ISSN: 0973-7510
E-ISSN: 2581-690X
To increase the expression of a native/foreign plant/bacterial gene, the complete network of cis-elements must be excavated to increase its biosynthetic yield, especially under industrial stress conditions. For selecting the best set of cis-elements for a foreign gene and aiding the workflow of researchers, often untrained in bioinformatics methodologies, we developed a modular PERL script for their identification and localization. The script is functional on any operating system. It localizes the cis element network of a gene. It aids an easy customization, as per the required analysis, and provides robust strategy, unlike the usually used databases where several applied calculations often become a tricky task. The script allows an uncomplicated analysis of multiplicity of cis elements along with their relative distances, making it easier for designing the more beneficial network of genes for directed evolution experiments. Through a batched scrutiny of several functionally similar genes, it would aid an easy extraction of their evolutionarily favored network of cis elements. It would be extremely helpful to develop the crop plants that are better adapted to the stressful conditions.
Cis element, transcription factor, directed evolution, promoter engineering
Plants and prokaryotic cells have been extensively deployed for the biosynthesis of several molecules including the antibodies, hormones, enzymes, and vaccines.1-6 An increased scale of bioproduction and bioaccumulation in the endoplasmic reticulum of plant cells lead to a substantially higher production of industrially important foreign genes, unlike the microorganisms or mammalian cells.7,8 Generally, in plants when a stress condition arises, the transcription factors play a key role at molecular level and bind to their recognition sequences, upstream to the stress responsive genes. For a highly versatile biosynthetic production of a broad spectrum of biomolecules, their industrial application has been booming up in the last two decades.9,10 However, the industrial stress conditions often leads to the improper growth in plants, and thus prove to be the major factor that decreases the overall yield.11 Natural adaptation of plants is time consuming and often fails to yield a gain-of-function mutation in terms of an enzymatic yield or turnover. Although both bacterial and plant systems have been extensively used for the industrial production of several biomolecules, the prokaryotic systems have been preferred over plants for their short doubling time and ease of batch scale-up, especially when proteins are biologically active in native forms, with no requirement of additional posttranslational modifications.4,6
Although the transcriptional rate is preliminarily aided by the strength of promoter and growth stage of a cell and cis- and trans-acting factors,12 the cis elements are found to be the key regulatory switches, acting as binding sites for one or more trans-acting factors.13,14 A trans-acting factor usually binds several different genes and controls their temporal and spatial expression patterns.15,16 The cis-acting regulatory segments are present in the introns and 5′ and 3′ untranslated segments, or coding regions of precursor RNAs and mature mRNAs, and are specifically recognized by atleast one trans-acting factor for selectively regulating the posttranscriptional gene expression.17 The cis elements are found to play a major role in the post-transcription and post-translation of prokaryotic systems.
As cis elements and length of spacers between them predominantly modulate the expression of a gene under varied levels of biotic stresses, a set of continuous/discontinuous set of cis elements often regulate the expression of a gene,18-20 and hence the cis elements must be extensively excavated.18-20 Besides the variant copy number and different sequence, the relative localization and mutual distance network of the cis elements, cognate to the gene, are highly variable, and it leads to a substantial change in the yield of the encoded protein.21 These segments are functionally diverse, and have multiple repeats in a gene,22 and are usually analyzed through sequence information of the conserved transcription factor binding sites and their unique organization within a gene. Due to differences in the set of cis elements, a tissue-specific gene expression profile has been observed at different developmental phases.23
Directed evolution strategy has been extensively used for many applications including the functional improvement of several plant/bacterial genes for producing vaccine or pharmaceuticals,24,25 strain improvement,26,27 and building variants with improved activity.28,29 The cis elements often function as an insulator, silencer, enhancer, and promoter in plants,17 gram-positive bacteria,30 thermophilic archaea,31 and photosynthetic bacteria.32 Although the synthetic plant/bacterial gene cassettes, with a customized minimal set of most productive cis elements, can be constructed and experimentally modulated under varied stress conditions to increase their expression level,33 their unambiguous identification is highly difficult, expensive and painstaking. Hence, the researchers often produce the functionally improved gene copies through the directed evolution algorithms without screening and optimizing the best promoter(s) sequences, applicable for a gene.
As the biologically favored cis element network would increase the expression level of cloned genes, promoter engineering should also be considered as the alternative strategy to map the best set of cis elements in the known set of functionally similar and evolutionarily closer genes. To simplify the computational methodology of extracting the naturally encoded set of cis elements in a large dataset of naturally available alternative gene copies, and to subsequently extract the most frequent set of cis elements and design its alternative copies, a PERL script is hereby developed to efficiently span the available sequence space, available in the genome databases. It would allow us to easily construct the functionally active protein variant with most active set of cis elements and would be significantly useful in reliably pacing up the computational promoter design methodologies.
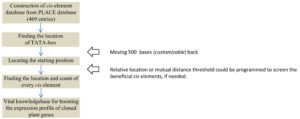
Besides the location and mutual distances, the number of repeats or multiplicity of cis elements in a gene is of prime interest for the selected gene to predict its expression level based on the strength of promoter. Although the cis elements are the significantly conserved motif segments, it is usually observed that the non-customizable and ill-programmed interface of the current servers do not allow batch scrutiny of the required scores for the input genes in a user-friendly manner. Moreover, the researchers are unable to customize the strength of the cis element database to restrict their search for only a few required entries. To resolve this issue, the most updated dataset of 469 cis elements (Supplementary Table 1) were retrieved from the PLACE database version 30.034 for define the source set in the in-house PERL script (Publicly available at Github; https://github.com/ashishr123/Cis-element-finding-script-and-dataset). The strategy, represented as a flowchart (Fig. 1) is used for an exemplary gene sequence AB022891.1, encoding the glucose-1-phosphate adenylyltransferase protein in Arabidopsis thaliana, to map the network of cis elements.
For the illustration, the glucose-1-phosphate adenylyl transferase gene AB022891.1 of Arabidopsis thaliana was selected. The programmed script identifies the cis elements and their multiplicity for the mentioned gene (Fig. 2). For AB022891.1, and a set of 21 cis elements, viz. SBOXATRBCS, WBOXATNPR1, OSE2ROOTNODULE, CURECORECR, GTGANTG10, ROOTMOTIFTAPOX1, CCAATBOX1, WBOXHVISO1, ACGTATERD1, MYB1LEPR, PYRIMIDINEBOXOSRAMY1A, ACGTABOX, WRKY71OS, CDA1ATCAB2, BIHD1OS, SURECOREATSULTR11, POLLEN1LELAT52, ARFAT, CAATBOX1, -10PEHVPSBD and DOFCOREZM, are found. While the first sixteen elements are orderly repeated only 1 time in the gene, the multiplicity of last element and the other four is found to be 3 and 2 respectively.
In comparison to the normally deployed servers like PLACE28 or PreCisIon,35 the programmed script allows the construction of a batched pipeline for several genes. The study would be useful to analyze the evolutionary relationship among the genes in terms of the type and multiplicity of different cis-elements. Moreover, the study will be useful to construct the most productive array of these elements, conserved across the several functionally similar genes, in a combinatorial cis-element based synthetic promoter, simplifying the previous methodologies.14-16 It will be extremely useful to design a reliable strategy for the construction of a more productive gene expression cassette, as has recently been used to generate a highly productive arrangement of cis-elements for an enhanced gene expression.36 The strategy has already been manually deployed in several recent articles,37,38 although such a modular script has not been published so far and its utility would be an added boost for improving the expression profile of a plant gene to attain an improved productivity under the natural/industrial stress environment. Subsequent engineering of gene sequence across its active site will then further boost its stability, turnover number, productivity, and lastly an increased crop-yield.
The discovery of the cis elements network in the promoter regions will aid an easy annotation of the genes encoding the putative transcription factors. The developed script locates TATA-box and every cis element within the customized boundary domain, which will assist to construct the batched pipelines for exploring the naturally encoded elements in the homologous set of genes, to figure out the evolutionarily favored set of cis elements under a specific environmental constraint. Computation of multiplicity and analysis of evolutionary relationship among the genes can thus be achieved through this simplified methodology. The fact that the script is based on previous experimental evidence, the prediction should be less prone to in-accuracies but the same also needs to be validated using the updated methodologies in-vitro. It will prove to be a handy tool to improve the expression levels of foreign genes under varied growth conditions or stress parameters and will be very handy in modulating the genetic expression.
Additional file: Additional Table S1.
ACKNOWLEDGMENTS
The authors would like to thank their universities for all the support provided throughout the study.
CONFLICT OF INTEREST
The authors declare that there is no conflict of interest.
AUTHORS’ CONTRIBUTION
AR, HV and VSR performed the experiments. DP, WS, NNVS and PH analyzed the data. HV and AR wrote the manuscript. All authors read and approved the final manuscript for publication.
FUNDING
None.
ETHICS STATEMENT
Not applicable.
AVAILABILITY OF DATA
All datasets generated or analyzed during this study are included in the manuscript and in the supplementary files.
- Madanala R, Gupta V, Pandey AK, et al. Tobacco Chloroplasts as Bioreactors for the Production of Recombinant Superoxide Dismutase in Plants, an Industrially Useful Enzyme. Plant Molecular Biology Reporter. 2015;33(4):1107-1115.
Crossref - Shaaltiel Y, Bartfeld D, Hashmueli S, et al. Production of glucocerebrosidase with terminal mannose glycans for enzyme replacement therapy of Gaucher’s disease using a plant cell system. Plant Biotechnol J. 2007;5(5):579-590.
Crossref - Sirko A, Vanek T, Gora-Sochacka A, Redkiewicz P. Recombinant cytokines from plants. Int J Mol Sci. 2011;12(6):3536-3552.
Crossref - Spadiut O, Capone S, Krainer F, Glieder A, Herwig C. Microbials for the production of monoclonal antibodies and antibody fragments. Trends Biotechnol. 2014;32(1):54-60.
Crossref - Perez-Perez DA, Pioquinto-Avila E, Arredondo-Espinoza E, Morones-Ramirez JR, Balderas-Renteria I, Zarate X. Engineered small metal-binding protein tag improves the production of recombinant human growth hormone in the periplasm of Escherichia coli. FEBS Open Bio. 2020;10(4):546-551.
Crossref - Rezaei M, Zarkesh-Esfahami SH. Optimization of production of recombinant human growth hormone in Escherichia coli. J Res Med Sci. 2012;17(7):681-685.
- Lopez J, Lencina F, Petruccelli S, Marconi P, Alvarez MA. Influence of the KDEL signal, DMSO and mannitol on the production of the recombinant antibody 14D9 by long-term Nicotiana tabacum cell suspension culture. Plant Cell Tiss Organ Cult. 2010;103(3):307-314.
Crossref - Martinez CA, Giulietti AM, Rodriguez Talou RJ. Expression of a KDEL-tagged dengue virus protein in cell suspension cultures of Nicotiana tabacum and Morinda citrifolia. Plant Cell Tiss Organ Cult. 2011;107:91-100.
Crossref - Eibl R, Meier P, Stutz I, Schildberger D, Huhn T, Eibl D. Plant cell culture technology in the cosmetics and food industries: current state and future trends. Appl Microbiol Biotechnol. 2018;102(20):8661-8675.
Crossref - Fowler MW. Plant-Cell Culture: Natural Products and Industrial Application. Biotechnol Genet Eng Rev. 1984;2(1):41-67.
Crossref - Feher A. Somatic embryogenesis – Stress-induced remodeling of plant cell fate. Biochim Biophys Acta. 2015;1849:385-402.
Crossref - Li WJ, Dai LL, Chai ZJ, Yin ZJ, Qu LQ. Evaluation of seed storage protein gene 3′-untranslated regions in enhancing gene expression in transgenic rice seed. Transgenic Research. 2012;21(3):545-553.
Crossref - Peremarti A, Twyman RM, Gomez-Galera S, et al. Promoter diversity in multigene transformation. Plant Mol Biol. 2010;73(4-5):363-378.
Crossref - Porto MS, Pinheiro MPN, Batista VGL, dos Santos RC, Filho Pde AM, de Lima LM. Plant promoters: an approach of structure and function. Mol Biotechnol. 2014;56(1):38-49.
Crossref - Wang D, Pan Y, Zhao X, Zhu L, Fu B, Li Z. Genome-wide temporal-spatial gene expression profiling of drought responsiveness in rice. BMC Genomics. 2011;12:149.
Crossref - Dutt M, Dhekney SA, Soriano L, Kandel R, Grosser JW. Temporal and spatial control of gene expression in horticultural crops. Hortic Res. 2014;1:14047.
Crossref - Yamaguchi-Shinozaki K, Shinozaki K. Organization of cis-acting regulatory elements in osmotic- and cold-stress-responsive promoters. Trends Plant Sci. 2005;10(2):88-94.
Crossref - Singh KB. Transcriptional Regulation in Plants: The Importance of Combinatorial Control. Plant Physiology. 1998;118(4):1111-1120.
Crossref - Wolberger C. Combinatorial transcription factors. Curr Opin Genet Dev. 1998;8(5):552-559.
Crossref - Remenyi A, Scholer HR, Wilmanns M. Combinatorial control of gene expression. Nat Struct Mol Biol. 2004;11(9):812-815.
Crossref - Venter M, Botha FC. Synthetic Promoter Engineering. In Pua EC, Davey MR (eds.) Plant Developmental Biology – Biotechnological Perspectives, Springer. 2010:393-414.
Crossref - Sharma N, Russell SD, Bhalla PL, Singh MB. Putative cis-regulatory elements in genes highly expressed in rice sperm cells. BMC Res Notes. 2011;4:319.
Crossref - Chavez-Barcenas AT, Valdez-Alarco’n JJ, Martınez-Trujillo M, et al. Tissue-Specific and Developmental Pattern of Expression of the Rice sps1 gene. Plant Physiology. 2000;124(2):641-654.
Crossref - Chartrain M, Peter MS, David KR, Barry CB. Metabolic engineering and directed evolution for the production of pharmaceuticals. Curr Opin Biotechnol. 2000;11(2):209-214.
Crossref - Whalen RG, Kaiwar R, Soong NW, Punnonen J. DNA Shuffling and Vaccines. Curr Opi Mol Ther. 2001;3(1):31-36.
- Lassnner M, Bedbrook J. Directed molecular evolution in plant improvement. Curr Opi Plant Biol. 2001;4(2):152-156.
Crossref - Linda AC, Daniel LS, Rebecca G, et al. Discovery and directed evolution of a glyphosate tolerance gene. Science. 2004; 304(5674):1151-1154.
Crossref - Rajendra RC, Reddivari M, Roger M, Poonam N. Improving the quality of industrially important enzymes by directed evolution. Mol Cell Biochem. 2001;224(1-2):159-168.
Crossref - Tian Y-S, Peng R-H, Xu J, et al. Mutations in two amino acids in phyI1s from Aspergillus niger 113 improve its phytase activity. World J Microbiol Biotechnol. 2010;26(5):903-907.
Crossref - Hueck CJ, Hillen W, Saier MH Jr. Analysis of a cis-active sequence mediating catabolite repression in Gram-positive bacteria. Res Microbiol. 1994;145(7):503-518.
Crossref - Condo I, Ciammaruconi A, Benelli D, Ruggero D, Londei P. Cis-acting signals controlling translational initiation in the thermophilic archaeon Sulfolobus solfataricus. Mol Microbiol. 1999;34(2):377-384.
Crossref - Asayama M. Regulatory System for Light-Responsive Gene Expression in Photosynthesizing Bacteria: Cis-Elements and Trans-Acting Factors in Transcription and Post-Transcription. Biosci Biotechnol Biochem. 2006;70(3):565-573.
Crossref - Bilas R, Szafran K, Hnatuszko-Konka K, Kononowicz AK. Cis-regulatory elements used to control gene expression in plants. Plant Cell Tiss Organ Cult. 2016;127:269-287.
Crossref - Kenichi H, Yoshihiro U, Masao I, Tomoko K. Plant cis-acting regulatory DNA elements (PLACE) database: 1999. Nucleic Acids Res. 1999;27(1):297-300.
Crossref - Elati M, Nicolle R, Junier I, et al. PreCisIon: PREdiction of CIS-regulatory elements improved by gene’s position. Nucleic Acids Res. 2013;41(3):1406-1415.
Crossref - Kiesenhofer DP, Mach RL, Mach-Aigner AR. Influence of cis Element Arrangement on Promoter Strength in Trichoderma reesei. Appl Environ Microbiol. 2018;84(1):e01742-17.
Crossref - Ali S, Kim WC. A Fruitful Decade Using Synthetic Promoters in the Improvement of Transgenic Plants. Front Plant Sci. 2019;10:1433.
Crossref - Yang Y, Lee JH, Poindexter MR, et al. Rational design and testing of abiotic stress-inducible synthetic promoters from poplar cis-regulatory elements. Plant Biotechnol J. 2021;19(7):1354-1369.
Crossref
© The Author(s) 2022. Open Access. This article is distributed under the terms of the Creative Commons Attribution 4.0 International License which permits unrestricted use, sharing, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.