ISSN: 0973-7510
E-ISSN: 2581-690X
Non-tuberculous Mycobacteria (NTM) are emerging as an important opportunistic pathogen. Since clinical presentation of NTM infection is similar to tuberculosis (TB), patients present as suspected TB or drug resistant TB. Presently in the National Tuberculosis Elimination Programme (NTEP ) NTM are not being speciated, but there is an urgent need to characterize the NTM so that appropriate treatment can be given as many species are multi-drug resistant. The purpose of the present study was to use Line Probe Assay (LPA) i.e. GenoType Mycobacterium CM/AS assay to characterize NTM for rapid early reporting and to know the pattern of NTM at Rajasthan. Sputum samples from 5000 TB and Multi Drug Resistant TB (MDRTB) suspect patients were processed and cultured on Mycobacterium Growth Indicator Tube (MGIT). Culture isolates found positive for mycobacteria in Ziehl Neelsen (ZN) staining and negative by MPT64 antigen test were then subjected for GenoType Mycobacterium CM/AS Among sputum samples from 5000 patients 1520 (30.4%) patient samples were positive for mycobacteria, among these 1488 (97.9%) were Mycobacterium tuberculosis (MTB) and 32 (2.1%) were NTM, among them 56.2% were Mycobacterium intracellulare, 21.8% Mycobacterium abscessus, 9.3% Mycobacterium fortuitum, 1% Mycobacterium simiae and 9.3% isolates showed invalid results. Incidence of NTM was very low (2.1%) among them M. intracellulare and M. abscessus were the most commonly isolated species, GenoType Mycobacterium CM/AS assay was found to be easy, rapid and reliable test giving valid results in 91% cases in 3-5 days of getting growth.
Line Probe Assay, Non-tuberculous Mycobacteria, CM/AS assay, Mycobacterium Growth Indicator Tube, Multi Drug-Resistant TB
Non-tuberculous Mycobacteria (NTM) are emerging as a significant pathogen causing disease in both immunocompromised and immunodeficient patients.1 There is reports worldwide of an increase in the incidence and prevalence of NTM lung disease. In India also the cases of pulmonary NTM infection have been increasing progressively over the last few years, ranging from 0.5% to 8.6%.2,3 Many of the NTM are resistant to the anti TB drugs so it’s important to speciate them so as to help in proper management of the patient.
Most of the Indian labs use traditional methods to identify mycobacteria such as sputum microscopy, culture on solid and liquid media and biochemical assays for characterization. Smear microscopy with Ziehl Neelsen (ZN) staining is a quick way to detect mycobacteria, however, it can’t differentiate between Mycobacterium tuberculosis (MTB) and NTM. Species identification of NTM by traditional methods requires an array of biochemical tests with periodic observation of growth rate and morphological characteristics. Besides, it is lengthy and labor-intensive and requires expertise.4 Other techniques for NTM identification to species level are chromatography (like Gas, Thin layer and High-Performance Liquid chromatography), sequencing, hybridization and other molecular techniques.
Recently, molecular techniques based on the hybridization assay and blotting have been introduced for NTM speciation. Among these, a Line Probe Assay (LPA) i.e. GenoType Mycobacterium CM/AS assay, (common mycobacterium/additional species) is the most used technique which gives results within 3- 5 days. It is a DNA strip assay developed by Hain Lifescience GmbH, Nehren, and Germany.5
The utilization of such molecular technologies can help in the rapid and early identification of various species of NTM which take long time by routine biochemical methods and results are not available in the clinically useful time period. The present study was carried out to know the prevalence of NTM in samples suspected of TB and MDRTB and to characterize the NTM isolated to know the profile of NTM prevalent in Rajasthan.
The present study was conducted at culture and Drug Susceptibility Testing (DST) lab, SMS Medical College, Jaipur, India. Specimens from TB and MDR TB suspect patients were received through NTEP from selected districts of Rajasthan, amongst them, sputum samples from 5000 patients received during April 2015 to December 2015 were included in the current study. Two sputum samples per patient were received as per NTEP policy. Out of 5000, 3940 (78.8%) patients were male and 1060 (21.2%) were female and belonged to different age groups. (See Table 1).
Table (1):
Age and Sex Wise Distribution of Patients.
S.No. |
Age |
No of Patients tested(%) |
No. of NTM identified in Male (%) |
No. of NTM identified in Female (%) |
Total No. of NTM identified (%) |
|---|---|---|---|---|---|
1 |
0-10 |
10 (0.2%) |
0/6 (0%) |
0/4 (0%) |
0 (0%) |
2 |
11-20. |
439(8.9%) |
0/238 (0%) |
1/201 (0.4%) |
1 (3.1%) |
3 |
21-30 |
1069 (21.4%) |
1/748 (0.1%) |
2/321 (0.6%) |
3 (9.3%) |
4 |
31-40 |
1105 (22.1%) |
2/906 (0.2%) |
2/199 (1.0%) |
4 (12.5%) |
5 |
41-50 |
991 (19.8%) |
4/842 (0.5%) |
3/149 (2.0%) |
7 (21.9%) |
6 |
51-60 |
835 (16.7%) |
8/720 (1.1%) |
1/115 (0.9%) |
9 (28.1%) |
7 |
61-70 |
443 (8.9%) |
6/381 (1.6%) |
1/62 (1.6%) |
7 (21.9%) |
8 |
71-80 |
97 (1.9%) |
1/91 (1.09%) |
0/6 (0%) |
1 (3.1%) |
9 |
Above 80 |
11 (0.2%) |
0/8 (0%) |
0/3 (0%) |
0 (0%) |
Total |
5000 |
3940 (78.8%) |
1060 (21.2%) |
32 (0.64%) |
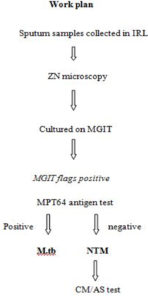
Upon receipt of the sputum specimen, they were processed for Acid Fast Bacilli (AFB) smear microscopy using the Ziehl-Neelsen (ZN) method,6 and for liquid culture using MGIT 960 by NALC-NaOH method.7 All culture-positive isolates were subjected to smear microscopy and AFB positive growth was subjected to MPT64 antigen by SD Bioline TB Ag MPT64 test kit (Standard Diagnostics, Yongin-si, Gyeonggi-do, Republic of Korea) to differentiate between MTB and NTM.8 All NTM were subjected to GenoType Mycobacterium CM/AS assay (ver 1.0 Hain Lifescience) for species identification as per manufacturer instructions.9 Briefly DNA of every isolate was extracted by Genolyse kit (ver 1.0 Hain Lifescience) as per manual given. After that DNA was amplified in the thermal cycler (2720, Applied biosystem) using master mix (given with the GenoType Mycobacterium CM/AS assay kit) then the amplified products were subjected to hybridization which was performed on Twincubator (Hain Lifescience, Germany). One positive (known MTB strain DNA was amplified and hybridized with each batch) and negative control each (PCR product without any bacterial DNA) was tested with each batch (Figure 1).
Among the 5000 patients sputum specimens, 1520 were positive for mycobacteria, from those 1488 (97.9%) were MTB that is positive for MPT64 antigen and only 32 (2.1%) were negative for MPT64 antigen (all of these were direct AFB smear positive). These were subjected to GenoType Mycobacterium CM/AS assay. Out of 32, 18 (56.2%) were identified as M. intracellulare, 7(21.8%) M. abscessus, 3(9.3%) M. fortuitum and 1(3.1%) M. simiae. Three isolates showed invalid results (Table 2). Time taken for LPA was 3- 5 days and the cost of per test was 1,815 INR (25 US$). Maximum no. of NTM 9(28.12%) were identified in age group 50-60. (Table 1).
Table (2):
Details of species identified using GenoType Mycobacterium CM/AS assay.
Species identified |
No. |
|---|---|
M. intracellulare |
18(56.2%) |
M. abscessus |
7(21.8%) |
M. fortuitum |
3(9.3%) |
M. simiae |
1(3.1%) |
Invalids |
3(9.3%) |
Total tested |
32 |
Since the clinical picture of NTM is similar to TB, as a result the infection due to NTM is mistaken as TB. Moreover, many NTM strains are resistant to the first line Anti Tubercular Treatment (ATT) drugs, so are commonly misidentified as Drug Resistant TB (DRTB). If such cases are not diagnosed timely, they will receive anti TB treatment which is not effective in NTM, moreover many NTM species are inherently drug resistant therefore it’s important to characterize them as treatment is species specific.10 In India, the prevalence of NTM varies from 0.5% to 27% in TB suspects.11,12 The most prevalent NTM species detected in the present study were M. intracellure, M.abcesess M. fortuitum, and M. simiae. M. intracellure has been reported as the predominant species in other studies from India, China, America10,13 and Japan also (33-65/100,000 cases in 2005).2 M. intracellure is a non-chromogenic, slow grower causing chronic pulmonary disease, local lymphadenitis, and disseminated opportunistic infections in HIV patients.14 It is widely found in tap water and has the ability to cause clinically serious illness.12
There are various methods used for the identification of NTM most common being the biochemical methods but they are time-consuming with low discrimination power. Similarly, though High Performance Liquid Chromatography (HPLC) is a rapid method but requires elaborate setup and trained personnel for interpretation of HPLC results.15,16 The GenoType Mycobacterium CM/AS assay was found to be reliable (91% valid results) and rapid method for identification of NTM, easy to perform and very suitable for a clinical laboratory as results are available within 3-5 days and treatment can be started timely.5,17 Though equipment and setup required for LPA is elaborate but all the labs under NTEP labs now have the molecular lab set up to do LPA as they are doing Genotype MTBDR and MTBDR plus LPA for detection of MDRTB and Extensively Drug Resistant TB reliable (XDRTB)18 Hence adopting this test is easy in the NTEP setting, there is no additional technical expertise required.19,20 Other molecular methods like sequencing are expensive and require technical expertise to perform molecular assays.21
The cost of LPA for NTM is its only limiting factor as there is no provision of identification of NTM in NTEP. The cost of LPA was found to be about Rs. 1,815 INR (25 US$). Similarly, other studies have reported the cost to be 25 US dollars and the cost of biochemical to be much cheaper at 2 dollars per test.22 Another limitation for its optimal use is that a minimum batch of ten samples have to be run at a time and maximum of 48 samples may be run at a time so in labs with low number of NTM samples either they have to wait to have an adequate number to available to test or waste reagents if results have to be given early. There were 3 isolates in our study that gave invalid results. These samples would require sequencing to identify the species.
For better discrimination of NTM it is anticipated that there should be a combination of two or more methods.16 Moreover, other methods like Matrix-Assisted Laser Desorption Ionization-Time of Flight Mass Spectrometry (MALDI-TOF MS) are now available, requiring expensive equipment as initial cost, but running cost is very low and rapid and easy to perform.23
Prevalence of NTM was low in our samples but should not be ignored, M intracellulare was the most frequent species isolated. Genotype Mycobacterium CM/AS assay was found to be a rapid, reliable, and easy to perform diagnostic tool and results are available in the clinically useful time frame. This test can be immediately used in NTEP setting due to the availability of infrastructure and technical expertise. The only limitation of the test is its cost which can sure be negotiable if implemented in the program setting.
ACKNOWLEDGMENTS
The authors would like to thank to the NTEP program, Foundation for Innovative Newer Diagnostics (FIND), India and SMS Medical College, Jaipur, India for their technical support and Hain Life sciences for providing kits to perform tests.
CONFLICT OF INTEREST
The authors declare that there is no conflict of interest.
AUTHORS’ CONTRIBUTION
SK performed the experiments and wrote the manuscript. BM conceived the research and edited the manuscript. WD edited the manuscript. SB helped in performing the experiments and in writing the manuscript. All authors read and approved the final manuscript for publication.
FUNDING
This study was supported by UGC (University Grants Commission) for providing research fellowship to Mrs. Shreya Khandelwal with Grant No.F.17-193/2014(SA-I)
ETHICS STATEMENT
This study was approved by the Institutional Ethics Committee, SMS Medical College Jaipur, India (Letter No: 3243 MC/EC/2017)
AVAILABILITY OF DATA
All data generated or analyzed during this study are included in the manuscript.
- Johnson MM, Odell JA. Nontuberculous mycobacterial pulmonary infections. J Thorac Dis. 2014;6(3):210-220.
Crossref - Jain S, Sankar MM, Sharma N, Singh S, Chugh TD. High prevalence of non-tuberculous mycobacterial disease among non-HIV infected individuals in a TB endemic country-experience from a tertiary center in Delhi, India. Pathog Glob Health. 2014;108(2):118-122.
Crossref - Patwardhan V, Chakravarti A. Non tubercular Mycobacteria (NTM) with special emphasis on drug resistance mechanisms, susceptibility testing and therapy. Jeevanu Times. 2014;14(1):22-25.
- Hawkey PM. The role of the polymerase chain reaction in the diagnosis of Mycobacterial infections. Rev Res Med Microbiol. 1994;5(1):27-32.
Crossref - Russo C, Tortoli E, Menichella D. Evaluation of the new GenoType Mycobacterium assay for identification of mycobacterial species. J Clin Microbiol. 2006;44(2):334-339.
Crossref - Kent PT, Kubica GP, Public Health Mycobacteriology: A Guide for the Level III Laboratory Atlanta, GA: US Department of Health and Human Services, Centers for Disease Control; 1985.
- Rishi S, Sinha P, Malhotra B, Pal N. A comparative study for the detection of Mycobacteria by BACTEC MGIT 960, Lowenstein Jensen media and direct AFB smear examination. Indian J Med Microbiol. 2007;25(4):383-386.
Crossref - Ribon W. Biochemical Isolation and Identification of Mycobacteria. In Jimenez-Lopez JC. (editor) Biochemical Testing: InTech 2012.
Crossref - Hain Lifescience. GenoType Mycobacterium CM Package Insert.Nehren, Germany: Hain Lifescience. 2011.
- Gopinath K, Singh S. Non-Tuberculous mycobacteria in TBendemiccountries: are we neglecting the danger? PLoS Negl Trop Dis. 2010;4(4):e615.
Crossref - Sharma SK, Sharma R, Singh BK, et al. A prospective study of non-tuberculous mycobacterial disease among tuberculosis suspects at a tertiary care centre in north India. Indian J Med Res. 2019; 150(5):458-467.
Crossref - Maurya AK, Nag VL, Kant S, et al. Evaluation of an immunochromatographic test for discrimination between Mycobacterium tuberculosis complex and non tuberculous mycobacteria in clinical isolates from extra-pulmonary tuberculosis. Ind J of Med Res. 2012;135(6):901-906. PMCID: PMC3410218
- Hoefsloot W, Van Ingen J, Andrejak C, al. The geographic diversity of nontuberculous mycobacteria isolated from pulmonary samples: an NTM-NET collaborative study. Eur Respir J. 2013;42(6):1604-1613.
Crossref - Greene JB, Sidhu GS, Lewin S, et al. Mycobacterium avium intracellulare: A cause of disseminated life-threatening infection in homosexuals and drug abusers. Ann Intern Med. 1982;97(4):53946.
Crossref - Butler WR, Guthertz LS. Mycolic Acid Analysis by High-Performance Liquid Chromatography for Identification of Mycobacterium Species Mycolic Acid Analysis by High-Performance Liquid Chromatography for Identification of Mycobacterium Species. Clin Microbiol Rev. 2001;14(4):704-726.
Crossref - Sebastian G, Nagaraja SB, Vishwanatha T, Hemalatha K, Vijayalakshmi N, Kumar P. Identification of Non-Tuberculous Mycobacterium by LPA (CM/AS) assay, HPLC and biochemical test: which is feasible for NTEP? Indian J Tuberc. 2018;65(4):329-334.
Crossref - Singh AK, Maurya AK, Umrao J, et al. Role of GenoType(®) Mycobacterium Common Mycobacteria/Additional Species Assay for Rapid Differentiation Between Mycobacterium tuberculosis Complex and Different Species of Non-Tuberculous Mycobacteria. J Lab Physicians. 2013;5(2):83-89.
Crossref - Chandak R, Malhotra B, Bhargava S, Goel S, Verma D, Tiwari J. Evaluation of MTBDRsl for detecting resistance in Mycobacterium tuberculosis to second-line drugs. Int J Tuberc Lung Dis. 2019;23(12):1257-1262.
Crossref - Lee AS, Jelfs P, Sintchenko V and Gilbert GL. Identification of non-tuberculous mycobacteria: Utility of the GenoType Mycobacterium CM/AS assay compared with HPLC and 16S rRNA gene sequencing. J Med Microbiol. 2009;58(7):900-904.
Crossref - Chihota VN, Grant AD, Fielding K, et al. Liquid vs. solid culture for tuberculosis: Performance and cost in a resource-constrained setting. Int J Tuberc Lung Dis. 2010;14(8):1024-1031. PMID: 20626948
- Philip K, Bottger EC. Species identification of mycobacteria using rDNA sequencing. Methods Mol Biol 1998;101:349-361.
Crossref - Couto I, Machado D, Viveiros M, Rodrigues L, Amarla L. Identification of nontuberculous mycobacteria in clinical samples using molecular methods: a 3-year study. Clin Microbiol Infect. 2010;16(8):1161-1164.
Crossref - Mediavilla-Gradolph MC, De Toro-Peinado I, Bermudez-Ruiz MP, et al. Use of MALDI-TOF MS for identification of nontuberculous mycobacterium species isolated from clinical specimens. Biomed Res Int. 2015;2015:854078.
Crossref
© The Author(s) 2022. Open Access. This article is distributed under the terms of the Creative Commons Attribution 4.0 International License which permits unrestricted use, sharing, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.