ISSN: 0973-7510
E-ISSN: 2581-690X
Chromium (VI) is a well-known pollutant that is present in industrially polluted soil and water, and has been reported to be mutagenic and carcinogenic. In the present study, we investigated the effective use of Leptolyngbya boryana (cyanobacterium) as an eco-friendly option to overcome Cr (VI) toxicity in tannery effluents. The main objective of this study was to identify the Cr reductase (ChrR) gene and its variability in the context of Cr (VI) stress. Industrial polluted soil samples were collected and processed according to standard protocols for ChrR variation and 16S rDNA gene analysis. Genomic DNA was isolated from the collected samples and the ChrR and 16S rDNA genes were amplified by PCR. Amplified 16S rDNA was sequenced and aligned with known sequences. In the present study, a strong correlation was established between the nucleotide sequences of the ChrR and 16S rDNA genes. The Minimum Inhibitory Concentration (MIC) was determined for Cr (VI), and pure strains of L. boryana were identified and isolated from soil samples. Cr (VI)-stressed conditions and their genetic variability were confirmed by sequencing. In conclusion, the L. boryana strain has been identified an eco-friendly option for overcoming Cr (VI) toxicity in tannery effluents.
Bioremediation, ChrR gene, Leptolyngbya Boryana, Genetic Variability, Chromium Reductase, Homology
Waste produced by different industries presents a big crisis, with the main challenge being to convert waste into eco-friendly compounds through sustained approaches. Many industrial wastes (effluents) consist of various toxic materials including metals, harmful volatile compounds, and large organic and inorganic residues. Due to developmental requirements, new strategies and novel sustained approaches are needed to enhance industrial waste management because of increasing urbanization.1 Several sources can be attribute to the presence of long-term industrial effluents in the atmosphere, and many infectious diseases, neurological disorders, metabolic abnormalities, and cancer have been recorded in recent years.2 Due to the liberation of these toxic materials, we have faced significant losses and challenges in all domains of society.3
Various studies have reported the presence of many toxic pollutants in industrial effluents, such as chromium, sulfides of metals, phenolic compounds, magnesium, sodium, potassium, and mercury.4 Chromium, an important toxic compound, is also an important micronutrient that is required for the growth of many microorganisms.5 Chromium in high concentrations is toxic in all ecosystems, i.e., air, water, and soil. Naturally, soil can have a chromium concentration in the range of 10–50 mg/kg.6 In a study, Indian tannery industries alone produced more than 2000-3000 tons of chromium in the environment with high chromium concentrations of more than 2000-5000 mg/L. Although, it has been found a safe recommended acceptable discharge limit is less than 2 mg/L.7
Many microbes, specifically blue-green algae, have been used in biological systems to reduce (biotransformation) Cr (VI) into Cr (III). Moreover, several studies have identified chromium-reducing microorganisms. Additionally, few fungi have also been studied for their Cr (VI) reducing properties.3,8 These properties of microbes to survive Cr (VI) metal exposure and regulate the detoxification mechanism into Cr (III) are used to rectify globally. Specific microbes have specific metal-tolerance capacities under the optimum environmental conditions. Plasmid-mediated removal of xenobiotics has been described by Bhatt et al.9 Some mechanisms of Cr (VI) reduction have been reported, e.g. segregation by a permeability barrier, active transport efflux pumps, intra-and extracellular appropriation, enzymatic methods, among this study.10 A bacterial species that can reduce toxic Cr (VI) to nontoxic Cr (III) has been reported previously.8,11 Similarly, a fungal species that has Cr (VI) bio-absorptive property has been reported. During biosorption, Cr (VI) is bound to the functional groups present on the surface of microbes and percolated inside.12 Several studies have reported on the significance of biofilm-mediated bioremediation, which has been used as a powerful tool for the removal of environmental pollutants.13
In nature, various species of cyanobacteria live on soil surfaces and might be morphologically and phylogenetically different.14 Leptolyngbya, a filamentous cyanobacterium, is characterized by the width of its cylindrical trichomes. Leptolyngbya spp. have been isolated from various industrial effluents in soil. Leptolyngbya boryana is phylogenetically coupled to Leptolyngbya sp.15 Although nitrogen fixation by L. boryana is well characterized, the ability to reduce Cr (VI) to nontoxic Cr (III) genomic analysis has not been reported previously. Recently, biosurfactants have been found to be powerful tools for bioremediation of heavy metals from contaminated soil.16
In our previous study, we reported the successful isolation and characterization of L. boryana from tannery soils.17 In the present study, we aimed to characterize the Cr reductase (ChrR) gene and investigate its variability in the context of Cr (VI) stress. The findings of this study may provide a more suitable, effective, eco-friendly, sustainable, and cost-effective biological treatment of wastewater from the leather industry. Therefore, in this study, we focused on the potential role of L. boryana in the biotransformation of Cr (VI) to Cr (III). This study is novel in the context of the bacterial species studied and the methodologies used for the identification and characterization of Cr (VI) toxicity in wastewater/soil.
Chemicals
All chemicals and reagents were of analytical grade and procured from Merck (Rahway, NJ, USA), HiMedia (Mumbai, India), and Qiagen (Hilden, Germany). A stock solution (1000 mg/L) of Cr (VI) was prepared using K2Cr2O7 in deionized water.
Collection of samples
Industrial effluent samples were collected from nearby industries in Kanpur (India) (26°26’59.7228″N 80°19’54.7356″E). Out of 100 samples collected from different places, L. boryana (ATCC-27894) was identified in 10 samples. We previously characterized these strains morphologically and biochemically.17 Pathogenic and antibiotic-treated L. boryana strains were excluded from this study.
Cyanobacteria and culture conditions
The screened L. boryana strains were grown in Erlenmeyer flasks containing liquid BG-II media under the following growth conditions: 16:8 light:dark cycle, 30 ± 2°C; and 6 irradiances of 3000–4000 lux (cool white light). Furthermore, the isolates were routinely subcultured every 20 days, as mentioned previously.17
Morphological and biochemical analysis
The morphology (colony morphology and color identification) of isolated L. boryana strains was characterized as described previously.17 For Cr reductase analysis, isolated and morphologically characterized L. boryana strains were inoculated in a large volume (5 L) of BG-II medium and maintained at 37°C for 10 days with constant agitation in at 100 rpm. L. boryana colonies were harvested by centrifugation (1000 rpm at 4°C for 30 min). Harvested colonies were lysed, the reduction of Cr (III) by the wild and stressed L. boryana strains was analyzed.18 Cr reductase activity was assessed for up to 14 days in the isolated and characterized strains. For analysis of minimum inhibitory concentration (MIC), cultures were inoculated with different concentrations (0–1000 mg/L) of chromium (VI) and tested at 7 days and 14 days.
Furthermore, biochemical analysis, including Gram’s staining, catalase activity, sucrose utility test, and indole test was performed for all strains as described previously.19
PCR amplification of Cr-responsive genes and In silico analysis
For genomic analysis, total genomic DNA was isolated from collected L. boryana cells using a Qiagen DNA isolation kit (Qiagen India Pvt. Ltd., New Delhi, India). The PCR reaction mixture (20 µL) contained 10 µL of master mix (GoTaq Master Mix, Promega, Madison, WI, USA), 1 µL each of forward and reverse primers, and 30 µg of DNA template and/or nuclease-free water. PCR was performed in a BioRad T100 Thermal Cycler (BioRad, Hercules, CA, USA) using the following cycling conditions: initial denaturation at 95°C for 5 min; followed by 35 cycles of 94°C for 30 s, 48°C for 30 s, 72°C for 1 min; and final extension at 72°C for 7 min. The amplified product was visualized by 1% agarose gel electrophoresis.
Further validation of the cyanobacterial isolate was performed using 16S rDNA sequencing. PCR analysis was performed on freshly grown culture as described by Yadav et al.17 Sequencing was performed by Chromous Biotech Pvt. Ltd. (Bengaluru, India). Briefly, the sequencing reaction (10 µL) contained 4 µL of the BigDye Terminator mix (version 3.1), 1 μL of primer (10 pmol) (Table 2), 1 μL of DNA template, and 3 μL of sterile water. Sequencing was carried out using an ABI 3500 Genetic Analyzer (Applied Biosystems, Waltham, MA, USA). A 50 cm capillary array column (capillary array; POP-7 polymer) was employed in the DNA sequencer. The analysis was performed using the protocol BDTv3-KB-Denovo-v 5.2 (Seq scape-v 5.2 software, Thermo Fisher Scientific, Waltham, MA, USA). The results were generated in ABI, PDF (Figure S4), and FASTA format, the 16S rDNA gene sequence was subjected to NCBI Blast to identify the similar sequences, and the phylogenetic tree was constructed using the MEGA5 software.20
Table (1):
Morphological, biochemical and molecular characteristics of Cr (VI) reluctant strains L. boryana, checked 100 different isolates and found 10 identical isolates.
Strain S.N. |
Morphological Appearance |
Motility |
Gram’s Straining |
Catalase test |
Indole Test |
Sucrose Utility Test |
16S rRNA test |
|---|---|---|---|---|---|---|---|
Strain (10) |
Thin filamentous Cylindrical trichomes colourless |
Solitary |
(+) ve |
(+) ve |
(+) ve |
(+) ve |
(+) ve |
Strain (90) |
Thin filamentous Cylindrical trichomes colourless |
Solitary |
(+) ve |
(+) ve |
(-) ve |
(+) ve |
(-) ve |
To check the expression of ChrR, RNA was isolated using an RNA isolation kit (Qiagen India Pvt. Ltd.), and the concentration was determined using a Nanodrop 2000 (Thermo Fisher Scientific). cDNA was synthesized from 1 µg of the isolated RNA using a high-capacity cDNA reverse transcription kit (Applied Biosystems) according to the manufacturer’s instructions. Relative gene expression was quantified using the qPCR SYBR Green PCR Master Mix (Applied Biosystems). Relative expression was calculated by a 2-ΔCt method21 using 16S rDNA as an endogenous control. The amplified product was visualized by 2% agarose gel electrophoresis. After purification, the band was subjected to gene sequencing. Primers used for 16S rDNA and ChrR genes are listed in Table 1. L. boryana strain was confirmed by 16S rDNA based on PCR results, and the sequences were aligned using Clustal W (https://www.ebi.ac.uk/Tools/msa/clustalo/). The results obtained after gene sequencing were subjected to homology analysis using the NCBI nucleotide database (https://blast.ncbi.nlm.nih.gov/Blast.cgi?PAGE_TYPE, BlastSearch).
Table (2):
Primers used in this study for amplification of 16S rDNA and ChrR genes in L. boryana.
| S.N. | Gene name | Sequences 5′→3′ | Tm (oC) | Amplicon size | Reference |
|---|---|---|---|---|---|
| 1. | 16S rDNAF | AGAGTTTGATCCTGGCTCAG | 50 | 1.5kb
|
12 |
| 2. | 16S rDNAR | TACGGTTACCTTGTTACGACTT | 50 | ||
| 3. | ChrR F | TCACGCCGGAATATAACTAC | 53 | 340bp
Used in qRT-PCR |
37 |
| 4. | ChrR R | CGTACCCTGATCAATCACTT | 54 | ||
| 5. | 16S rDNA F | CACACTGGGACTGAGACAC | 56 | 190bp
Used in qRT-PCR |
38 |
| 6. | 16SrDNAR | CTGCTGGCACGGAGTTAG | 56 |
Statistical Analysis
In the present study, MIC and dissimilar variables were assessed using the SPSS software version 22 (IBM, Armonk, NY, USA).22 Data are presented as mean ± standard deviation of three independent experiments.
Selection and identification of the cyanobacteria based on morphological and biochemical analysis
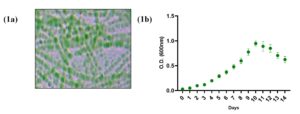
In the present study, 10 out of 100 samples collected from different locations contained only L. boryana and were subjected to biochemical, morphological, and genetic analysis. The genus Leptolyngbya is a simple filamentous cyanobacteria that exhibits slight morphological differences between species. The same bacterial strain was isolated from 10 locations (Figure 1a). The isolated L. boryana strains were observed for up to 14 days. Culture growth increased from the 6th day to the 10th day of culture and start decreasing from the 11th day of culture compared with that on the 1st day of culture (Figure 1b). Biochemical tests characterized the cultures as thin filamentous bacteria with cylindrical trichomes, gram-positive, catalase-positive, indole test positive, and sucrose test positive (Table 1).
Figure 1. (1a) Representative micrograph depicts morphological characteristics of L. boryana (10X). (1b) Graph represents the growth curve of isolated culture observed up to 14 days. Each dot represents the mean value of O.D. at 600 nm of three independent culture sets
Genetic screening of the selected strain
Genomic DNA was isolated from the morphologically and biochemically characterized L. boryana (Figure S1a). In the present study, bacterial genotypic confirmation was performed using 16S rDNA sequencing (Figure S1b). The obtained 16S rDNA sequence of L. boryana strain was homologous to sequences available in the NCBI database (Figure S1f). The strain exhibited 98% sequence identity with known sequences (Figure S1c–e).
Expression of ChrR gene in the selected strain under Cr (VI) stress
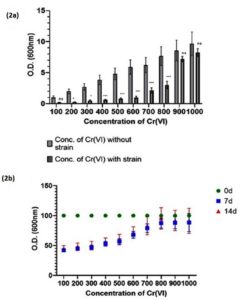
The optimized level of tolerance for Cr (VI) of the selected strain was found to be 800 mg/L of K2Cr2O7 while other isolates did not grow well at Cr (VI) concentrations above 800 mg/L (Figure 2a). Cr (VI) stress tolerance was also assessed for up to 14 days. No significant differences were observed between 7 and 14 days (Figure 2b). Furthermore, ChrR gene was isolated and amplified from L. boryana. In the present study, five isolates were confirmed to express the ChrR gene (Figure S2a) when cultured with 800 mg/L of Cr (VI), and the band size of ChrR gene was found to be approximately 340 bp (confirmed with a DNA ladder) (Figure 3a). The sequenced ChrR gene was aligned with known sequences (Figure S2b, c).
Figure 2. (2a) Histogram represents the optimum MIC of Cr (VI) mg/L in the growth culture of L. boryana. (2b) Histogram represents the Cr (VI) mg/L stress in the growth culture of L. boryana observed at 0 day, 7 days and 14 days. Values represented as mean ±SD after three independent experiments. ***p<0.001, *p<0.05, and ns=not significant
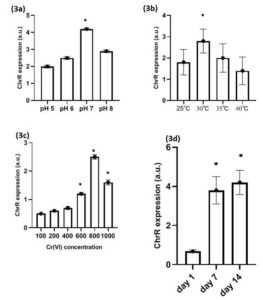
Figure 3. Expression of ChrR genes in different physiological conditions of L. boryana culture, (3a) ChrR genes expression at pH 5-8, (3b) ChrR genes expression at temp 25-40 °C (3c) ChrR genes expression at Cr (VI) concentration 100-1000mg/L, (3d) ChrR gene expression observed at 0 day, 7 days and 14 days in L. boryana. Values represented as mean ±SD after three independent experiments *p<0.05
High ChrR expression was observed in L. boryana at an optimum pH of 7 and at an optimum temperature of 37°C (Figure 3a, b). High ChrR expression was also observed in L. boryana strains when cultured with 800 mg/L of Cr (VI) (p < 0.05) and after 7 days of culture (p < 0.05); however, no significant difference was observed between 7 and 14 days (Figure 3c, d) and (Figure S3 a–d).
The present study focused on assessing the genetic variability and expression of ChrR gene in L. boryana isolated from tannery soils with respect to Cr (VI) reductase activity. Similar work has been done by Ilias et al.23 and Camargo et al.24 who reported that Cr (VI)-resistant bacteria degrade for Cr (VI) at concentrations between 500–2000 mg/L. The optimum MIC of Cr (VI) was reported to be approximately 740 mg/L, and above this concentration, the bacterium was not found to be suitable for Cr (VI) reduction. A similar finding was obtained in our study; the optimum MIC was found to be approximately 800 mg/L (Figure 2a).
Our findings are supported by the findings of Chaturvedi25 and Poornima et al.26 who reported Cr (VI) tolerance in Pseudomonas putida and found positive results for in catalase, indole, and sucrose utility tests. Similar results were also obtained in the present study; L. boryana exhibited positive tests for catalase, indole, and sucrose utility (Table 1).
There are limited genetic studies that have analyzed 16S rDNA and ChrR genes in microbes with relation to Cr (VI) reduction. In the present study, both 16S rDNA and ChrR genes were found out and showed 98% homology identity with 16S rRNA gene and 98% homology identity with ChrR gene in L. boryana under Cr (VI) stress state (Figure S1 and S2). A similar study was performed by Baldiris et al.,27 who showed the presence of ChrR gene in cyanobacteria and reduction of Cr (VI). In another study by Rocco et al., which used S. maltophilia, a crucial property for the binding of metals, such as Hg, Co, Zn, and Cd in tannery effluents was demonstrated.28 The 16S rDNA (Figure S1) and ChrR partial gene sequence homology was obtained by comparison with available sequences in NCBI databases (Figure S2). A similar finding was obtained by Rathnayake et al.,29 for Cr (VI) biotransformation in Phormidesmis molle in the presence of 16S rDNA and ChrR genes. Additionally, the findings of Sundar et al.,30 also support the presence of 16S rDNA and ChrR genes with 99% homology in Bacillus Cereus strains for Cr (VI) reduction.
In the recent past, Cr (VI)-reducing bacteria on biosolids from wastewater were isolated by Velez et al.,31 and the data generated supported the findings of our study. In the literature, a partial (268 bp) chromate reductase gene has been identified in three gram-positive bacterial isolates from soils contaminated with Cr from tannery effluents. In the present study, we obtained a 340 bp (Figure S2) partial gene sequence for L. boryana. Similarly, Denget al.,32 obtained a 321 bp (partial) Cr reductase gene in gram-positive bacteria. This confirmed the presence of the ChrR gene in the DNA of these two bacteria, reaffirming their chromium-reducing property. Thus, our findings confirm the presence of the ChR gene in L. boryana and strengthen the ability to reduce Cr (VI) to Cr (III).33 The present study is also consistent with the findings of Deshpande et al.,34 in relation to the reduction of Cr (VI) to Cr (III). In the literature, biofilm formation by xenobiotic-degrading microorganisms was reported by Bhatt et al.,35 which supported by the findings of our study in terms of Cr (VI) reduction to Cr (III).
In Cr (VI)-resistant bacteria, ChrR gene catalyzes the reduction of Cr (VI) to Cr (III) with the transfer of electrons from the electron donor NADPH to Cr (VI), resulting in the production of reactive oxygen species.36 The ChrR gene belongs to the chromate ion transport (ChrR) super-family and has been widely acknowledged in Archaea, bacteria, and Eukarya.37 The data obtained in the present study was very similar to the findings of Mishra et al.,16 in the context of Cr (VI) reduction. In the present study, the ChrR gene was obtained from genomic DNA of L. boryana (Figure S1). Similar findings have been previously reported in the literature. Some researchers have reported that the Cr (VI) resistance gene is different from that associated with Cr (VI) reduction, and the location of both genes may be different in different microbes.27 To determine the exact location of these genes, there is a need for more studies on such microbes to explore the mechanisms of Cr (VI) reduction.
This is the first study to report on 16S rDNA and ChrR genes in relation with Cr (VI) reduction in L. boryana. The ChrR gene obtained in this study exhibited 98% homology with known sequences in the NCBI database, and genetic variability has been experimental in stressed L. boryana. There is a need for the identification and characterization of the enzymes (protein encoded by ChR gene) and to determine the location of this gene in microbes. The findings of this study can be further verified with total genome sequencing methods and comparative genomic approaches to elucidate Cr (VI) reductase activity. Additionally, such studies are required to determine the strength and exact nature of gene functions to establish the role of Cr (VI) biotransformation in L. boryana. The data generated from this study may be helpful for young researchers to plan their research and help society obtain safe drinking water and ecosystems.
Additional file: Additional Figure S1-S4.
ACKNOWLEDGMENTS
The authors would like to thank Director of Rama Institute of Engineering and Technology, Department of Biotechnology, Mandhana, Kanpur, Indian Institute of Technology, Kanpur for providing necessary facilities to carry out the present study and for instrumental facility and Chromous Biotech Pvt Ltd, Bangaluru, India for 16S rDNA based bacterial identification.
CONFLICT OF INTEREST
The authors declare that there is no conflict of interest.
AUTHORS’ CONTRIBUTION
AK, SK, VD and APSY conceived and designed the experiments. APSY performed the experiments. APSY, AK, VD and SK contributed reagents, materials, analysis tools or data. VD and APSY analyzed and interpreted the data. APSY wrote the manuscript. All authors read and approved the final manuscript for publication.
FUNDING
None.
DATA AVAILABILITY
All datasets generated or analyzed during this study are included in the manuscript and/or in the supplementary files.
ETHICS STATEMENT
Not applicable.
- Evelyne JR, Ravisankar V. Bioremediation of chromium contamination-a review. Int J Res Earth Environ Sci. 2014;1(6):20-26.
- Megharaj M, Avudainayagam S, Naidu R. Toxicity of hexavalent chromium and its reduction by bacteria isolated from soil contaminated with tannery waste. Curr Microbiol. 2003;47(1):51-54.
Crossref - Deepali. Bioremediation of Chromium (VI) from Textile Industry’s Effluent and Contaminated Soil Using Pseudomonas putida, Iranica J Energy Environ. 2011;2(1):24-31.
- Yadav APS, Dwivedi V, Kumar S, Kushwaha A, Goswami L, Reddy BS. Cyanobacterial Extracellular Polymeric Substances for Heavy Metal Removal: A Mini Review. J Compos Sci. 2021;5(1):1.
Crossref - Thacker U, Madamwar D. Reduction of Toxic Chromium and Partial Localization of Chromium Reductase Activity in Bacterial Isolate DM1. World J Microbiol Biotechnol., 2005;21(6):891-899.
Crossref - Pechova A, Pavlata L. Chromium as an essential nutrient: a review. Vet Med (Praha). 2007;52(1):1-18.
Crossref - Belay AA. Impacts of Chromium from Tannery Effluent and Evaluation of Alternative Treatment Options. J Environ Prot. 2010;1(1):53-58.
Crossref - Jayalakshmi R, Ramachandra CSV. Isolation, Screening and Molecular Characterization of Chromium Reducing Cr (VI) Pseudomonas Species. J Chem Bio Phy Sci Sec. 2013;3(1):297-304.
- Bhatt P, Bhandari G, Bhatt K, et al. Plasmid-mediated catabolism for the removal of xenobiotics from the environment. J Hazard Mater. 2021;420:126618.
Crossref - Bruins S, Kapil F, Oehme W. Microbial resistance to metals in the environment. Ecotoxicol Environ Safety. 2000;45(3):198-207.
Crossref - Qian LI, Yang Z, Chai L, Wang B, Xiong S, Liao Y, Zhang S. Optimization of Cr (VI) bioremediation in contaminated soil using indigenous bacteria. J Cent South Univ. 2013;20:480-487.
Crossref - Noorjahan CM. Physicochemical Characteristics, Identification of Fungi and Biodegradation of Industrial Effluent. J Environ Earth Sci. 2014;4:32-39.
- Sandhya M, Huang Y, Li J, et al. Biofilm-mediated bioremediation is a powerful tool for the removal of environmental pollutants. Chemosphere. 2022;294:133609.
Crossref - Joo MH, Hur SH, Han YS, Kim JY. Isolation, Identification, and Characterization of Bacillus strains from the Traditional Korean Soybean-fermented Food, Chungkookjang. J Appl Biol Chem. 2007;50(4):202-210.
- Anagnostidis K, Komarek J. Modern approach to the classification system of cyanophytes. 3-Oscillatoriales. Arch Hydrobiol Algol Stud. 1988;50(53):327-472.
- Mishra S, Lin Z, Pang S, Zhang Y, Bhatt P, Chen S. Biosurfactant is a powerful tool for the bioremediation of heavy metals from contaminated soils. J Hazard Mater. 2021;418:126253.
Crossref - Yadav APS, Dwivedi V, Kumar S. Leveraging the biosorption potential of Leptolyngbya boryana for Cr (VI) from aqueous solution. Cleaner Engineering and Technology. 2021;4:100198.
Crossref - Huang J, Cao Y, Shao Q, Peng X, Guo Z. Magnetic nanocarbon adsorbents with enhanced hexavalent chromium removal: Morphology dependence of fibrillarvs. Particulate structures. Ind Eng Chem Res. 2017;56:10689-10701.
Crossref - Sundari GD, Kumar M. Simultaneous Cr (VI) reduction and phenol degradation using Stenotrophomonas sp. isolated from tannery effluent contaminated soil. Environ Sci Pollut Res. 2013;20:6563-6573.
Crossref - Kushwaha SK, Vetukuri RR, Grenville-Briggs LJ. Draft Genome Sequence of the Mycoparasitic Oomycete Pythium periplocum Strain CBS 532.74. Genome Announc. 2017b;5(12):17-57e.
Crossref - Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001.
Crossref - Corp, IBM., Released 2011, IBM SPSS Statistics for Windows, Version 20.0., Armonk, NY, IBM, Corp.
- Ilias M, Rafiqullah IM, Debnath BC, Mannan KSB, Hoq MM. Isolation and characterization of chromium (VI) reducing bacteria from tannery effluents. Indian J Microbiol. 2011;51:76-81.
Crossref - Camargo FAO, Bento FM, Okeke BC, Frankenberger WT. Chromate reduction by chromium-resistant bacteria isolated from soils contaminated with dichromate. J Environ Qual. 2003;32:1228-1233.
Crossref - Chaturvedi MK. Studies on chromate removal by chromium-resistant Bacillus sp. Isolated from tannery effluent. J Environ Protect. 2011;2(1):76-82.
Crossref - Poornima K, Karthik L, Swadhini SP, Mythili S, Sathiavelu A. Degradation of chromium by using novel strains of Pseudomonas species. J Microb Biochem Technol. 2010;2(4):95-99.
Crossref - Baldiris R, Acosta-Tapia N, Montes A, Hernandez J, Vivas-Reyes R. Reduction of Hexavalent Chromium and Detection of Chromate Reductase (ChrR) in Stenotrophomonas maltophilia. Molecules., 2018;23(2):406-436.
Crossref - Rocco F, De Gregorio E, Colonna B, Di Nocera PP. Stenotrophomonas maltophilia genomes: A start-up comparison. Int J Med Microbiol. 2009;299(8):535-546.
Crossref - Rathnayake IVN, Megharaj M, Krishnamurti GSR, Bolan NS, Naidu R. Heavy metal toxicity to bacteria-are the existing growth media accurate enough to determine heavy metal toxicity. Chemosphere. 2013;90(3):1195-1200.
Crossref - Sundar K, Vidya R, Mukherjee A, Chandrasekaran N. High chromium tolerant bacterial strains from Palar River basin: Impact of tannery pollution. Res J Environ Earth Sci. 2010;2:112-117.
- Velez JA, Quiroz LF, Ruiz OS, Montoya OI, Turrion MB, Orduz S. Hexavalent chromium-reducing bacteria on biosolids from the San Fernando Wastewater Treatment Plant in Medellin (Colombia). Revista Colombiana de Biotecnologia. 2021;23(1):32-45.
Crossref - Deng P, Tan X, Wu Y, Bai Q, Jia Y, Xiao H. Cloning and sequence analysis demonstrate the chromate reduction ability of a novel chromate reductase gene from Serratia sp. Exp Ther Med. 2015;9(3):795-800.
Crossref - Tang YW, Graevenitz AV, Waddington MG, et al. Identification of Coryneform Bacterial Isolates by Ribosomal DNA Sequence Analysis. J Clin Microb. 2000;38(4):1676-1678.
Crossref - Deshpande K, Cheung S, Rao MS, Dave BC. Efficient sequestration and reduction of hexavalent chromium with organosilica sol-gels. J Mater Chem. 2005;15(29):2997-3004.
Crossref - Bhatt P, Bhatt K, Huang Y, Li J, Wu S, Chen S. Biofilm formation in xenobiotic degrading microorganisms. Crit Rev Biotechnol. 2022:1-21.
Crossref - Thatoi H, Das S, Mishra J, Rath BP, Das N. Bacterial chromate reductase, a potential enzyme for bioremediation of hexavalent chromium: A review. J Environ Manag. 2014;146:383-399.
Crossref - Pimentel BE, Moreno-Sanchez R, Cervantes C. Efflux of chromate by Pseudomonas aeruginosa cells expressing the ChrA protein. FEMS Microbiol. Lett. 2002;212(2):249-254.
Crossref - Patra RC, Malik S, Beer M, Megharaj M, Naidu R. Molecular characterization of chromium (VI) reducing potential in Gram positive bacteria isolated from contaminated sites. Soil Biol Biochem. 2010;42(10):1857-1863.
Crossref - Pinto F, Pacheco CC, Ferreira D, Moradas-Ferreira P, Tamagnini P. Selection of suitable reference genes for RT-qPCR analyses in cyanobacteria. PLoS One. 2012;7(4):e34983.
Crossref
© The Author(s) 2023. Open Access. This article is distributed under the terms of the Creative Commons Attribution 4.0 International License which permits unrestricted use, sharing, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.