ISSN: 0973-7510
E-ISSN: 2581-690X
Vibrio vulnificus is a Gram-negative, halophilic bacterium that mainly inhabits marine environments. It is responsible for causing gastroenteritis upon consuming contaminated seafood or exposure of an open wound to seawater. In addition, it has the ability to cause wound infection and septicaemia. It is also known to be an opportunistic organism that targets immunocompromised patients and those with liver disease. In the present study, 362 seawater samples were collected from 17 different locations along the coastal areas of the Eastern Province of Saudi Arabia and were analyzed for the presence of V. vulnificus. There were 65 (17.95%) positive samples and 234 isolates of V. vulnificus. All positive isolates were tested for pathogenicity using PCR to detect the hemolysin-cytolysin (vvhA) gene, which was found in 52 (22%) of the isolates. The antibiotic susceptibility test indicated high resistance to ampicillin (96%), cephalothin (73%), rifampicin (63%), and amoxicillin-clavulanic acid (56%). The MAR index was calculated for all antibiotics and revealed significant values (>0.2) for 34.6% of V. vulnificus isolates. Isolates positive for the vvhA gene were genotyped by using Enterobacterial Repetitive Intergenic Consensus (ERIC-PCR) DNA fingerprinting. ERIC-PCR fingerprints of 52 isolates of V. vulnificus generated high similarity scores ranging from 85 to 100%, indicating significant genetic relatedness between the isolates. This study is the first to report the isolation of V. vulnificus positive for the vvhA gene from the coastal water in the Eastern Province of Saudi Arabia.
Vibrio vulnificus, Hemolysin-cytolysin gene (vvhA), Antibiotic resistance, ERIC-PCR
Humans have a permanent relationship with marine systems, including different activities such as fishing for food, transportation, and entertainment. This has had an impact on the development of human life. However, it has a poor effect on coastal and estuarine water environments. The quality of water in developing countries has been declining throughout the years. Economic development, industrial growth, and social progress are changing the balance of the ecosystem1. Pollution is the first factor that results from contaminating water by different waste, including oil, agricultural chemicals, sewage, and factory waste. All these environmental modifications lead to increased incidence of emerging diseases2.
Emerging pathogens that are causing new infections and antimicrobial resistance have had increasing incidence in the human population in the past two decades and are expected to increase in the future.2 One of the emerging organisms is bacteria in the genus Vibrio, which belongs to the Vibrionaceae family. According to the Centers for Disease Control and prevention (CDC), Vibrio infections are classified as cholera and vibriosis3. The CDC has established a national system to monitor infections by V. cholerae and other vibriosis cases, which is known as Cholera and Other Vibrio Illness Surveillance (COVIS)3. The CDC initiated COVIS in collaboration with the Food and Drug Administration (FDA), and this system is used for reporting human infections with pathogenic Vibrio species. The program is also interested in testing data on antibiotic susceptibility and the route of transmission of these pathogens. In the United States, COVIS reported an increase of Vibrio infection between 1996 and 2010, and the analysis determined that the most cases of death were from Vibrio vulnificus 3,4.
V. vulnificus is a halophilic bacterium that is naturally found in brackish and fresh water. It is an opportunistic organism that mainly targets immunosuppressed patients and those with liver disease5. The most common disease caused by V. vulnificus is gastroenteritis, which results from the consumption of contaminated seafood. V. vulnificus has the ability to cross the intestinal wall and enter the blood stream, which results in aggressive infection and primary septicemia. Direct contact between an open wound and sea water may lead to infection with a high mortality rate6,7. V. vulnificus is classified into different biotypes depending on genetics, serology, chemical reactions, and type of host. The essential virulence genes are well defined and affect motility, polysaccharides, the potential for neutralization by acid, the iron acquisition system, and hemolysin8.
Laboratory identification with the conventional culture method is done on proper media, followed by chemical and serology identification, as recommended by the FDA9. Molecular methods for detection are considered to be the most sensitive and specific. Infection by V. vulnificus is rare with approximately 100 cases occurring per year in the USA and Europe. It is responsible for 95% of deaths related to foodborne pathogens in the USA10. Increased infections by V. vulnificus are expected in the future11. The abundance of V. vulnificus has seasonal variability because of the effects of temperature and salinity on proliferation12. We conducted a study on the occurrence of V. vulnificus in seawater samples by testing the isolates for the hemolysin-cytolysin (vvhA) gene and antimicrobial susceptibility. In addition, all the positive isolates for the vvhA gene were genotyped by ERIC-PCR.
Study site
The Arabian Gulf is a subtropical marine system that is very shallow with a maximum depth of around 60 m. Many studies report that the gulf environment is under different stresses that lead to alteration of the surroundings. The most significant natural alterations are the warming of the climate and salinity13. Due to the impacts of development in Arabian Gulf countries, the water is polluted by industrial wastes such as heavy metals, oil, and gases, as well as sewage discharge. The Arabian Gulf consider has high pollution due to the semi-enclosed ecosystem with low water exchange1.
In this study, seawater samples were collected from 17 different locations that were divided into two or three sites, which were coordinated using GPS. The locations are along the coastline of the Arabian Gulf, and recreational areas were targeted, as shown in Fig. 1. The locations are Alaziziyah Beach (AZB), Corniche Tiba Jubail (CTJ), Dammam Corniche (DMC), Dammam Marina Front (DMF), Fanateer Corniche (FNC), Half-Moon Beach (HMF), Alkobar Corniche (KBC), Alkobar Marina Front (KBF), Albuhairah Beach (LAK), Almorjan Island (MOI), Palm Beach Jubail (PBJ), Qatif Corniche (QTC), Ras Tanura Corniche (RTC), Saihat Corniche (SEC), Alshibaly (SHB), Tarout Corniche (TRC), and Imam Abdulrahman Bin Faisal University (IAU).
Water sample collection and transportation
A total of 362 seawater samples were collected from the surfaces of the different locations between February 2015 and February 2016. The samples were collected using sterile 500-ml screw-cap bottles (Fischer, UK) and kept in insulated coolers. All samples were immediately transported to the microbiology research laboratory at Imam Abdulrahman Bin Faisal University and analyzed for the isolation of V. vulnificus.
Sample treatment and cultivation
All seawater samples were enriched with alkaline peptone water (APW) containing 1% NaCl, as described in the Bacteriological Analytical Manual (BAM) of the FDA9. Briefly, 25 ml of the samples were added to 225 ml of prepared APW and then incubated for 18-24 hours at 37°C. After the enrichment step, full loops of enriched seawater samples were streaked onto thiosulfate-citrate-bile salts-sucrose (TCBS) agar (Oxoid, UK) and Vibrio chromogenic agar (CHROM Agar, France) and incubated at 37°C for 18-24 hours. Since V. vulnificus varies in fermenting sucrose, the colonies appeared on TCBS in yellow and green colors. On the Vibrio chromogenic agar, the colonies appeared as turquoise. Around 3 to 5 typical colonies were selected and sub-cultured on tryptic soy agar (TSA) enriched with 3% NaCl and incubated at 37°C.
Biochemical identification
After the culture and isolation of pure colonies, presumptive V. vulnificus was confirmed by a series of biochemical tests. The confirmation includes Gram staining, oxidase tests, and Kligler’s iron agar (KIA). For typical V. vulnificus, the results must be Gram negative rods and produce a purple color on strips that indicate a positive oxidase test. When the selected colonies are emulsified in bile salt on a clean slide, they appear viscous. Pulling the loop gently produces a mucoid string mass, which indicates a positive string test, and the colonies have a K/A pattern with no gas or H2S production. All the isolates of typical V. vulnificus according to biochemical tests were reconfirmed by API 20E strip tests (Bio Mérieux, France). The API suspension was prepared using 2% saline, and V. vulnificus ATCC 27562 strain was used as a positive control. The pure identified colonies of V. vulnificus were preserved in tryptic soya broth supplemented with 3% NaCl and glycerol and stored at -80°C.
Genomic DNA Extraction
For genomic DNA extraction, one loop full of the preserved presumptive V. vulnificus was sub-cultured on Luria-Bertani (LB) broth supplemented with 3% NaCl and then incubated at 37°C over night. The DNA was extracted according to Silvester et al. ( 2015) with little modification. Briefly, 1.5 ml of the incubated LB broth was centrifuged at 10,621 x g for 2 minutes, and the supernatant was decanted. The pellets were then suspended in sterile distilled water, followed by vortexing for 2 minutes. Finally, all sample suspensions were boiled in a water bath at 100°C for 15 minutes to lyse the bacterial cells and free the DNA. After boiling, all samples were kept at -20oC to be used for detection of the vvhA gene and ERIC-PCR genotyping.
Detection of vvhA gene in Vibrio vulnificus
PCR amplification was performed to detect the vvhA gene as described elsewhere14. The total volume of the reaction mixture was 25 µl, which was composed of 1 µl of diluted DNA (1:10) template, 5 µl of 10X buffer (Promega, USA), 4 µl of 25 mM MgCl2 solution (Promega, USA), 0.5 µl of 2.5 mM deoxynucleotide triphosphate (dNTP) solution (Promega, USA), 0.25 µl of Taq DNA polymerase (Promega, USA), 1 µl of forward primer 52 TTCCAACTTCAAACC GAACTATGA-32, 1 µl of reverse primer 52 ATTCCAGTCGATG CG AATACGTTG-32 , and 12.25 µl of deionized water (Promega, USA). V. vulnificus ATCC 27562 was used as a positive control, while V. alginolyticus ATCC 17749 was used as a negative control.
The reaction program began at 94°C for initial denaturation for 3 minutes, followed by 30 cycles of amplification including 1 minute at 94°C for denaturation, 1 minute at 65°C for annealing, and 1 minute at 72°C for extension. The 30 amplification cycles were followed by a final extension step at 72°C for 5 minutes. After that, 10 µl of the PCR reaction mixture was loaded on 1.5% agarose electrophoresis gel with 1 µl of ethidium bromide dye (Promega, USA) added to make it visible under UV light. The buffer used was 1X Tris Borate EDTA. The positive vvhA gene was determined after comparison with the positive control, which has a 205-bp amplicon.
Enterobacterial repetitive intergenic consensus (ERIC) PCR
ERIC-PCR was performed according to a previously described method15. The total volume of the reaction mixture was 25 µl, which was composed of 1 µl of diluted DNA (1:10) template, 5 µl of 10X buffer (Promega, USA), 4 µl of 25 mM MgCl2 solution (Promega, USA), 0.5 µl of 2.5 mM dNTPs solution (Promega, USA), 0.25 µl of Taq DNA polymerase (Promega, USA), 2 µl of ERIC primer, and 12.25 µl of distilled water. The amplification reaction was 4 minutes at 94°C for pre-denaturation, followed by 35 cycles of 45 seconds at 94°C for denaturation, 1 minute at 52°C for annealing, an extension step at 65°C for 8 minutes, and a final extension at 65°C for 10 minutes. After that, 10 µl of the PCR reaction mixture was loaded on 1% agarose electrophoresis gel with 1 µl of ethidium bromide dye added (Promega, USA) to make it visible under UV light. The buffer used was 1X Tris Borate EDTA.
Fingerprinting analysis
All gels were scanned for DNA bands, and the images were captured using a gel documentation system (Syngene G: Box). The gel images were analyzed using GelJ software to analyze the DNA fingerprint16. The dendrogram tree was constructed based on the unweighted average pair group method (UPGMA) using a Dice coefficient. The numerical index of discrimination (D) was calculated empirically using a previously described formula17:
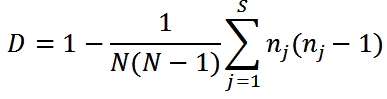
where D is the discriminatory index, N is the total number of strains in the sample population, s is the total number of types described, and nj is the number of strains belonging to the jth type.
Antibiotic susceptibility testing
The isolates of V. vulnificus recovered from sea water samples that were positive for the vvhA gene were tested against 24 antibiotics (Oxoid, England). The disc diffusion method was performed according to the CLSI protocols18. The following antimicrobial agents were tested: ampicillin (AMP: 25 µg), amoxicillin-clavulanic acid (AMC: 30 µg), piperacillin (PRL: 100 µg), piperacillin-tazobactam (TZP: 110 µg), aztreonam (ATM: 30 µg), cephalothin (KF: 30 µg), cefoxitin (FOX: 30 µg), cefotaxime (CTX: 30 µg), ceftazidime (CAZ: 30 µg), ceftriaxone (CRO: 30 µg), cefepime (FEP: 30 µg), imipenem (IPM: 10 µg), meropenem (MEM: 10 µg), amikin (AK: 30 µg), gentamicin (GN: 10 µg), streptomycin (S: 10 µg), tetracycline (TE: 30 µg), doxycycline (DO: 30 µg), ciprofloxacin (CIP: 5 µg), levofloxacin (LEV: 5 µg), chloramphenicol (C: 30 µg), trimethoprim-sulfate (SXT: 25 mcg), nalidixic acid (NA: 30 µg), and rifampicin (RD: 5 µg). The reference strain Escherichia coli ATCC 1175 was used as a control while performing the antibiotic susceptibility testing.
Multiple antibiotic resistance index
The Multiple Antibiotic Resistance (MAR) index was calculated as a/b, where a is the number of multiple antibiotics to which the isolates are resistant, and b is the total number of multiple antibiotics to which particular isolates have been exposed19.
Multidrug resistance
The categories of antibiotics used in this study were penicillins, cephalosporin, carbapenem, aminoglycosides, tetracycline, fluoroquinolone, amphenicol, folate inhibitor, quinolone, and rifampin. The strains with resistance to at least one antibiotic from three or more different classes of antimicrobial were considered as having multidrug resistance (MDR)20.
This study aimed to determine the occurrence and characterization of V. vulnificus isolated from seawater samples collected from the eastern costal environment of Saudi Arabia (Fig. 1). V. vulnificus is a marine bacterium that naturally inhabits estuarine and coastal waters worldwide. Infection by V. vulnificus includes self-limited gastroenteritis, primary septicemia, and wound infection with a 50% mortality rate6. Several studies report that approximately 95% of cases occur in the subtropical Western Pacific and Atlantic coastal regions, including Korea, Japan, Taiwan, and the United States from the Gulf of Mexico21. In this study, V. vulnificus was isolated from 65 (17.95%) of the 362 samples examined and yielded 234 isolates (Table 1).
Table (1):
Total number of seawater samples and V. vulnificus isolates recovered from different locations.
Location |
No. of samples |
No. of positives samples |
No. of isolates |
No. of isolates positive for vvhA gene by using PCR |
|---|---|---|---|---|
Alaziziyah Beach (AZB) |
29 |
6 |
25 |
6 |
Corniche Tiba Jubail (CTJ) |
11 |
3 |
9 |
2 |
Dammam Corniche (DMC) |
25 |
5 |
23 |
6 |
Dammam marina Front (DMF) |
14 |
2 |
6 |
2 |
Fanateer Corniche (FNC) |
16 |
3 |
8 |
1 |
Half-Moon Beach (HMF) |
36 |
2 |
5 |
1 |
Alkobar Corniche (KBC) |
13 |
5 |
11 |
3 |
Alkobar Marina Front (KBF) |
5 |
1 |
3 |
2 |
Albuhairah Beach (LAK) |
101 |
17 |
82 |
27 |
Almorjan Island (MOI) |
15 |
2 |
5 |
0 |
Palm Beach Jubail (PBJ) |
15 |
4 |
13 |
2 |
Qatif Corniche (QTC) |
12 |
2 |
4 |
0 |
Ras Tanura Corniche (RTC) |
19 |
3 |
9 |
0 |
Saihat Corniche (SEC) |
3 |
1 |
2 |
0 |
Alshibaly (SHB) |
8 |
2 |
5 |
0 |
Tarout Corniche (TRC) |
30 |
5 |
18 |
0 |
Imam Abdulrahman Bin Faisal University (IAU) |
10 |
2 |
6 |
0 |
Total |
362 |
65 (17.95 %) |
234 |
52 (21.8%) |
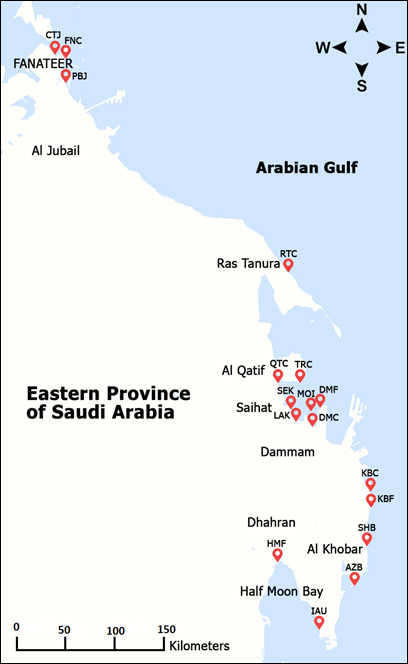
Fig. 1. Sampling sites and locations in Eastern Province of Saudi Arabia
V. vulnificus was isolated and identified according to procedures recommended by BAM from the FDA9. The presumptively identified colonies on CHROMagar Vibrio (Paris, France) were phenotypically confirmed using API20E strips (BioMérieux, France). All the isolates of V. vulnificus (n=234) were tested for the presence of the vvhA virulence gene using PCR, and 52 (21.8%) of the isolates tested positive for this gene (Fig. 2). As shown in Table 2, the vvhA gene was detected in 10 out of 17 sample locations, and the highest number of positive isolates were from LAK.
Table (2):
Results of antibiotic susceptibility testing performed on 52 V. vulnificus isolates.
| Antibiotics class | Antimicrobial agents | Antibiogram pattern of V. vulnificus | ||
|---|---|---|---|---|
| Resistant (%) | Intermediate (%) | Susceptible (%) | ||
| Penicillins | Ampicillin | 52 (100%) | 0 | 0 |
| Amoxicillin- clavulanic acid | 32 (61.5%) | 12 (23%) | 8 (15.4%) | |
| Piperacillin | 4 (8%) | 14 (27%) | 34 (65%) | |
| Piperacillin-Tazobactam | 1 (2%) | 2 (4%) | 49 (94%) | |
| Monobactams | Aztreonam | 2 (4%) | 7 (13%) | 43 (83%) |
| Cephalosporins | Cephalothin | 38 (73%) | 12 (23%) | 2 (4%) |
| Cefoxitin | 8 (15%) | 37 (71%) | 7 (14%) | |
| Cefotaxime | 0 | 2 (4%) | 50 (96%) | |
| Ceftazidime | 0 | 0 | 52 (100%) | |
| Ceftriaxone | 0 | 4 (8%) | 48 (92%) | |
| Cefepime | 0 | 0 | 52 (100%) | |
| Carbapenem | Imipenem | 0 | 0 | 52 (100%) |
| Meropenem | 0 | 0 | 52 (100%) | |
| Aminoglycosides | Amikacin | 12 (23%) | 24 (46%) | 16 (31%) |
| Gentamicin | 0 (0%) | 9 (17%) | 43 (83%) | |
| Streptomycin | 23 (44%) | 27 (52%) | 2 (4%) | |
| Tetracycline | Tetracycline | 0 | 0 | 52 (100%) |
| Doxycycline | 0 | 0 | 52 (100%) | |
| Quinolones | Ciprofloxacin | 0 | 21 (40%) | 31 (60%) |
| Levofloxacin | 0 | 4 (8%) | 48 (92%) | |
| Amphenicol | Chloramphenicol | 0 | 0 | 52 (100%) |
| Folate inhibitor | Trimethoprim- sulfate | 0 | 0 | 52 (100%) |
| Quinolone | Nalidixic acid | 0 | 0 | 52 (100%) |
| Rifampin | Rifampicin | 34 (65%) | 17 (32.7%) | 1 (4%) |
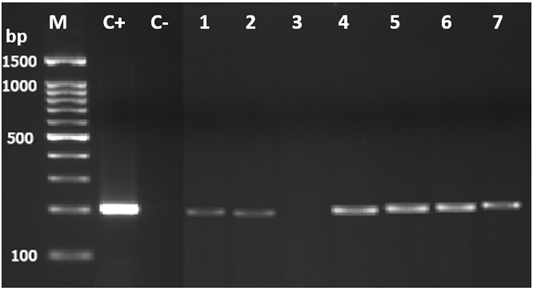
Fig. 2. PCR agarose gel electrophoresis analysis showing the vvhA gene in V. vulnifi-cus by using PCR. Lane M: molecular weight marker (100 bp DNA ladder; Promega); C+, V. vulnifi-cus (positive control); C-, negative control; 1 to 7, representative isolates of V. vulnificus iso-lated in this study
Although it has high abundance in nature, infection by V. vulnificus is rare. This can be explained by most of the strains being non-pathogenic to humans. The identification of pathogenic markers and classifying the strains as pathogenic and non-pathogenic is very hard for environmental bacteria due to horizontal gene transfer22. The reported cases of V. vulnificus have been increasing in past four decades with rising mortality rates21. The cases of V. vulnificus infection had the highest hospitalization rate of 79% among all cases of vibrio according to COVIS, and they had the highest mortality rate of 18%3.
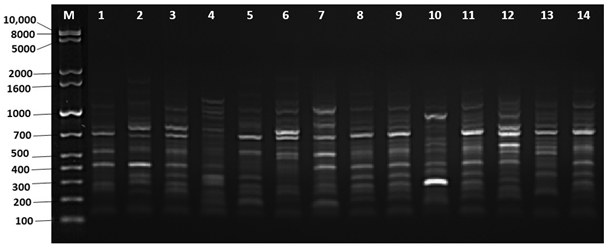
Fig. 3. Amplified DNA fingerprints generated by ERIC-PCR using ERIC1R primer. Lane M: 1kb plus ladder; lane 1 to 14: representative isolates of V. vulnificus isolated in this study
Out of 52 isolates of V. vulnificus that were positive for the vvhA gene, only 51 isolates were typed using ERIC-PCR to find out the genetic relationship among the isolates. All isolates generated 6 to 16 fingerprint bands ranging in size from 180 to 1100 bp by using the ERIC primer. The fingerprints of the isolates were grouped based on the similarities generated using the UPGMA algorithm, as shown in Fig. 4. The genetic similarities were calculated by using 1% optimization for the Dice coefficient and 1% for the position tolerance. As shown in Fig. 4, the dendrogram generated by the ERIC primer clustered the 51 isolates with 90% identical genetic similarity. However, the dendrogram fingerprint result revealed that there was no correlation between the locations and months of samples, and all the V. vulnificus isolates were identical and clustered together (Fig. 4). We used ERIC-PCR for genetic typing because it has multiple advantages, such as speed, simplicity, no need for advanced devices, ease, not requiring an expensive setup, and needing a small amount of DNA template23.
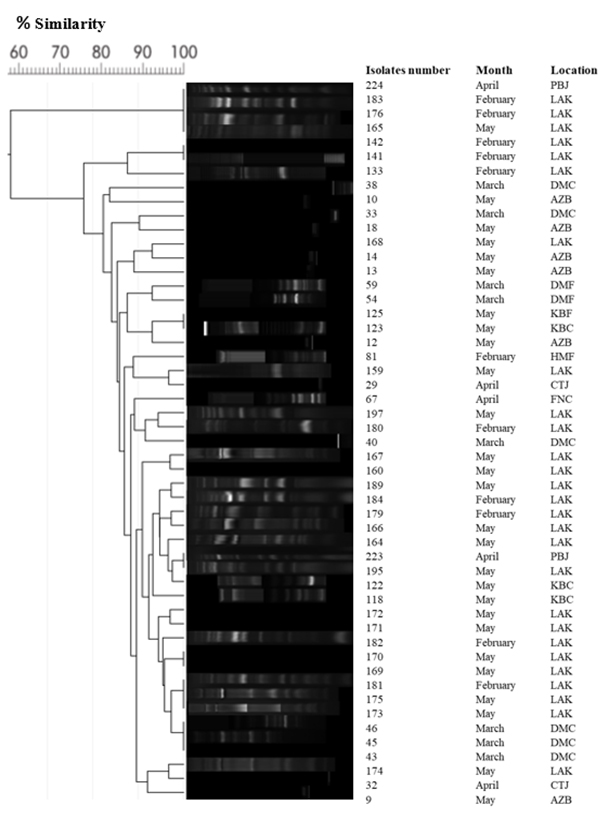
Fig. 4. Cluster generated by ERIC-PCR fingerprints of 51 isolates of V. vulnificus grouped by the ERIC1R primer according to their genetic similarities using the UPGMA algorithm. The fingerprints genetic similarities between the isolates were calculated using the Dice coefficient
V. vulnificus showed high degrees of resistance to ampicillin (100%), cephalothin (73%), rifampicin (65.4%), amoxicillin with clavulanic acid (61.5%), and streptomycin (44%) (Table 2). The lowest resistance occurred for amikacin (23%), cefoxitin (15%), piperacillin (8%), aztreonam (4%), and piperacillin with tazobactam (2%). As presented in Table 2, nine antibiotics were effective (100%) for all V. vulnificus isolates: ceftazidime, cefepime, Imipenem, meropenem, tetracycline, doxycycline, chloramphenicol, trimethoprim-sulfate, and nalidixic acid.
The results of antibiotic susceptibility testing showed that 100% of the V. vulnificus isolates were resistant to ampicillin, which has been reported previously24-26. As shown in Table 2, high resistance of V. vulnificus isolates was also found for cephalothin (73%), rifampicin (65%), amoxicillin-clavulanic acid (61.5%), and streptomycin (44%). Similar resistance to these antibiotics was reported by other researchers10, 25-27.
Table (3):
Antibiotic resistance profile and multiple antibiotic resistance (MAR) index of V. vulnificus.
Antibiotic Resistance Profile |
No. of isolates |
Multiple antibiotic resistance index (MAR) |
|---|---|---|
AMP, S |
1 |
0.08 |
AMP, KF |
1 |
0.08 |
AMP, AMC |
5 |
0.08 |
AMP, S, RD |
1 |
0.13 |
AMP, KF, RD |
6 |
0.13 |
AMP, AMC, S |
1 |
0.13 |
AMP, AMC, KF |
4 |
0.13 |
AMP, AMC, RD |
2 |
0.13 |
AMP, KF, S, RD |
1 |
0.17 |
AMP, AMC, KF, S |
1 |
0.17 |
AMP, AMC, S, RD |
1 |
0.17 |
AMP, AMC, KF, RD |
8 |
0.17 |
AMP, AMC, KF, FOX |
1 |
0.17 |
AMP, AMC, PRL, FOX |
1 |
0.17 |
AMP, KF, AK, S, RD |
6 |
0.21 |
AMP, TZP, KF, AK, S |
1 |
0.21 |
AMP, AMC, KF, S, RD |
4 |
0.21 |
AMP, AMC, KF, FOX, S |
1 |
0.21 |
AMP, AMC, PRL, AK, RD |
1 |
0.21 |
AMP, KF, FOX, AK, S, RD |
2 |
0.25 |
AMP, ATM, KF, FOX, AK, S, RD |
1 |
0.29 |
AMP, AMC, PRL, ATM, KF, FOX, S |
1 |
0.29 |
AMP, AMC, PRL, KF, FOX, AK, S, RD |
1 |
0.33 |
Interestingly, there no V. vulnifcus isolates have been reported to be resistant to antibiotics recommeded by the CDC in the USA for treating V. vulnificus infection, such as doxycycline and third-generation cephalosporin (ceftazidime)3. The MAR index ranged from 0.08 to 0.33, and nine different antibiotic resistance patterns had a significant MAR index value >0.2. (Table 3). The majority of them were isolated from LAK (Figure 1 and Table 1). A MAR index that exceeds 0.2 suggests there are high-risk sources of contamination where the use of antimicrobial agents is common19,25. Accordingly, the present study found that 41 out of 45 (86.5%) isolates of V. vulnficus had MDR, and most of them were isolated from LAK as well (Table 3). MDR has previously been reported in V. vulnificus isolates28. All 52 isolates of V. vulnificus that tested positive for the vvhA gene were multi-drug resistant (Table 3).
Our findings are also in agreement with other published studies that reported the spread of MDR in V. vulnificus isolated from coastal water environments29-31. In the present study, V. vulnificus was mostly isolated from samples collected from LAK (Fig. 1). The LAK location is an artificial lake that is linked with the Arabian Gulf by a canal, which feeds it with seawater. A potential reason for the LAK site being the most contaminated location is that it is a closed area with poor water exchange and possible contamination by sewage. Some hypothesize that the deterioration of water quality and pollution are the causes of the increase in pathogenic strains in the environment7,32. Reasons for the spread of the resistant bacteria are selective pressure from antibiotics and excessive use of antimicrobials in agriculture, which are mostly discharged into marine water. Other reasons are the ability of bacteria to be reservoirs of resistance by transferring plasmids through conjugation, transformation, and integrons33. The multidrug resistant strains of bacteria are a major threat to human life and becoming an international health crisis34. The Food and Agriculture Organization set up an action plan to minimize the spread of antibiotic resistance by increasing the awareness of resistance to antibiotics, developing programs to monitor the resistance, and improving the systems of using antimicrobials in agriculture35.
The results of this study confirmed the presence of V. vulnificus in the Arabian Gulf coast of the Eastern Province of Saudi Arabia. In addition, this study is the first report of the presence of the vvhA gene in isolates of V. vulnificus, which may have a potential to cause human infection, especially in populations with a high percentage of diabetes mellitus and among hematological patients with high serum iron. The antibiogram results showed a high percentage of MDR of V. vulnificus to clinically important antibiotics. This constitutes a possible risk for humans from the consumption or handling of contaminated seafood or from the exposure of open wounds to seawater.
ACKNOWLEDGMENTS
This project was funded by the National Science, Technology, and Innovation Programs (NSTIP) of King Abdulaziz City for Science and Technology (KACST), Kingdom of Saudi Arabia (award number: 10-ENV1337-46).
CONFLICT OF INTEREST
The authors declare that there is no conflict of interest.
- Naser HA. Assessment and management of heavy metal pollution in the marine environment of the Arabian Gulf: a review. Mar Pollut Bull, 2013; 72(1), 6-13.
- Sharma S, Sachdeva P, Virdi JS. Emerging water-borne pathogens. App Microbiol Biotechnol, 2003; 61(5), 424-428.
- CDC. Cholera and Other VibrioIllness Surveillance (COVIS), Center for Disease Control and Prevention. https://www.cdc.gov/vibrio/surveillance.html, Accessed 18 April, 2018.
- Newton A, Kendall M, Vugia DJ, Henao OL, Mahon BE. Increasing rates of vibriosis in the United States, 1996-2010: review of surveillance data from 2 systems. Clin infect Dis, 2012; 54 Suppl(5), S391-S395.
- Oliver JD. Wound infections caused by Vibrio vulnificus and other marine bacteria. Epidemiol Infect, 2005; 133(03), 383-391.
- Horseman MA, Surani S. A comprehensive review of Vibrio vulnificus: an important cause of severe sepsis and skin and soft-tissue infection. Int J Infect Dis, 2011; 15(3), 157-166.
- Li M, Zhao L, Ma J, Zhao N, Luo J, Wang C, Chen L, Ma G, Wang Y, He H. Vibrio vulnificus in aquariums is a novel threat to marine mammals and public health. Transbound Emerg Dis, 2018; doi: 10.1111/tbed.12967.
- Strom MS, Paranjpye RN. Epidemiology and pathogenesis of Vibrio vulnificus, Microbes Infect, 2000; 2(2), 177-188.
- FDA. 2004. Bacteriological Analytical Manual (BAM) Chapter 9: Vibrio. Food and Drug Administration, Bacteriological Analytical Manual, Washington DC.
- Igbinosa EO, Okoh AI. Emerging Vibrio species: an unending threat to public health in developing countries. Res Microbiol, 2008; 159(7), 495-506.
- Baker-Austin C, Trinanes JA, Taylor NGH, Hartnell R, Siitonen A, Martinez Urtaza J. Emerging Vibrio risk at high latitudes in response to ocean warming. Nat Clim Change, 2012; 3(1), 73-77.
- Baker-Austin C, Trinanes J, Gonzalez-Escalona N, Martinez-Urtaza J. Non-Cholera Vibrios: The Microbial Barometer of Climate Change. Trends Microbiol, 2017; 25(1), 76-84.
- Sheppard C, Al-Husiani M, Al-Jamali F, Al-Yamani F, Baldwin R, Bishop J, Benzoni F, Dutrieux E, Dulvy NK, Durvasula SR, Jones DA, Loughland R, Medio D, Nithyanandan M, Pilling GM, Polikarpov I, Price AR, Purkis S, Riegl B, Saburova M, Namin KS, Taylor O, Wilson S, Zainal K. The Gulf: A young sea in decline. Mar Pollut Bull, 2010; 60(1), pp. 13-38.
- Panicker G, Myers ML, Bej AK. Rapid Detection of Vibrio vulnificus in Shellfish and Gulf of Mexico Water by Real-Time PCR. Appl Environ Microbiol, 2004; 70(1), 498-507.
- Versalovic J, Koeuth T, Lupski JR. Distribution of repetitive DNA sequences in eubacteria and application to fingerprinting of bacterial genomes. Nucleic Acids Re,1991; 19(24), 6823-6831.
- Heras J, Domínguez C, Mata E, Pascual V, Lozano C, Torres C, Zarazaga M, 2015. GelJ a tool for analyzing DNA fingerprint gel images. BMC Bioinformatics, 2015; 16, 270.
- Hunter PR, Gaston MA, 1988. Numerical index of the discriminatory ability of typing systems: an application of Simpson’s Index of Diversity. J Clin Microbiol, 1988; 26(11), 2465-2466.
- CLSI. 2012. Methods for antimicrobial dilution and disk susceptibility testing of infrequently isolated or Fastidious Bacteria; Approved Guideline, Second Edition. CLSI document M45-A2. Wayne, PA, Clinical and Laboratory Standards Institute.
- Krumperman PH. Multiple antibiotic resistance indexing of Escherichia coli to identify high-risk sources of fecal contamination of foods. Applied and Environmental Microbiology, 1983; 46(1), 165-170.
- Magiorakos AP, Srinivasan A, Carey RB, Carmeli Y, Falagas ME, Giske CG, Harbarth S, Hindler JF, Kahlmeter G, Olsson-Liljequist B, Paterson DL, Rice LB, Stelling J, Struelens MJ, Vatopoulos A, Weber JT, Monnet DL. Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: an international expert proposal for interim standard definitions for acquired resistance. Clin Microbiol Infect, 2012; 18(3), pp. 268-281.
- Huang KC, Weng HH, Yang TY, Chang TS, Huang TW, Lee MS. Distribution of Fatal Vibrio vulnificus Necrotizing Skin and Soft-Tissue Infections: A Systematic Review and Meta-Analysis. Medicine, 2016; 95(5), 2627.
- Quirke AM, Reen FJ, Claesson MJ, Boyd EF. Genomic island identification in Vibrio vulnificus reveals significant genome plasticity in this human pathogen. Bioinformatics, 2006; 22(8), 905-910.
- Cheah YK, Tay LW, Aida AA, Son R, Nakaguchi T, Nishibuchi M. Molecular characterization of Escherichia coli isolated from different food sources. Int Food Res J, 2015; 22(1), 31.
- Zanetti S, Spanu T, Deriu A, Romano L, Sechi LA, Fadda G. In vitro susceptibility of Vibrio spp. isolated from the environment. Int J Antimicrob Agents, 2001; 17(5), pp. 407-409.
- Elhadi N. Antibiotic Resistance and Plasmid Profiling of Clinically Significant Vibrio vulnificus Isolated from Coastal Water in Eastern Province of Saudi Arabia. Br J Pharmacol Toxicol, 2012; 3(2), pp. 93-97.
- Sudha S, Mridula C, Silvester R, Hatha AAM. Prevalence and antibiotic resistance of pathogenic Vibrios in shellfishes from Cochin market. Indian J Geomarine Sci, 2014; 43(5), 815-824.
- Wang RX, Wang JY, Sun YC, Yang BL, Wang AL. Antibiotic resistance monitoring in Vibrio spp. isolated from rearing environment and intestines of abalone Haliotis diversicolor. Mar Pollut Bull, 2015; 101(2), 701-706.
- Heng SP, Letchumanan V, Deng CY, Ab Mutalib NS, Khan TM, Chuah LH, Chan KG, Goh BH, Pusparajah P, Lee LH. Vibrio vulnificus: An Environmental and Clinical Burden. Front. Microbiol, 2017; 8:997. doi: 10.3389/fmicb.2017.00997
- Kim JH, Choresca Jr CH, Shin SP, Han JE, Jun JW, Park SC. Occurrence and Antibiotic Resistance of Vibrio Vulnificus in Seafood and Environmental Waters in Korea: Isolation of Vibrio vulnificus in Korea. Journal of Food Safety, 2011; 31(4), 518-524.
- Baker-Austin C, McArthur JV, Lindell AH, Wright MS, Tuckfield RC, Gooch J, Warner L, Oliver J, Stepanauskas R. Multi-Site Analysis Reveals Widespread Antibiotic Resistance in the Marine Pathogen Vibrio vulnificus. Microbial Ecology, 2009; 57(1):151-159.
- Roig FJ, Llorens A, Fouz B, Amaro C. Spontaneous Quinolone Resistance in the Zoonotic Serovar of Vibrio vulnificus. Appl Environ Microbiol, 2009; 75(8), 2577-2580.
- Tantillo GM, Fontanarosa M, Pinto AD, Musti M. Updated perspectives on emerging vibrios associated with human infections. Lett App Microbiol, 2004; 39(2).117-1126.
- Raissy M, Moumeni M, Ansari M, Rahimi E. Antibiotic resistance pattern of some Vibrio strains isolated from seafood. Iran J Fish Sci, 2012; 11(3)618-626 2012.
- 34. WHO. Antimicrobial resistance global report on surveillance. World Health Organization, 2014. http://www.who.int/drugresistance/documents/ surveillancereport/en/, Accessed 22 April, 2018.
- FAO. 2016. The FAO Action Plan on Antimicrobial Resistance 2016-2020. Food and Agriculture Organization of the United Nation, Rome.
© The Author(s) 2018. Open Access. This article is distributed under the terms of the Creative Commons Attribution 4.0 International License which permits unrestricted use, sharing, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.


