal R.S. Enhanced production of poly (g-glutamic acid) from Bacillus licheniformis NCIM 2324 by using metabolic precursors. Appl. Biochem. Biotechnol., 2009; 159: 133-141.
- Richard A. & Margaritis A. Rheology, oxygen transfer, and molecular weight characteristics of poly(glutamic acid) fermentation by Bacillus subtilis. Biotechnol Bioeng; 2003; 82: 299–305.
- Birrer G. A., Cromwick A.-M. & Gross R.A. c-Poly (glutamic acid) formation by Bacillus licheniformis 9945a: physiological and biochemical studies. Int J. Biol. Macromol., 1994; 16: 265–275.
- Mabrouk M., Abou-Zeid D. and Sabra W. Application of Plackett–Burman experimental design to evaluate nutritional requirements for poly (g-glutamic acid) production in batch fermentation by Bacillus licheniformis A13. Microbiol. Res., 2012; 1: 6-18.
- Sirisansaneeyakul S., Cao M., Kongklom N., Chuensangjun C., Shi Z., Chisti Y. Microbial production of poly-g-glutamic acid. World J. Microbiol. Biotechnol., 2017; 33: 173.
- Zhang C., Wu D., Qiu X. Stimulatory effects of amino acids on g-polyglutamic acid production by Bacillus subtilis. Sci. Rep., 2018; 8: 17934.
- Bovarnick M. The formation of extracellular d (-)-glutamic acid polypeptide by Bacillus subtilis. J. Biol. Chem., 1942; 145: 415-424.
- Huang J., Du Y., Xu G., Zhang H., Zhu F., Huang L. and Xu Z. High yield and cost-effective production of poly (g-glutamic acid) with Bacillus subtilis. Eng. Life Sci., 2011; 11: 291-297
- Zhang H., Zhu J., Zhu X., Cai J., Zhang A., Hong Y., Huang J., Huang L. and Xu Z. High-level exogenous glutamic acid-independent production of poly-(g-glutamic acid) with organic acid addition in a new isolated Bacillus subtilis C10. Bioresour. Technol., 2012; 116: 241-246.
- Manocha B. and Margaritis A. A novel Method for the selective recovery and purification of g polyglutamic acid from Bacillus licheniformis fermentation broth. Biotechnol. Prog., 2010; 26: 734-742.
- Mהntele W., Deniz E. UV–VIS absorption spectroscopy: Lambert-Beer reloaded.
- Ho G., Ho T., Hsieh K., Su Y., Lin P., Yang J., Yang K. and Yang S. g-Polyglutamic Acid Produced by Bacillus Subtilis (Natto): Structural Characteristics, Chemical Properties and Biological Functionalities. J. Chin. Chem Soc., 2006; 53: 1363-1384.
- Pretsch E., B hlmann P. Badertscher M. Structure determination of organic compounds. 4 th ed., Berlin: Springer-Verlag Berlin Heidelberg, 2009.
- Borbely M., Nagasaki Y., Borbיly J., Fan K., Bhogle A. and Sevoian M. Biosynthesis and chemical modification of poly (2009-glutamic acid). Polym. bull., 1994; 32: 127-132.
- Ashiuchi M., Kamei T., Baek D.H., Shin S.Y., Sung M.H., Soda K., Yagi T., Misono H Isolation of Bacillus subtilis (chungkookjang), a poly-g-glutamate producer with high genetic competence. Appl Microbiol. Biotechnol., 2001; 57: 764–769.
- Birrer G.A., Cromwick A.M., Gross R.A. g-Poly (glutamic acid) formation by Bacillus licheni-formis 9945a: physiological and biochemical studies. Int. J. Biol. Macromol., 1994; 16: 265–275.
- Cao M.F., Geng W.T., Liu L., Song C.J., Xie H., Guo W.B., Jin Y.H., Wang S.H. Glutamic acid independent production of poly-g-glutamic acid by Bacillus amyloliquefaciens LL3 and cloning of pgsBCA genes. Bioresour. Technol., 2011; 102: 4251–4257.
- Shi F., Xu Z.N., Cen P.L. Efficient production of poly-g-glutamic acid by Bacillus subtilis ZJU-7. Appl Biochem Biotechnol., 2006; 133: 271–281.
- Wei X.T., Ji Z.X., Chen S.W. Isolation of halotolerant Bacillus licheniformis WX-02 and regulatory effects of sodium chloride on yield and molecular sizes of poly-g-glutamic acid. Appl. Biochem. Biotechnol., 2010; 160: 1332–1340.
- Zhang H.L., Zhu J.Z., Zhu X.C., Cai J., Zhang A.Y., Hong Y.Z., Huang J., Huang L., Xu Z.N. High-level exogenous glutamic acidindependent production of poly-(g-glutamic acid) with organic acid addition in a new isolated Bacillus subtilis C10. Bioresour Technol., 2012; 116: 241–246.
- Nishikawa M., Ogawa K.I. Distribution of microbes producing antimicrobial e-poly-L-lysine polymers in soil microflora determined by a novel method. Appl. Environ. Microbiol., 2002; 68: 3575-81.
- Zeng W., Lin Y., Qi Z., He Y., Wang D., Chen G., Liang Z. An integrated high-throughput strategy for rapid screening of poly (g-glutamic acid)-producing bacteria. Appl. Microbiol. Biotechnol., 2013; 97: 2163-72.
- Chatterjee P.M., Datta S., Tiwari D.P., Raval R., Dubey A.K. Selection of an effective indicator for rapid detection of microorganisms producing g-polyglutamic acid and its biosynthesis under submerged fermentation conditions using Bacillus methylotrophicus. Biotechnol. Appl. Biochem., 2018; 185: 270-88.
- Yamaguchi F., Ogawa Y., Kikuchi M., Yuasa K., Motai H. Detection of g-polyglutamic acid (g-PGA) by SDS-PAGE. Biosci. Biotechnol. Biochem., 1996; 60: 255-8.
- Myhr B.C., Foss J.G. Polyglutamic acid–acridine orange complexes. Cotton effects in the random coil region. Biopolymers: Original Research on Biomolecules., 1966; 4: 949-52.
- Yu X., Wang M., Wang Q.H., Wang X.M. Biosynthesis of polyglutamic acid and its application on agriculture. Adv Mat Res., 2011; 183: 1219-1223). Trans Tech Publications.
- Zhang L., Gao D., Li J., Fang N., Wang L., Shi Y. Effects of poly-g-glutamic acid on soil nitrogen and carbon leaching and CO2 fluxes in a sandy clay loam soil. Can. j. soil sci., 2017; 97: 319-28.
- Yu Y., Yan F., Chen Y., Jin C., Guo J.H., Chai Y. Poly-g-glutamic acids contribute to biofilm formation and plant root colonization in selected environmental isolates of Bacillus subtilis. Front microbiol., 2016; 10: 1811.
- Jiang Y., Liu L., Luo G., Zhang Z., Davidmark J., Huang Y. Effect of g-PGA coated urea on N-release rate and tomato growth. Wuhan University Journal of Natural Sciences., 2014; 19: 335-40.
- Pereira A.E., Sandoval-Herrera I.E., Zavala-Betancourt S.A., Oliveira H.C., Ledezma-Pיrez A.S., Romero J., Fraceto L.F. g-Polyglutamic acid/chitosan nanoparticles for the plant growth regulator gibberellic acid: Characterization and evaluation of biological activity. Carbohydr Polym., 2017; 157: 1862-73.
- Xu Z., Wan C., Xu X., Feng X., Xu H. Effect of poly (g-glutamic acid) on wheat productivity, nitrogen use efficiency and soil microbes. J. Plant Nutr. Soil Sci., 2013; 13: 744-55.
- Campos V., Fernandes A.R., Medeiros T.A., Andrade E.L. Physicochemical characterization and evaluation of PGA bioflocculant in coagulation-flocculation and sedimentation processes. J. Environ. Chem. Eng., 2016; 4: 3753-60.
- Taniguchi M., Kato K., Shimauchi A., Ping X., Nakayama H., Fujita K.I., Tanaka T., Tarui Y., Hirasawa E. Proposals for wastewater treatment by applying flocculating activity of cross-linked poly-g-glutamic acid. J. Biosci. Bioeng., 2005; 99: 245-51.
- Carvajal-Zarrabal O., Nolasco-Hipףlito C., Barradas-Dermitz D.M., Hayward-Jones P.M., Aguilar-Uscanga M.G., Bujang K. Treatment of vinasse from tequila production using poly-glutamic acid. J. Environ. Manage., 2012; 95: S66-70.
- Mark S.S., Crusberg T.C., DaCunha C.M., Iorio A.A. A heavy metal biotrap for wastewater remediation using poly g-glutamic acid. Biotechnol. Prog., 2006; 22: 523-31.
- Bhat A.R., Irorere V.U., Bartlett T., Hill D., Kedia G., Morris M.R., Charalampopoulos D., Radecka I. Bacillus subtilis natto: a non-toxic source of poly-g-glutamic acid that could be used as a cryoprotectant for probiotic bacteria. AMB Express., 2013; 3: 36.
- Lim S.M., Kim J., Shim J.Y., Imm B.Y., Sung M.H., Imm J.Y. Effect of poly-g-glutamic acids (PGA) on oil uptake and sensory quality in doughnuts. Food Sci. Biotechnol., 2012; 21: 247-52.
- Sato S., Satokawa H., Iwasaki T., Tomiyama Y., Ishii K., Seguro K. Taste improver. International Patent Application: Publication No. WO2008/146491, 2008.
- Yamashiro D., Yoshioka M., Ashiuchi M. Bacillus subtilis pgsE (Formerly ywtC) stimulates poly g glutamate production in the presence of zinc. Biotechnol. Bioeng., 2011; 108: 226-30.
- Mitsuiki M., Mizuno A., Tanimoto H., Motoki M. Relationship between the antifreeze activity and the chemical structures of oligo- and poly (glutamic acid)s. J. Agric. Food Chem., 1998; 46: 891–895.
- Tanimoto H., Mori M., Motoki M., Torii K., Kadowaki M. & Noguchi T. Natto mucilage containing poly-c-glutamic acid increases soluble calcium in the rat small intestine. Biosci. Biotechnol. Biochem., 2001; 65: 516–521.
- Tanimoto H., Fox T., Eagles J., Satoh H., Nozawa H., Okiyama A., Morinaga Y. & Fairweather-Tait S. J. Acute effect of poly-cglutamic acid on calcium absorption in post-menopausal women. J. Am. Coll. Nutr., 2007; 26: 645–649.
- Tanimoto H., Sato H., Karasawa M., Iwasaki K., Oshima A., Adachi S. Feed composition containing poly-g-glutamic acid. European Patent Application: Publication No. EP0826310, 1998.
- Li C., Yu D.F., Newman R.A., Cabral F., Stephens L.C., Hunter N., Milas L., Wallace S. Complete regression of well-established tumors using a novel water-soluble poly (L-glutamic acid)-paclitaxel conjugate. Cancer research. 1998; 58(11): 2404-9.
- Singer J.W. Paclitaxel poliglumex (XYOTAX, CT-2103): a macromolecular taxane. J. Control Release, 2005; 109: 120–126.
- Ryu M., Nakazawa T., Akagi T., Tanaka T., Watanabe R., Yasuda M., Himori N., Maruyama K., Yamashita T. & other authors. Suppression of phagocytic cells in retinal disorders using amphiphilic poly(c-glutamic acid) nanoparticles containing dexamethasone. J. Control Release, 2011; 151: 65–73.
- Tsao C.T., Chang C.H., Lin Y.Y., Wu M.F., Wang J.L., Young T.H., Han J.L. & Hsieh K.H. Evaluation of chitosan/cpoly(glutamic acid) polyelectrolyte complex for wound dressing materials. Carbohydr Polym., 2011; 84: 812–819.
- Ben-Zur N. & Goldman D.M. c-Poly glutamic acid: a novel peptide for skin care. Cosmetics Toiletries 2007; 122, 65–74.
- Ho G.H., Yang T.H., Yang J., inventors; Tung Hai Biotechnology Corp, assignee. Moisturizers comprising one or more of gamma-polyglutamic acid (gamma-pga, h form), gamma-poly-glutamates and gamma-polyglutamate hydrogels for use in cosmetic or personal care products. United States patent application US 12/260,503, 2009.
- Ashiuchi M., Fukushima K., Oya H., Hiraoki T., Shibatani S., Oka N., Nishimura H., Hakuba H., Nakamori M., Kitagawa M. Development of antimicrobial thermoplastic material from archaeal poly-g-L-glutamate and its nano-fabrication. ACS Appl. Mater. Interfaces., 2013; 19(5): 1619-24.
© The Author(s) 2019. Open Access. This article is distributed under the terms of the Creative Commons Attribution 4.0 International License which permits unrestricted use, sharing, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.