ISSN: 0973-7510
E-ISSN: 2581-690X
Pyridoxal kinase encoded by pdxK gene, is the important key enzyme in the salvage pathway of vitamin B6 biosynthesis. The enzyme catalyzes the phosphorylation of the 5′ alcohol groups of free form vitamin B6 into their 5′-phosphate forms that requires metal ion and ATP. Pyridoxal kinase have been reported in many organisms except in the thermophilic bacterium. Therefore, this study aimed to clone, express and characterize pyridoxal kinase of Geobacillus sp. H6a isolated from the hot spring in the North of Thailand. The GhpdxK gene (810 base pairs) was inserted into pET28a(+) plasmids at restriction site of NdeI and BamHI and transformed into E.coli BL21(DE3). The expressed pyridoxal kinase of this bacterium exhibits a homodimer, in which each subunit had a molecular mass of about 32 kDa when examined by SDS-PAGE and gel filtration. The enzyme showed maximal activity at 70°C and at pH 8.0. The expressed enzyme obtained in this study was found to be more active (>50%) in the broad pH range (6.0 – 9.0) than those previously reported. This enzyme prefers Mg2+ and also accepts other cations to the less extent. Under optimal conditions, the expressed enzyme has higher affinity toward PN (20 ± 1.35 µM), while it showed the same affinity to pyridoxal (100 ± 0.76 µM) and pyridoxamine (100 ± 1.21 µM). The Km value for ATP and 4-amino-5-hydroxymethyl-2-methylpyridine were 8.99 ± 1.76 µM and 19 ± 0.85 µM, respectively. With high activity at high temperature and active in the broad pH range, it could be considered as a potential candidate for future application particularly bioconversion of vitamin B6.
pyridoxal kinase, salvage, thermophile, vitamin B6
Vitamin B6 is a group of pyridine derivative related vitamers, including pyridoxal (PL), pyridoxine (PN), pyridoxamine (PM), and their 5′-phosphorylated forms. Among the B6 group, pyridoxal 5′-phosphate (PLP) and pyridoxamine 5′-phosphate (PMP) are the most important coenzymes, involved in various biochemical reactions, including the metabolisms of amino acids, carbohydrates, fatty acids, lipids, nucleic acids, hemoglobin, serotonin, dopamine, noradrenaline, and gamma-aminobutyric acid (GABA) neurotransmitters.1
Eukaryotes obtain vitamin B6 from food. Microorganisms and plants can newly synthesize vitamin B6 by using a deoxyxylulose 5′-phosphate (DXP)-dependent pathway or a DXP-independent pathway, while a salvage pathway is found in most organisms. A role of salvage pathway is to interconvert the six different forms of vitamin B6 by using four enzymes. Pyridoxal kinase (PLK; EC 2.7.1.35) phosphorylates the 5′-alcohol groups of all three unphosphorylated B6 vitamers into their phosphate forms. Pyridoxine 5′-phosphate oxidase (PNPO; EC 1.4.3.5) catalyzes the oxidation of PMP and PNP to PLP. Dephosphorylation of PNP, PMP and PLP is catalyzed by phosphatase. The reduction of PL to PN is catalyzed by PL reductase (EC 1.1.1.65).2,3
Pyridoxal kinase belongs to the ribokinase superfamily. It transfers a phosphate group at ϒ position of ATP to the C5 position of free form vitamin B6 in a metal (mono and divalent cations)-dependent manner. Besides PN, PL and PM, PLK can catalyze the mono-phosphorylation of 4-amino-5-hydroxymethyl-2-methylpyridine (HMP) involving thiamine biosynthesis.4 This enzyme has been found in various sources. PLK is encoded by 2 kinase genes. A basic kinase gene (pdxK) has found in eukaryotes and prokaryotes. The other (pdxY) found in Escherichia coli, is able to phosphorylate only PL as substrate.2,5
Pyridoxal kinase from the pdxK gene is a homodimer. Molecular mass of each monomer has approximately 35.4 kDa. The amino acid sequence of mammal PLK is highly conserved with >80% identity, while that of PLKs showed approximately 40% identity in the group of microorganisms, plants, and mammals.6 Bioinformatics assays showed the comparison between Leishmania infantum PLK and Trypanosoma protein sequence had a low similarity, with identity percentage of 61.04 – 66.11%.7 These data indicate PLK has high variation.
Pyridoxal kinase has been purified and characterized from Arabidopsis thaliana,8 human,9 Bacillus subtilis,10 Plasmodia falciparum,11 E. coli,12 Bombyx mori,13 Trypanosoma brucei,14 Pseudomonas aeruginosa,5 Salmonella typhimurium,15 however, purification and characterization of PLK from thermophilic bacteria have not been reported.
Our previous study showed that Geobacillus sp.H6a (Gh) could produce vitamin B6 outside the cell.16 This thermophilic bacterium grows at a range of temperature of 30 – 70°C with the highest growth rate at 60°C. The objectives of this study were to clone, express the pdxK gene in E. coli and characterize the recombinant PLK of Geobacillus sp.H6a.
Materials
A Genomic DNA mini kit, High-Speed Plasmid Mini Kit and a Gel/PCR DNA fragment extraction kit were purchased from Geneaid Biotech Ltd. (Taipei, Taiwan). A Taq DNA Polymerase was purchased from RBC Bioscience (Taipei, Taiwan). A Ni-nitrilotriacetic acid (NTA) resin column was purchased from Novagen, UK. A Superdex 200 and Vivaspin® 20 were purchased from GE Healthcare Bio-Science AB (Sweden). All others were of analytical grade chemicals.
Cloning of GhpdxK gene
The genomic DNA of Geobacillus sp. H6a was isolated by Genomic DNA mini kit. The primers FpdxKNde (5΄-AAACATATGATGCCAA AAGCGTTAACGATCGCC -3΄) with a NdeI recognition site (underlined) and 3pdxKBam (5΄-AAAGGATCCTTATTTGTTCGCCGTAGCGG-3΄)17 with a BamHI recognition site (underlined) were used to amplify GhpdxK gene (accession number FJ497251). Amplification was operated in a thermal-cycler as follows: 30 cycles of 1 min at 95°C, 1 min at 62°C, 2 min at 72°C and final 10 min at 72°C. The annealing temperature was decreased by 2°C per cycle until 58°C. The PCR product was checked by agarose gel electrophoresis and was subsequently purified before digestion with NdeI and BamHI.
The pET28a(+) vector was isolated using High-Speed Plasmid Mini Kit and subsequently digested with the same restriction enzymes. The resulting digested PCR product and the digested pET28a(+) vector were purified by using a Gel/PCR DNA fragment extraction kit and were ligates by using T4 ligase enzyme. The resulted constructs designated as pET28a-GhpdxK was transformed into E. coli BL21(DE3) by using heat shock method. Colony PCR was used to check the transformants. DNA sequencing was carried out by First Base Laboratories (Malaysia).
Expression optimization of recombinant GhPLK (rGhPLK)
The bacterial cells harboring pET-28a-GhpdxK were cultured in Luria – Bertani (LB) medium with 50 µg ml-1 kanamycin, until the absorbance at 600 nm of the culture increased to 0.5. The expression of rGhPLK was induced by adding different concentrations of isopropyl-1-thio-D-galactopyranoside (IPTG) (0 – 1 mM), different incubation temperatures (25 °C, 30 °C, 37 °C) and times (0, 1, 3, 6 h).
The cells were collected by centrifugation at 10,000 x g, 4°C for 10 min. The cell pellet was resuspended in 5 ml of ice-cold 1x phosphate buffer saline (PBS), pH 7.4, containing 1 mM phenylmethylsulfonyl fluoride (PMSF). The suspension was lysed by ultrasonication and the resulting cell lysate was centrifuged at 15,000×g for 10 min at 4°C. Protein concentration was determined by the Bradford assay,18 using bovine serum albumin as a standard protein.
Purification of recombinant GhPLK
The rGhPLK protein was purified by Ni-nitrilotriacetic acid (NTA) resin column which was equilibrated with 20 mM potassium phosphate buffer (KPB) pH 8.0, containing 0.5 M NaCl, 10% glycerol, 1 mM 2–mercaptoethanol (2-ME) and 10 mM imidazole and collected at 1 ml per fraction at flow rate of 0.25 ml/min. The column was washed with 20 mM KPB pH 8.0, containing 0.5 M NaCl, 10% glycerol, 1 mM 2–ME and 90 mM imidazole until absorbance at 280 nm was less than 0.1. The rGhPLK protein was eluted with the same buffer, containing 500 mM imidazole. The fractions were tested activity and determined absorbance at 280 nm. The eluted fractions displaying enzyme activities, were pooled for dialysis against 20 mM KPB pH 8.0, containing 10% glycerol and 1 mM 2-ME. The dialysis protein was concentrated by Vivaspin® with 10 kDa cutoff and was subjected further analysis.
Enzyme assay of recombinant GhPLK
The rGhPLK activity was measured following to the method of Huang13 with some modifications. The assay activity was performed at 70°C. The reaction mixture of 1 ml contained 70 mM KPB pH 8.0, 0.4 mM PL, 3 mM ATP, 10 mM MgCl2. The assay activity was started by adding 0.3 µg purified rGhPLK and then the reaction was incubated for 30 min. PLP concentration was determined by monitoring the absorbance at 388 nm (e 388 = 4900 M-1cm-1 in phosphate buffer, pH 7.0) comparing with a standard curve of different concentrations of PLP standard against A388. One unit of enzyme was described as the amount of the enzyme required to produce 1 µmol of PLP per min.
To study the effect of pH, temperature and metal ions, the formed product (PLP) in enzyme reaction mixture using PL as a substrate was counted using the method of Wada and Snell.19 The reaction mixture contained 70 mM KPB pH 8.0, 0.4 mM PL, 3 mM ATP, 10 mM MgCl2, 0.3 µg purified rGhPLK and then incubated for 30 min at 70°C. A 8% trichloroacetic acid (TCA) was added and the reaction was leaved on-ice for 10 min. The centrifugation was then performed at 10,000×g for 1 min. The supernatant was added with 0.1% phenylhydrazine reagent and left for 10 min at room temperature. The concentration of PLP was determined by an increase of an absorbance at 410 nm which was compared to that of PLP standard (0 – 400 nmol).
To study kinetic, the activity of purified rGhPLK was performed using a pyruvate kinase – lactate dehydrogenase coupled system.10 The reaction mixture contained 70 mM KPB pH 8.0, 0.4 mM PL, 3 mM ATP, 10 mM MgCl2 and 0.3 µg purified rGhPLK. The reaction was incubated at 70°C for 30 min, then stored on ice. One mM of phosphoenolpyruvate, 0.2 mM of NADH, 8 units of each pyruvate kinase and lactate dehydrogenase were added in the previous reaction. The reaction was then incubated at 37 °C for 10 min. The activity was determined by measuring the decrease in absorbance at 340 nm resulted from the oxidation of NADH. One unit of activity means the oxidation of 1 µmol of NADH per min using an extinction coefficient for NADH (A 340 = 6220 M-1cm-1) to determine rate of ADP production.
Molecular mass determination
The molecular mass and the purity of the rGhPLK protein were determined by using 12% SDS-PAGE.20 Both SDS-PAGE and Native-PAGE were stained with Coomassie Brilliant Blue R-250. The native form of recombinant enzyme was determined by using gel filtration column (1 X 30 cm) manually packed with Superdex 200. The gel filtration standards (Bio-Rad) composes of thyroglobulin (670 kDa), g-globulin (158 kDa), ovalbumin (44 kDa), myoglobin (17 kDa), and Vitamin B12 (1.35 kDa) were also loaded on the column and were used for determination of the molecular weight of rGhPLK.21
Kinetic of recombinant GhpdxK
The enzyme kinetic (Km and Vmax) were determined by incubating the purified rGhPLK with varying concentrations of substrates. The data were calculated by using GraphPad Prism software version 6.01 (GraphPad Software, Inc).21
Effects of pH and temperature
The activity of rGhPLK was studied in pH range 3.0 – 10.0 with 70 mM of each buffer: pH 3.0 glycine–HCl buffer; pH 4.0 – 5.0 acetate buffer; pH 6.0 phosphate buffer; pH 7.0 – 8.0 HEPES buffer; pH 9.0 Tris–HCl buffer; pH 10.0 glycine-NaOH buffer. For pH stability, rGhPLK was incubated in different pH for 1 h at 30°C then measured activity by phenylhydrazine method.19 Effect of temperature was studied at 25 – 80°C with 70 mM KPB pH 8.0. For temperature stability, rGhPLK was incubated in different temperatures (30 – 80°C) and then the activity was measured by using phenylhydrazine method.19
Effects of metal ions
Metal ions effect was assayed using a variety of monovalent and divalent cations (Cs+, Zn2+, Ca2+, Mg2+, Mn2+, Cu2+, Ni2+ and Fe2+) in 20 mM KPB pH 8.0 at 70°C. The enzymatic activity was assayed with 0.6, 3, and 6 mM of each metal ions. The relative percentage activity was calculated on the basis of the measured activity in the highest activity of metal ion.21
Cloning and expression of recombinant GhPLK
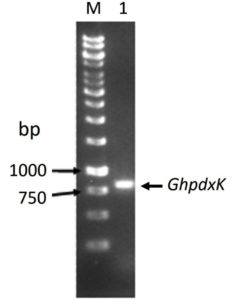
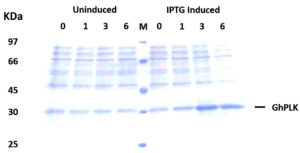
The PCR product of pdxK gene from Geobacillus sp.H6a was 810 base pairs (Fig. 1). Its open reading frame (ORF) encoded 269 amino acids with a predicted molecular weight of 29.06 kDa and theoretical isoelectric point of 5.56.17 The expression of rGhPLK was not found in an uninduced crude extract. The low expression of rGhPLK was found upon induction with 1 mM IPTG at 37°C. In order to increase the rGhPLK expression level in crude extract, different induction temperatures of 25 and 30°C, and IPTG concentrations of 0 – 1 mM, and induction time (1, 3 and 6 h) were examined. The result showed that using 0.5 mM IPTG with an induction time of 6 h at 30°C was the optimum condition to increase activity of crude rGhPLK (Fig. 2).
Fig. 1. Electrophoresis of the PCR product of pdxK gene from Geobacillus sp.H6a. Lane M is 1 kb DNA Ladder marker (Thermo Fisher Scientific); Lane 1 is GhpdxK gene.
Fig. 2. SDS-PAGE (12%) analysis of GhpdxK expression in E. coli BL21(DE3). Total proteins of BL21 (DE3) harboring pET-28a-GhpdxK induced by 0.5 mM IPTG at 30°C for 0, 1, 3 and 6 h and uninduced fractions at different time point were included as a control.
Purification of recombinant GhPLK
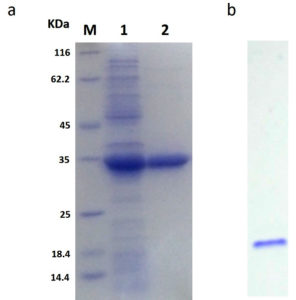
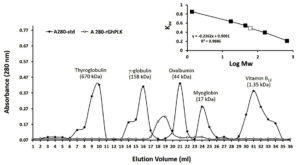
The rGhPLK contained N-terminal hexa-histidine tag sequence, was purified from crude lysates by using Ni NTA column. The profile of purification was shown in Table 1. In Fig. 3a, SDS-PAGE showed single band of approximately 32 kDa in molecular weight. Molecular weight of single subunit of rGhPLK was similar to PLK reported from other organisms: 35 kDa from E. coli,22 37 kDa from T. brucei,23 35 kDa from A. thaliana,8 and 33.9 kDa from B. mori,13 The rGhPLK showed single band on Native PAGE (Fig. 3b), while gel filtration showed single peak of rGhPLK with molecular weight calculated approximately of 60 kDa (Fig. 4). These data suggested that the rGhPLK was a dimer with two identical subunits, which was the same as other PLKs.5,8,13,15,22,23
Table (1):
Purification of recombinant GhPLK produced by E. coli BL21 (DE3).
Fraction |
Total Protein (mg) |
Total activity (mmol/min) |
Specific Activity (mmol/min/mg) |
Purification (fold) |
Yield (%) |
|---|---|---|---|---|---|
Crude extract |
378.9 |
3276.9 |
8.65 |
1.00 |
100 |
Ni-NTA affinity |
39.48 |
498.4 |
12.62 |
1.46 |
15.21 |
Fig. 3. Analysis of the purification of rGhPLK by SDS-PAGE (12%) and Native PAGE (6%). (a) Lane M is protein molecular weight marker (Thermo Fisher Scientific); Lane 1 is crude extract of E. coli BL21 (DE3) harboring pET-28a-GhpdxK; Lane 2 is purified rGhPLK. (b) Native PAGE of purified rGhPLK.
Fig. 4. Gel filtration chromatography of recombinant GhPLK. Inset, Kav of peaks of standard (black square) and peak rGhPLK (white square) in logarithmic scale.
Effects of pH and temperature
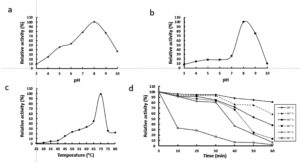
The activity of rGhPLK was found in a pH range of 4 – 10. It gradually increased at a pH range of 3 – 8 and showed the highest activity at pH 8.0 (Fig. 5a). The rGhPLK was found to be active in the broad pH range when compared with PLK from A. thaliana,8 S. typhimurium15 and B. mori13 which had activities in the narrow pH range of 5.5 – 7.5, 6.2 – 6.6 and 5.5 – 6.0, respectively. The stability of rGhPLK between pH 3 and 10 was showed in Fig. 5b. The rGhPLK was stable (>70% activity) between pH 8 and 9, and its activity extremely lost at a pH range of 3 – 7 after incubation for 1 h at 30°C.
Fig. 5. Effects of temperature and pH on activity and stability of the recombinant GhPLK. (a) pH optimum (b) pH stability (c)Temperature optimum (d) Thermal stability.
The rGhPLK has an optimal temperature at 70°C (Fig. 5c). For temperature stability, the enzyme activity was decreased when the time was increased (Fig. 5d). It retained >50 % activity after storage at 30°C – 70°C for 30 min and at 30°C – 40°C for 60 min. The loss of activity at 30 – 70°C is somewhat strikingly because G. H6a could grow at a range of temperature of 30 – 70°C and the optimal growth temperature is 60°C.16 It is possible that the enzyme is not in the desire environment as it is present in a cell. Recombinant expression might affect folding of enzyme structure and influence the stability of enzyme.
Effects of metal ions
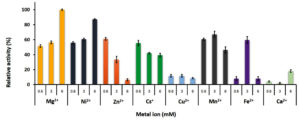
Both mono and divalent cations are involved the function of PLK. Fig. 6 showed the effect of metal ions on the rGhPLK activity using three concentrations of 0.6, 3 and 6 mM. The highest activity was found when used 6 mM of Mg2+. The activity of rGhPLK was increased when increased the concentrations of Mg2+ and Ni2+ from 0.6 mM to 6 mM. While Cs+, Zn2+ and Cu2+ showed decrease of the activity when increasing concentration of the metals. Mn2+ and Fe2+ had positive effect on the enzyme activity at 3 mM concentration. While Ca2+ had little effect to the activity of rGhPLK. Navarro et al.24 observed that the effect of metal ion on the activity of PLK is highly dependent on the relative concentrations of metal and nucleotide used. Our results obviously showed this effect when Fe2+ was at equimolar concentrations with the ATP concentration (3 mM). However, the effect of Fe2+ on the enzyme activity at 3 mM still less than that of Mg2+ at 6 mM concentration. Mg2+ has found to be the preferred divalent ion in PLK from S. typhimurium15 and E. coli9 under physiological conditions in cells. While PLK from other organisms prefers Zn2+ such as B. mori,13 A. thaliana8 and human.24
Fig. 6. Effects of metal ions on the activity of recombinant GhPLK. The assay mixture contained 20 mM KPB at pH 8, 0.4 mM PL, 3 mM ATP and 0.3 µg of rGhPLK. Assays were performed in triplicate.
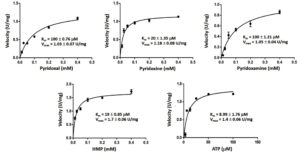
Kinetic of recombinant GhpdxK
The Km and Vmax values were examined by incubating the rGhPLK with varying concentrations of substrates, which explained by Michaelis-Menten plots (Fig. 7). Among vitamin B6 substrates (PL, PN and PM), rGhPLK has higher affinity toward PN (20 µM), while it showed the same affinity (100 µM) to PL and PM. These data show the affinity to vitamin B6 substrates similar with PLK from human (PN, 20 µM; PL, 30 µM; PM, 35 µM) and E. coli9 (PN, 25 µM; PL, 100 µM; PM, 30 µM). However, the Km value for PL of rGhPLK is about 2 times lower than that of the enzyme of B. subtilis10 (46.6 µM), B. mori13 (44.1 µM) and S. typhimurium15 (44 µM). The Km value of rGhPLK for ATP was 8.99 µM, which is about 10 times higher than the Km value reported for S. typhimurium15 (100 µM) and P. aeruginosa5 (102 µM). The Km value for HMP (19 µM) was higher than those determined for the B. subtilis10 (2,030 µM) and S. aureus25 (1,988 µM).
This is the first report describing the characterization of PLK from thermophilic bacterium. The rGhPLK is a dimer with two identical subunits (32 kDa). The enzyme can phosphorylate all B6 vitamers (PN, PM, PL) and HMP. The preferred substrate is PN. The rGhPLK prefers Mg2+ and also accepts other cations to the less extent. The rGhPLK had optimum temperature at 70°C, and found to be active in the broad pH range. It could be considered as a potential candidate for future application particularly bioconversion of vitamin B6. Future research will be focused on development of a more effective thermal stability and activity of this enzyme.
ACKNOWLEDGMENTS
None.
CONFLICT OF INTEREST
The authors declare that there is no conflict of interest.
AUTHORS’ CONTRIBUTION
YT designed and directed the project. JP performed the experiments. CC and YT wrote the manuscript. All authors read and approved the final manuscript for publication.
FUNDING
This work was granted by Khon Kaen University (Grant No. 540025) Research Fund, and Khon Kaen University under the Incubation Researcher Project, Khon Kaen University, Thailand.
ETHICS STATEMENT
This article does not contain any studies with human participants or animals performed by any of the authors.
AVAILABILITY OF DATA
All datasets generated or analyzed during this study are included in the manuscript.
- Shtyrlin YG, Petukhov AS, Strelnik AD, et al. Chemistry of Pyridoxine in Drug Design. Russ Chem Bull. 2019;68(5):911-945.
Crossref - di Salvo ML, Contestabile R, Safo MK. Vitamin B6 Salvage Enzymes: Mechanism, Structure and Regulation. Biochim Biophys Acta BBA – Proteins Proteomics. 2011;1814(11):1597-1608.
Crossref - Ramos RJ, Albersen M, Vringer E, et al. Discovery of Pyridoxal Reductase Activity as part of Human Vitamin B6 Metabolism. Biochim Biophys Acta BBA – Gen Subj. 2019;1863(6):1088-1097.
Crossref - Cea PA, Araya G, Vallejos G, et al. Characterization of Hydroxymethylpyrimidine Phosphate Kinase from Mesophilic and Thermophilic Bacteria and Structural Insights into their Differential Thermal Stability. Arch Biochem Biophys. 2020;688:108389.
Crossref - Kim MI, Hong M. Crystal Structure and Catalytic Mechanism of Pyridoxal Kinase from Pseudomonas aeruginosa. Biochem Biophys Res Commun. 2016;478(1):300-306.
Crossref - Shi R, Zhang J, Jiang C, Huang L. Bombyx mori Pyridoxal Kinase cDNA Cloning and Enzymatic Characterization. J Genet Genomics. 2007;34(8):683-690.
Crossref - Oliveira-da-Silva JA, Machado AS, Ramos FF, et al. Evaluation of Leishmania infantum Pyridoxal Kinase Protein for the Diagnosis of Human and Canine Visceral Leishmaniasis. Immunol Lett. 2020;220:11-20.
Crossref - Lum HK, Kwok F, Lo S. Cloning and Characterization of Arabidopsis thaliana Pyridoxal Kinase. Planta. 2002;215(5):870-879.
Crossref - di Salvo ML, Hunt S, Schirch V. Expression, Purification, and Kinetic Constants for Human and Escherichia coli Pyridoxal Kinases. Protein Expr Purif. 2004;36(2):300-306.
Crossref - Park JH, Burns K, Kinsland C, Begley TP. Characterization of Two Kinases Involved in Thiamine Pyrophosphate and Pyridoxal Phosphate Biosynthesis in Bacillus subtilis: 4-Amino-5-Hydroxymethyl-2-Methylpyrimidine Kinase and Pyridoxal Kinase. J Bacteriol. 2004;186(5):1571-1573.
Crossref - Wrenger C, Eschbach ML, Muller IB, Warnecke D, Walter RD. Analysis of the Vitamin B6 Biosynthesis Pathway in the Human Malaria Parasite Plasmodium falciparum. J Biol Chem. 2005;280(7):5242-5248.
Crossref - Safo MK, Musayev FN, di Salvo ML, Hunt S, Claude JB, Schirch V. Crystal Structure of Pyridoxal Kinase from the Escherichia coli pdxK Gene: Implications for the Classification of Pyridoxal Kinases. J Bacteriol. 2006;188(12):4542-4552.
Crossref - Huang SH, Ma W, Zhang PP, Zhang JY, Xie YF, Huang LQ. Recombinant Expression, Purification and Characterization of Bombyx mori (Lepidoptera: Bombycidae) Pyridoxal Kinase. Eur J Entomol. 2011;108(1):25-34.
Crossref - Jones DC, Alphey MS, Wyllie S, Fairlamb AH. Chemical, Genetic and Structural Assessment of Pyridoxal Kinase as a Drug Target in the African Trypanosome. Mol Microbiol. 2012;86(1):51-64.
Crossref - Deka G, Kalyani JN, Jahangir FB, Sabharwal P, Savithri HS, Murthy MRN. Structural and Functional Studies on Salmonella typhimurium Pyridoxal Kinase: the First Structural Evidence for the Formation of Schiff Base with the Substrate. FEBS J. 2019;286(18):3684-3700.
Crossref - Anutrakunchai C, Niamsanit S, P. Wangsomnuk P, Trongpanich Y. Isolation and Characterization of Vitamin B6-Producing Thermophilic Bacterium, Geobacillus sp. H6a. J Gen Appl Microbiol. 2010;56(3):273-279.
Crossref - Waithayawanthiti C, Anutakunchai C, Trongpanich Y. Bioinformatics and Expression Analysis of Pyridoxal Kinase in Vitamin B-6-Producing Thermophilic Bacterium, Geobacillus sp. H6a. KKU Sci J. 2020;48(3):307-317.
- Bradford MM. A Rapid and Sensitive Method for the Quantitation of Microgram Quantities of Protein Utilizing the Principle of Protein-Dye Binding. Anal Biochem. 1976;72(1-2):248-254.
Crossref - Wada H, Snell EE. The Enzymatic Oxidation of Pyridoxine and Pyridoxamine Phosphates. J Biol Chem. 1961;236(7):2089-2095.
Crossref - Manns JM. SDS-Polyacrylamide Gel Electrophoresis (SDS-PAGE) of Proteins. Curr Protoc Microbiol. 2011;22(1):A.3M.1-A.3M.13.
Crossref - Sinthusiri A, Champasri C, Trongpanich Y. Recombinant Expression Purification and Characterization of Pyridoxal 5ʹ-phosphate Synthase from Geobacillus sp. H6a Thermophilic Bacterium Producing Extracellular Vitamin B6. Iran J Biotechnol. 2021;19(4):70-82.
Crossref - White RS, Dempsey WB. Purification and Properties of Vitamin B6 Kinase from Escherichia coli B. Biochemistry. 1970;9(21):4057-4064.
Crossref - Scott TC, Phillips MA. Characterization of Trypanosoma brucei Pyridoxal Kinase: Purification, Gene Isolation and Expression in Escherichia coli. Mol Biochem Parasitol. 1997;88(1-2):1-11.
Crossref - Navarro F, Ramirez-Sarmiento CA, Guixe V. Catalytic and Regulatory Roles of Species Involved in Metal-Nucleotide Equilibriums in Human Pyridoxal Kinase. Bio Metals. 2013;26(5):805-812.
Crossref - Nodwell MB, Koch MF, Alte F, Schneider S, Sieber SA. A Subfamily of Bacterial Ribokinases Utilizes a Hemithioacetal for Pyridoxal Phosphate Salvage. J Am Chem Soc. 2014;136(13):4992-4999.
Crossref
© The Author(s) 2022. Open Access. This article is distributed under the terms of the Creative Commons Attribution 4.0 International License which permits unrestricted use, sharing, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.