ISSN: 0973-7510
E-ISSN: 2581-690X
Azo dyes such as red 40 are characterized by an R1-N=N-R2 structure and are widely used in the food industry, but their anaerobic degradation in the gut microbiota by azoreductases originate toxic or mutagenic aromatic amines, so their use has been restricted in many countries. The aim of this study was to submit the red dye 40 to degradation by enterobacteria and partially characterize the products of its anaerobic reduction. Two enterobacteria isolated from an infant together with the reference strains: Escherichia coli DH5a, Escherichia coli E2348/69, Citrobacter rodentium DBS13 and Enterobacter cloacae were used to inoculate samples of 30 ppm of red dye 40. The degradation percentage was estimated by UV/Vis spectroscopy and the products of anaerobic reduction were characterized by HPLC. Enterobacter sp. and E. coli isolates from the infant fecal sample yielded a degradation of 60% and 49% respectively. With respect of the reference strains, the degradation percentages were high up more than 70%. The degradation of red dye 40 by the different types of bacteria used in this research revealed a formation of compounds analogous to 1-naphthol and aromatic amines.
Red dye, degradation, enterobacteria, anaerobic reduction.
Azo dyes are characterized by having one or more R1-N = N-R2 bonds and they are widely used in the textile, cosmetic, pharmaceutical and food industry due to their ease of synthesis and their chemical stability1,2. With regard to the textile industry they are the most used organic dyes, and therefore constitute the largest group available on the market3. Its toxicity is due to the production of aromatic amines during their natural degradation process4,5,6,7. When aromatic amines enter the human body through food chains, they are transformed into acyloxyamines which denature the DNA (8). Therefore, azo dyes chemical reduction is not recommended although it achieves discoloration.
Humans are exposed to these compounds through ingestion, inhalation or physical contact; children are the most vulnerable group since it is associated with hyperactivity and attention deficit9. The anionic dye red 40 (2 -naphthalenesulfonic acid, 6-hydroxy-5 – ((2 -methoxy-5 -methyl-4 -sulfophenyl) azo) -, disodiumsalt, and disodium 6 -hydroxy-5 – ((2 -methoxy -5 –methy l-4 -sulfophenyl) azo) -2 naphthalenesulfonate), also known as allura red or food red 17, is a monoazo acid dye with the molecular formula C18H14N2Na2O8S2 and has a molecular mass of 496.42 g/mol. It has acidic properties and an aromatic structure consisting of three benzene rings (Figure 1)10. This dye is widely used in beverages, bakery products, meat, powder desserts, candies, cereals, medicines, cosmetics and tattoo inks11 and its use has been restricted or eliminated in Nigeria, Switzerland, Canada and countries of the European Union as Denmark, Belgium, France, Germany, Switzerland, Sweden, Austria and Norway for the reasons mentioned above12.
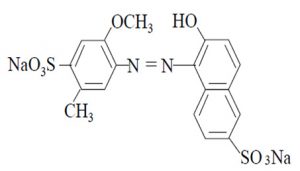 Fig. 1. Chemical structure of the anionic dye Red 40
Fig. 1. Chemical structure of the anionic dye Red 40Some bacteria can metabolize azo dyes by their enzymatic systems13 and its degradation products result to be toxic and/or mutagenic14, 15. Biodegradation of azo dyes by bacteria can be performed by anaerobic and/or aerobic processes (16). During an anaerobic degradation the intestinal microbiota is involved in the reduction of the azo bond by azoreductases, using the cofactors nicotinamide adenine dinucleotide (NAD), flavin mononucleotide (FMN) and flavin adenine dinucleotide (FAD) as electron donors. Although there are reports of the use of the intestinal microbiota bacteria in the removal of dyes in wastewater17, there is little information on the interaction of intestinal microorganisms with food colorants.
Enterobacteria were isolated and identified from a fecal sample of a 2 year and 6 month old child following the recommendations described by Prats (18), the sample was transported in Stuart medium to keep it fresh and was processed in less than 24 hours. Suspicious colonies were isolated and identified by routine laboratory tests; later, this isolated and identified strains were stored in LB broth supplemented with glycerol at -70°C for later use. The following nonpathogenic strains were used for reference experiments: Escherichia coli DH5-Alpha (DH5±), enteropathogenic Escherichia coli (EPEC) E2348/69, Citrobacter rodentium DBS 13 (Citrobacter DBS 13) and Enterobacter cloacae.
The degradation of red dye 40 with each of the enterobacteria isolated from the intestinal microbiota and the reference strains was performed as follows: a microbial culture was growth to the logarithmic phase then adjusted to a concentration of 1.5 x 109 CFU/ml in a 0.85% saline solution. Once the microbial population was adjusted, an inoculum was deposited on a nutrient broth tube containing 30 and 50 ppm of the dye. Following the methodology described by Isik et al (19), fermentation was carried out for 10 days by measuring the absorbance every 24 hours to determine the percentage of removal. The maximum of absorbance of the red dye 40 dissolved in nutrient broth was determined by UV/Vis spectroscopy.
The degradation products of a 30 ppm solution of red dye 40 were characterized by HPLC (UFLC-Prominence, Shimadzu) using a PS/DVB chromatographic column (Nucleogel® Sugar 810H, Macherey-Nagel) and a mobile phase of 5 mM sulfuric acid/water with a flow rate of 0.65 ml/min.
Standard solutions of 1-naphthol 0.051M were prepared and chromatograms were obtained using wavelengths of 285 and 315 nm, similarly, aniline dilutions with water (1:1000) were measured at 260 and 285 nm for 20 minutes each. Chromatograms of the culture medium added with dye were used as control and the samples were processed at a 1:10 dilution.
The maximum absorbance of red dye 40 was at 504 nm which is consistent with the technical specifications of the Food Drug and Cosmetic Act (FD & C Act) and confirms the purity of the product used during this investigation. Enterobacter sp. and Escherichia coli were identified from the fecal sample of the infant.
The percentages of decolorization of 30 ppm of red dye 40 by different strains of this study are shown in figure 2, Escherichia coli DH5± had the highest percentage of decolorization with 83.2%, for the case of Citrobacter DBS 13 its percentage was of 82%, for Escherichia coli E2348/69 was 77% and for Enterobacter cloacae was of 65%. Colonies isolated from the stool sample are also shown on the same graph, for Enterobacter sp.* the percentage of decolorization was 60% and for Escherichia coli was 49%*.
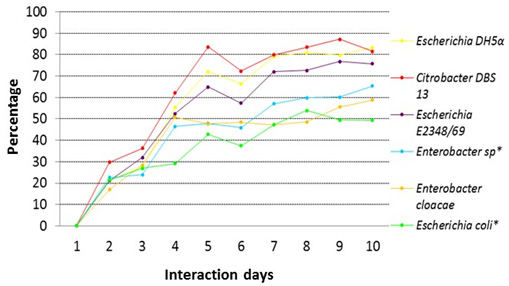 Fig. 2. Comparison of the percentage of decolorization for a solution of 30 ppm of red dye 40 for the different strains of this study
Fig. 2. Comparison of the percentage of decolorization for a solution of 30 ppm of red dye 40 for the different strains of this study*Note: Strains from the intestinal microbial
For the solution of 50 ppm of red dye 40, discoloration percentages were as follows: 84.29% for Escherichia coli E2348/69, 65% for Escherichia coli DH5±, 94% for Citrobacter DBS13 and 35% for Enterobacter cloacae. For the stool samples, the percentage were 34% for Enterobacter sp* and 43% for Escherichia coli * (Fig. 3).
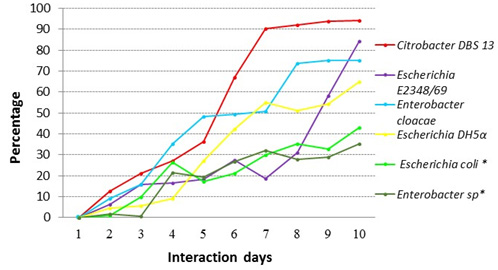 Fig. 3. Comparison of the percentage of decolorization of red dye 40 for a 50 ppm solution for the different strains in this study
Fig. 3. Comparison of the percentage of decolorization of red dye 40 for a 50 ppm solution for the different strains in this study*Note: Strains from the intestinal microbial
Several authors like Oforka and Oranusi20 have reported decolorization of azo dyes like Ponceau 4R dye and carmoisine by Escherichia coli, the strain used in their study was originally obtained from the Department of Microbiology at the University of Port Harcourt in Nigeria and was isolated from the natural human intestinal flora. In another study reported by Oranusi21, Escherichia coli isolated from the intestinal microbiota was capable of degrading a solution of 20 ppm of the azo dye tartrazine with a decolorization percentage of 60.38% over a period of 5 days. Wang et al,22 reported that Citrobacter CK3 isolated from activated sludge of a textile factory was able to decolorize 96% of the textile dye reactive red 180 in a concentration of 50 ppm under anaerobic conditions. Besides Citrobacter, species of the genus Enterobacter have been used in the degradation of azo dyes, Keharia et.al.23 reported that Enterobacter agglomerans was able to degrade methyl red; Moutaouakkil et.al24, 25 reported that Enterobacter aerogenes is capable of degrading the dye orange II. Kalyanee et al26 reported that Enterobacter sp is capable of degrading 91% of the reagent red CI195 when it was cultivated anaerobically in a concentration of 30 ppm.
In the degradation of dyes not only the genus Citrobacter and Enterobacter have been employed, Isik19 worked on the degradation of direct black 38 and congo red using a concentration of 30 ppm by Escherichia coli under anaerobic conditions, resulting in a degradation of 98% and 72% respectively.
Decolorization studies have identified a number of genes that confer the ability to degrade azo dyes. Chang et al,27 reported decolorization of azo dyes using the azo reductase genes of a wild strain of Pseudomonas luteola. In other word Chang28 cloned and expressed a fragment of Rhodococcus sp. genomic DNA. in Escherichia coli, obtaining a recombinant strain capable to decolorize azo dyes.
Partial characterization of the degradation products
The retention times (RT) obtained for the control red dye 40, standard 1-naphthol, standard aniline and the studied samples are shown in tables 1 and 2.
Table (1):
Retention times (RT) for the control red dye 40, standard 1-naphtol and samples at different wavelengths.
Sample |
285 nm RT (min) |
315 nm RT (min) |
|---|---|---|
Control (red 40) |
2.643 5.545 5.667 12.038 |
2.704 5.545 5.676 |
Standard 1-naphtol |
5.585 5.816 |
5.591 |
Escherichia coli DH5α |
5.599 |
5.595 |
Escherichia coli E2348/69 |
5.573 |
5.551 |
Citrobacter DBS 13 |
5.591 |
5.591 |
Enterobacter sp* |
5.559 |
5.553 |
Escherichia coli* |
5.553 |
5.548 |
Enterobacter cloacae |
5.522 12.28 |
5.595 |
*Note: Strains from the intestinal microbial
Table (2):
Retention times (RT) for the control red dye 40, standard aniline and samples at different wavelengths.
Sample |
260 nm RT (min) |
285 nm RT (min) |
|---|---|---|
Control (red 40) |
2.411 4.691 8.251 12.053 |
2.413 4.545 8.226 |
Standard aniline |
3.145 4.089 5.581 |
4.093 5.586 |
Escherichia DH5α |
3.425 5.573 |
5.586 |
Escherichia E2348/69 |
4.127 5.564 |
4.130 5.591 |
Citrobacter DBS 13 |
5.554 |
4.882 5.578 |
Enterobacter sp* |
4.132 5.538 |
4.134 5.560 |
Escherichia coli* |
3.306 5.537 |
5.591 |
Enterobacter cloacae |
5.336 |
5.548 |
*Note: Strains from the intestinal microbial
The chromatograms of the degradation products of Escherichia coli DH5± at 285 and 315 nm are shown in figure 4 and 5, they were used for comparison with 1-naphthol chromatograms.
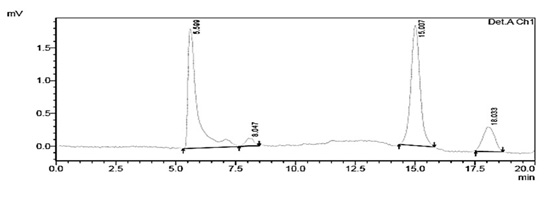
Fig. 4. Chromatogram of the degradation products of Escherichia coli DH5α at a wavelength of 285 nm showing a RT of 5.599 min
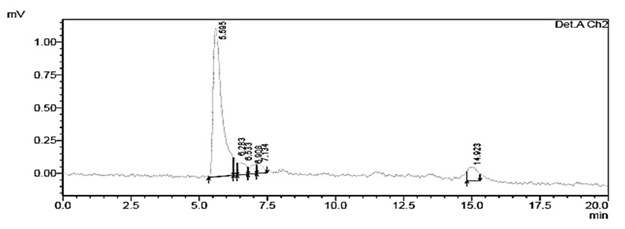
Fig. 5. Chromatogram of degraded red 40 from Escherichia coli DH5α at a wavelength of 315 nm with an RT of 5.595 min
The degradation products from the reference strains Citrobacter DBS 13, Escherichia coli DH5±, Escherichia coli E2348/69, Enterobacter cloacae and the intestinal microbiota Enterobacter sp showed a very similar RT in comparison with the control 1-naphthol at 285 and 315nm. With respect to the control aniline, all strains showed similarity in their RT at 260 and 285 nm. In this case we must highlight the peaks at RT of 5.326-5.581.
Regarding the degradation products obtained with the strain Escherichia coli DH5±, similarity exists between the chromatograms of 1-naphthol at 285 nm and 315 nm with RT of 5.599 minutes and 5.595 minutes respectively. The standard aniline showed the same RT at 285.
It has been reported that the degradation by chemical reduction of azo dyes produces aromatic amines which are toxic and carcinogenic, so this process is not recommended although the discoloration is carried out. Its pathway of reduction involves azo-reductases in anaerobic conditions29.
Degradation of a 30 ppm solution of red dye 40 by the different types of bacteria used in this study showed evidence for the formation of analogous compounds to 1-naphthol and an aromatic amine.
Furthermore, Van der Zee et al, (30) reported that the azo dye acid orange 7 was biologically degraded in anaerobic conditions to sulfanilic acid and 1-amino-2-naphthol. The red dye 40 (Fig. 1) has a similar structure to the azo dye acid orange 7 (Fig.6) so the degradation products could be similar.
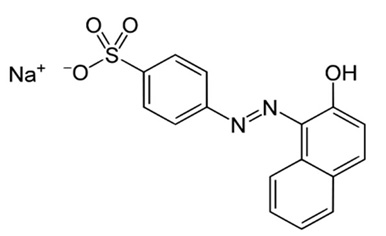 Fig. 6. Azo dye acid orange 7
Fig. 6. Azo dye acid orange 7The strains isolated from the infant fecal sample, Escherichia coli and Enterobacter sp., were able to interact with the food dye red 40, degrading it 49.38% and 58.8% respectively.
The strain Citrobacter DBS 13 gave the best decolorization percentages (above 80%), suggesting greater capability for degrading this colorant compared with the strains isolated from the fecal sample, Escherichia coli and Enterobacter sp., and the reference strains.
HPLC characterization studies demonstrated that an enzymatic reaction was carried out during the decolorization of the red dye 40, suggesting the formation of degradation products that were analogous to 1-naphthol and an aromatic amine.
Finally, the results of this research showed that the reference strains and the intestinal microbiota strains of Enterobacteriaceae, isolated from a child of the State of Puebla, Mexico, are capable of degrading the red dye 40 probably through mechanisms of azo-reductases, producing substances that are harmful to health.
ACKNOWLEDGMENTS
Thanks are due to Programa para el Desarrollo Profesional Docente (PRODEP)
914–922.
- Ashfaq, N. and Masud T. Surveillance on artiûcial colors in diûerent ready to eat foods. Pak.J.Nutr. 2002; 1, 223–225.
- Board, N. The Complete Technology Book on Dyes & Dye Intermediates, ed. National Institute of Industrial Research: Dehli, 2003; pp. 730.
- Sandhya, S. Biodegradation of Azo Dyes Under Anaerobic Condition: Role of Azoreductase. In Biodegradation of Azo Dyes. The Handbook of Environmental Chemistry, 2010; 9, 39-57.
- Golka, K., Kopps, S. and Myslak, Z. Carcinogenicity of azo colorants: influence of solubility and bioavailability. Toxicol., 2004; 151(1), 203-210.
- Gnanamani, A., Bhaskar, M., Ganga, R., Sekaran, G., Sadulla, S. Chemical and enzymatic interactions of Direct Black 38 and Direct Brown 1 on release of carcinogenic amines. Chemosphere, 2004; 56(9), 833-841.
- Weiburger, S. H. Comments on the history and importance of aromatic and heterocyclic amines in public health. Mutat Res., 2002; 9(20), 506-507.
- Takahashi, H. y Hashimoto, Y. Formaldehyde-mediated modification of deoxyguanosine with amines: one-pot cyclization as a molecular model for genotoxicity. Bioorg Med Chem Lett., 2011; 11(5), 729–731.
- Arnold, LE., Lofthouse N., Hurt E. Artificial food colors and attention-deficit/hyperactivity symptoms: conclusions to dye for. Neurotherapeutics, 2012; 9(3): 599-609.
- Srinivasan, A. and Viraraghavan, T. Decolorization of dye wastewaters by biosorbents: a review. In: Journal of environmental management. 2010; 91(10): 1915-1929.
- Soylak, M., UnsaL, Y.E. and TUzen, M. Spectrophotometric determination of trace levels of allura red in water samples after separation and preconcentration. In: Food and chemical toxicology. 2009; 49(5): 1183-11877.
- Kobylewski, S. and Jacobson M F. Toxicology of food dyes. Int J. Occup Environ Health. 2012; 18(3) 220-46.
- Saratale, R B., Saratale G.D., Chang J.S., Govingdwar S. P. Bacterial decolorization and degradation of azo dyes. Journal of the Taiwan Institute of Chemical Engineers, 2011; 42, 138-157.
- Chen Huizhong. Recent Advances in Azo Dye Degrading Enzyme Research. Current Protein and Peptide Science, Division of Microbiology, National Center for Toxicological Research, U.S. 2006; 7(2):101-111.
- Weisburger JH. Comments on the history and importance of aromatic and heterocyclic amines in public health. Mutat Res, 2002; 506-507: 9-20.
- Dos Santos AB and Traverse J. Enhancing the electron transfer capacity and subsequent colour removal in bioreactors by applying thermophilic anaerobic.artículos treatment and redox mediators. Biotechnol. 2005; 42-52.
- Yang Q, Wang J, Wang H, Chen X, Ren S, Li X, Xu Y, Zhang H, Li X. Evolution of the microbial community in a full-scale printing and dyeing wastewater treatment system. Bioresour Technol. 2012; 117:155-63.
- Prats, Guillem. Microbiología Clínica. Buenos aires, Editoral Medica Panamericana 363p; Parte II Técnicas Microbiológicas, 2005; 33-74p.
- Isik Mufasa., and Sponza Delia. Effect oxygen on decolorization of azo dye by Escherichia coli and Pseudomonas sp. and fate of aromatic amines. Process Biochemistry, 2003; 38: 1183-1192.
- Oforka, N. C., Oranusi, N. A. Decolourisation of Azo based food colorants (carmoisine and Ponceau 4R by Escherichia coli. Biology. 2004; 6: 45 -51.
- Oranusi, N A and Njoku,H O. Biotransformation of Food Dyes by Human Intestinal Bacteria (Streptococcus faecalis, Eschericia coli) All rights reserved J. Appl. Sci. Environ, 2006; 10(2) 159-163.
- Wang Hui, Su Jian Qiang, Zheng Xiao Wei. Bacterial decolorization and degradation of the reactive dye Reactive Red 180 by Citrobacter sp. CK3. International Biodeterioration & Biodegradation, 2009; 63(4):395–399.
- Keharia H and Madamwar D. Bioremediation concepts for treatment of dye containing wastewater: A review, Indian Journal of Experimental Biology, 2003; 41(9): 1068-1075.
- Moutaouakkil A, Zeroual Y, Zohra Dzayri F, Talbi M, Lee K, Blaghen M. Purification and partial characterization of azoreductase from Enterobacter agglomerans. Arch Biochem Biophys. 2003; 1; 413(1):139-46.
- Moutaouakkil A, Zeroual Y, Dzayri FZ, Talbi M, Lee K, Blaghen M. Decolorization of azo dyes with Enterobacter agglomerans immobilized in different supports by using fluidized bed bioreactor. Curr Microbiol; 2004; 48(2):124-9.
- Kalyanee Jirasripomgpun, Rujikan Nasanit, Jongjira Niruntasook and Boonsiri Chotikasatian. Decolorization and degradation of C.I reactive red 195 by Enterobacter sp. Thammasat.J. Sc. Tch. 2007; 12(4) 6-11.
- Chang, Jo-Shu and Chou, Chien. Kinetic characteristics of bacterial azo-dye decolorization by Pseudomonas luteola. Elsevier Science Ltd. All rights reserved Printed in Great Britain. 2001a; 35: 2841–2850.
- Chang, Jo-shu and Lin, Chia Yu. Decolorization kinetics of a recombinant Escherichia coli strain harboring azo dye decolorizing determinants from Rhodococcus sp. Biotechnology Letters. 2001b; 23(8): 631-636.
- Stolz, A. Basic and applied aspects in the microbial degradation of azo dyes. Appl. Microbiol. Biotechnol. 2001; 56: 69-80.
- Van der Zee, F.P., Lettinga, G. and Field, J.A. The role of (auto)catalysis in the mechanism of anaerobic azo dye reduction. Water Sci. Technol. 2000; 42: 301-308.
© The Author(s) 2016. Open Access. This article is distributed under the terms of the Creative Commons Attribution 4.0 International License which permits unrestricted use, sharing, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.


