ISSN: 0973-7510
E-ISSN: 2581-690X
In developing countries diarrheal illness are causing severe health problem in children up to five years of age group and it also results in causing the morbidity and mortality. Worldwide among all the bacterial pathogens diarrheagenic Escherichia coli is responsible for epidemic and endemic diarrhoea. E. coli rarely cause disease in healthy individuals and most of E. coli strains live harmlessly in the intestines. The objective of the study was to detect six categories of DEC in one set of Multiplex PCR reactions. Total number of 155 in and outdoor children stool sample with complain of diarrhoea were collected in Rama Medical College, Kanpur, India. Target genes used for amplification by Multiplex PCR were eaeA and bfpA for EPEC, elt and stla for ETEC, CVD432 for EAEC and hlyA for EHEC. Out of 155 isolates examined, 71 were E.coli isolates, out of 71 isolates 28 were ETEC, 02 were EHEC. elt gene of ETEC and hlyA gene of EHEC diarrheagenic E.coli were detected. Antibiotic sensitivity tests for diarrheagenic strains were done by Kirby Bauer Method. Antibiotics used in the study were Ampicillin, Cotrimoxazole, Ciprofloxacin, Norflox, Nalidixic acid, Cefotaxime, Chloramphenicol, Imipenem, Azithromycin, Gentamicin. In most cases ETEC and EHEC diarrheagenic isolates were found resistant to Ampicillin, Gentamicin, Cotrimoxazole and Azithromycin. Multiplex PCR, a rapid method for detecting virulent genes of DEC at a time and it also provide a platform in understanding the diarrheal diseases in a more improved manner.
E. coli, Morbidity, Mortality, Antibiotic sensitivity, ETEC, EHEC, Endemic.
Globally diarrheal diseases in low- to middle- income countries and children under five are main cause of morbidity and mortality and also results in 0.5 million deaths1. Diarrhoea is caused by various numbers of parasites, bacteria and viruses2. E. coli is facultative anaerobic and non-pathogenic member of the human gastrointestinal microbiota3. Association with certain serotypes the diarrheagenic strains of E. coli is divided into five main categories: enterotoxigenic E. coli, enterohemorrhagic E. coli, enteropathogenic E. coli, enteroaggregative E. coli and enteroinvasive E. coli and are based on specific virulence determinants, distinct epidemiological and clinical features4. E. coli is normal flora and plays pathogenic role in human host. Worldwide Diarrheagenic E. coli (DEC) is reported as one of the main causes of gastrointestinal disorders5-8. Enterotoxigenic E. coli (ETEC) causes traveller’s diarrhoea and in population and it is a significant pathogenic form of E. coli9-11. Amplification of two marker genes elt and stla that encodes heat stable and heat labile secretory enterotoxins confirms ETEC in the stool specimen.
Gastrointestinal infections is caused by Enterohemorrhagic E. coli (EHEC), a subgroup of Shiga toxin- producing E.coli and it secretes toxins so EHEC colonizes large intestine which may lead to severe complications like (HUS)haemolytic uremic syndrome12.
Polymerase chain reaction (PCR) provides a rapid, profitable and sensitive method in identification of enteric pathogen in developing countries. This study was designed to calculate the preponderance of DEC in paediatric group below 5 years of age suffering from diarrhoea by multiplex PCR as they are present as commensal in non-diarrhoeal stool samples of healthy children and to determine the aetiology of infantile diarrhoea by examining the pathogenic E. coli strains acridity property.
Sampling
155 diarrheal patients aged between 0 to 5 years with ailment of abdominal pain, vomiting, fever, three or more loose stools/day and mucus or blood in stool, admitted or opd to Rama Medical College Hospital, Kanpur were collected.
Identification and isolation of E. coli Isolates
All faecal specimens were first processed for wet mount for screening of parasites as an exclusion criterion. Then gram staining was done for screening of gram negative bacteria and presence of pus cells. Gram negative bacteria cultured on Eosin Methylene Blue (EMB) appear as green with black metallic sheen colony, are again sub cultured on fresh EMB agar plates to obtain presumptive E. coli isolates. These presumptive E. coli isolates were confirmed by the confirmatory biochemical test (IMViC) for E. coli.
Isolation of E. coli O157:H7
The confirmed E. coli isolates were sub cultured on Sorbital MacConkey Agar plates (SMAC), and its colonies appears colourless on SMAC were presumptive method of E. coli O157.
Molecular Characterization
All gram negative lactose fermenting growths of E. coli were confirmed by molecular characterisation method. DNA of all lactose fermenting growth of E. coli strains was extracted by Qiazen DNeasy Ultra clean microbial kit.
E. coli colonies for the virulence genes were confirmed by Multiplex PCR. The primers (Table 1) selected for six different virulence genes (elt, eaeA, stla, bfp, CVD432 and hlyA) concurrently in a single reaction. EHEC and ETEC were identified by the appearance of the elt and hlyA genes, which are present only in ETEC and EHEC. Total volume of 25μl in a reaction mixture is performed by adding 12.5μl PCR master mix 2x, manufactured by Quantitect multiplex PCR kit containing 400 mM deoxy-nucleoside triphosphates, 0.05 U/μlTaq DNA polymerase, 4 mM MgCl2, 5.5μl sterile double distilled water, 1μl from each primers, and 5μl DNA template. Cycling parameter used are as follows: 94°C for 4 min for initial denaturation of the DNA, then 30 cycles for 30 sec at 94°C, 30sec at 59°C, 1.30 min at 72°C, and lastly final single prolonged extension at 72°C for 10 min for elt genes, and 3 min at 95°C, then 35 cycles for 30sec at 94°C, 30 sec at 55°C, 30 sec at 72°C, and finally a single prolonged elongation at 72°C for 10 min. The amplified product were visualized by gel electrophoresis in 1.5% agarose gel containing ethidium bromide for 45 min at 100 V and then it is visualized under UV light of gel documentation system.
Table (1):
Biochemical test applied for screening of E. coli in paediatric patients.
S. No. |
Indole |
MR |
TSI |
Citrate |
Urease |
Mannitol |
Total (N) |
Positive |
|---|---|---|---|---|---|---|---|---|
1 |
Positive |
Positive |
A/A |
Negative |
Negative |
Positive |
155 |
71 |
Antibiotic Sensitivity testing
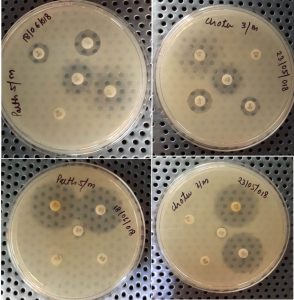
Antibiotic sensitivity test for the isolates that yielded positive results in the Multiplex PCR assay were cultured on Mueller-Hinton agar by the Kirby- Bauer method, following Clinical and Laboratory Standard Institute (CLSI)13. Ten antibiotic discs (Himedia) of different classes that are generally used in medical diagnostic laboratories were selected. The discs used were Nalidixic acid (NA, 30μg), gentamicin (G, 10μg), ampicillin (AMP, 10μg), Cefotaxime (CTX, 10μg), Cotrimoxazole (SXT, 25μg), Imipenem (IPM, 10mcg), Azithromycin (AZM, 15μg), Norflox (NOR, 10μg), ciprofloxacin (CIP, 5μg), chloramphenicol (C, 30μg).
Ethical consideration
Institutional ethics review board of Rama Medical College, Kanpur approved the study. Before enrollment into the study, written consent was taken from parents/guardians of the children.
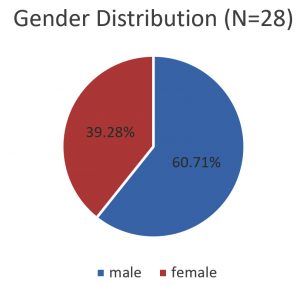
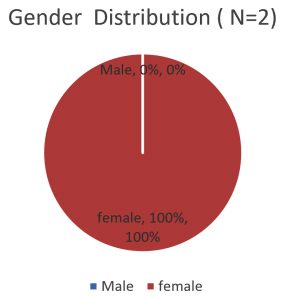
Out of 71 E. coli isolates (five-six colonies from each stool sample) which were identified by biochemical tests (Table 1), 30 isolates were identified as diarrheagenic E. coli by Multiplex PCR. Out of which 28 (39.4%) were ETEC (elt positive) and 02(2.8%) were EHEC (hlyA positive) as shown in Fig. 1, 2 and 3. Gender distribution was 17(60.71%) female and 11(39.28%) male in case of ETEC strains and 0 (00%) male and 2 (100%) female in case EHEC strains shown in Fig. 4 and 5. The sex distribution was 17 (60.71%) male and 11 (39.28%) female in case of ETEC strains.
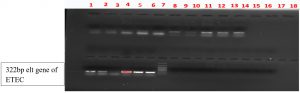
Fig. 1. Multiplex PCR of pediatric samples. Lane 2-7 and 11-14 ETEC ( amplicon size 322bp) and Lane 9-10 EHEC (amplicon size534bp) Lane 8: Ladder (100bp -1000bp).
Fig. 2. Lane: 1, 2, 4, 5 and 6 contains elt gene of 322bp of ETEC gene and Lane 7 contain ladder (100bp-1000bp).
Fig. 3. Lane: 1-7 and 9-14(13samples) contains elt gene of 322bp of ETEC gene and Lane 8: Ladder (100bp-1000bp).
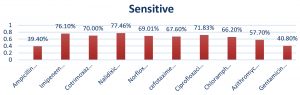
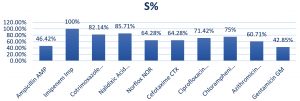
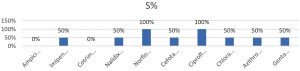
Antimicrobial susceptibility testingThe result of antibiotic sensitivity testing of ten antibiotics for 71 E. coli isolates isolated from diarrheal patients are shown in Fig. 6 and 7. The most common sensitive profile of antibiotics found were Imipenem, Nalidixic acid and ciprofloxacin. Susceptible rate of 100% Imipenem was found to be the most effective antibiotic in 28 strains of ETEC strains (Fig. 8). Of the 2 EHEC strains tested, cefotaxime and chloramphenicol was found to be most sensitive with susceptible rate of 100%
(Fig. 9).
Worldwide diarrheagenic E. coli strains are found to be the most important paediatric enteropathogens14. Bacterial and parasitic pathogens with rotavirus, viral, diarrheagenic E. coli, Shigella spp. and Salmonella spp are the cause of diarrhoea in developing countries15. In the current research work blood was found in diarrheal patients with the EHEC patho type, but two of them belonged to the O157:H7 serotype. In previous studies it is found that EHEC was less frequently isolated than other of diarrheagenic E. coli16. Concurrence with the low profile of EHEC infection was noted in the current study. Vilchez et al. found few EHEC strains in diarrheal patient in developing countries17. In the present study, diarrhoea causing E. coli has the highest rate of ETEC (39.4%) strains. ETEC strains were detected significantly more (60.71%) in adult man with diarrhoea. In some studies in developing countries it was observed that EHEC was less frequently isolated than ETEC or EPEC strains18. The most important pathogen responsible for traveller’s diarrhoea is ETEC and it causes approx 700,000 childhood deaths per year, mostly in paediatric group19. Widespread use of antibiotics is the cause of high incidence of antibiotic-resistant in isolates of DEC. Diverse resistance construction to usual antibiotic results is due to transfer of resistance genes between species. The ecological and pathogenic characters of bacterial species are changed by these transfers20. In our study, the rate of antibiotic-resistant is high in isolates of EHEC was seen to Ampicillin and Cotrimoxazole (100%). The drug sensitivity pattern of our isolates demonstrated a good response to Norflox and Ciprofloxacin (100%). There are consistent reports from other authors that frequent resistance to common antibiotics such as tetracycline, ampicillin and trimethoprim-sulfamethoxazole was detected in ETEC strains21-26.
The detection of ETEC and EHEC in this region suggested that this strain is main common source of diarrhoea among paediatric age group. Education on diarrheal infections management should be taught to the parents/guardians of the children affecting with diarrhoea as well as the implementation of more stringent policies governing the availability of antibiotics should be given. Heavy clinical use of antibiotics, and have a low cost leads to high-level resistance to antibiotics. The indiscriminate use of antibiotics needs to be avoided and the guidelines should be made for proper use of antibiotics for treatment of diarrhoea in the region needs to be established.
ACKNOWLEDGMENTS
The study was conducted with the support of expertise staff of the Department of Microbiology, Rama Medical College, Kanpur .The authors are thankful for their immense co-operation and assistance in the collection of specimen and processing.
CONFLICT OF INTEREST
There are no conflicts of interest.
AUTHORS’ CONTRIBUTION
All authors listed have made equal substantial and intellectual contribution to the work, and approved it for publication.
FUNDING
None.
ETHICS STATEMENT
Study is ethically approved by Rama University, Kanpur U.P. with reference no. 90 Res/Reg/RUK/2017.
AVAILABILITY OF DATA
All datasets analysed in the study are included in the manuscript and presented as tables and figures.
- Diarrhoeal disease. [Cited 6 May 17]. Available from: http://www.who.int/mediacentre/factsheets/fs330/en/.
- Hodges Kzim, Gill R. Infectious diarrhea Cellular and molecular mechanisms. Gut Microbes: Landes Bioscience, 2010; 1(1): 4-21.
Crossref - Trung Vu Nguyen,Phung Le Van, Chinh Le Huy, Khanh Nguyen Gia,Andrej Weintraub. Detection and Characterization of Diarrheagenic Escherichia coli from Young Children in Hanoi, Vietnam. Journal Of Clinical Microbiology, 2005; 43(2): 755–760.
Crossref - Jafari A, Aslani MM, Bouzari S. A brief review of diarrheagenic pathotypes and their role in diarrheal diseases in Iran. Iranian Journal of Microbiology,2012; 4(3): 102-117.
- Frank C, Werber D, Cramer JP, Jakob P. Cramer, M.D., Mona Askar, M.D., Mirko Faber, an der Heiden M, Bernard H, Fruth A, Prager R, Spode A, Maria Wadl, Zoufaly A. Epidemic profile of Shiga- toxin– producing Escherichia coli O104: H4 outbreak in Germany. N Engl J Med, 2011; 365: 1771-1780.
Crossref - Radosavljevic V, Finke EJ, Belojevic G. Analysis of Escherichia coli O104: h4 Outbreak in Germany in 2011 Using Differentiation Method for Unusual Epidemiological Events. Cent Eur J Public Health, 2016; 24(1): 9-15.
Crossref - Guzman-Hernandez R, Contreras-Rodriguez A, Hernandez-Velez R, Martinez Perez I Merino Lopez, Zaidi B M, Gracia- ET. Mexican unpasteurised fresh cheeses are contaminated with Salmonella spp., non- O157 Shiga toxin producing Escherichia coli and potential uropathogenic E. coli strains: a public health risk. Int J Food Microbiol, 2016; 237: 10-16.
Crossref - Ochi S, Shah M, Odoyo E, Bundi M, Miringu G, Guyo S, Wandera E, Kathiiko C, Kariuki S, Karama M, Tsuji T, Ichinose Y. An Outbreak of Diarrhea in Mandera, Kenya, Due to Escherichia coli Serogroup O- Nontypable Strain that had a Coding Gene for Enteroaggregative E. coli Heat- Stable Enterotoxin 1. ASTMH. 2016; 96(2):457-464.
Crossref - Dutta S, Guin S, Ghosh S, Pazhani PG, Rajendran K, Bhattacharya KM, Takeda Y, Nair B, Ramamurthy T, Gordon VS. Trends in the prevalence of diarrheagenic Escherichia coli among hospitalized diarrheal patients in Kolkata, India. PloS One, 2013; 8(2): e56068.
Crossref - Kaper JB, Nataro JP, Mobley HL. Pathogenic Escherichia coli .Nature Rev Microbiol., 2004; 2: 123-140.
Crossref - Campos LC, Franzolin MR, Trabulsi LR. Diarrheagenic Escherichia coli categories among the traditional enteropathogenic E. coli O serogroups: a review. Mem Inst Oswaldo Cruz, 2004; 99(6): 545-552.
Crossref - Y Nguyen, Sperandio V. Enterohemorrhgic E.coli (EHEC) Pathogenesis. Front Cell Infect Microbiol, 2012; 2: 90.
Crossref - Mohamed SM Nassar, Walaa A Hazzah, Wafaa MK Bakr. Evaluation of antibiotic susceptibility test results: how guilty a laboratory could be? Journal of the Egyptian Public Health Association, 2019; 94: 4.
Crossref - Ochoa JT, Contreras AC. Enteropathogenic E. coli (EPEC) infection in children. Curr Opin Infect Dis, 2011; 24(5): 478–483.
Crossref - Saeed A, Abd H, Sandstrom G. Microbial aetiology of acute diarrhoea in children under five years of age in Khartoum, Sudan. J Med Microbiol, 2015; 64(4): 432–437.
Crossref - Bouzari S, Farhang E, Hosseini MS, Alikhani YM. Prevalence and antimicrobial resistance of shiga toxin-producing Escherichia coli and enteropathogenic Escherichia coli isolated from patients with acute diarrhea. Iran J Microbiol, 2018; 10(3): 151–157.
- Vilchez S, Reyes D, Paniagua M, Bucardo F, Mollby R, Weintraub A. Prevalence of diarrhoeagenicEscherichia coli in children from Leon, Nicaragua. J Med Microbiol, 2009; 58: 630–637.
Crossref - Nguyen TV, Le Van P, Le Huy C, Gia KN, Weintraub A. Etiology and epidemiology of diarrhea in children in Hanoi, Vietnam. Int J Infect Dis,2006; 10: 298–308.
Crossref - WHO Future directions for research on enterotoxigenicEscherichia coli vaccines for developing countries. Wkly Epidemiol Rec, 2006; 81: 97.
- Ochman H, Lawrence JG, Groisman EA. Lateral gene transfer and the nature of bacterial innovation. Nature, 2000; 405: 299–304.
Crossref - Jordi Vila, Tibor Pal. Update on Antibacterial Resistance in Low-Income Countries: Factors. The Open Infectious Diseases Journal, 2010, 4: 38-54.
Crossref - Estrada-Garcia T, Cerna JF, Paheco-Gil L, Velazquez RF, Ochoa TJ, Javier Torres, Herbert L. DuPont. Drug-resistant diarrheagenicEscherichia coli, Mexico. Emerg Infect Dis,2005; 11(8): 1306–1308.
Crossref - J Ayatollahi, S H Shahcheraghi, R Akhondi, SS Soluti. Antibiotic Resistance Patterns of Escherichia coli Isolated from Children in Shahid Sadoughi Hospital of Yazd Iran. J Ped Hematol Oncol, 2013; 3(2): 78–82.
- Al-Gallas N, Abbassi SM, Hassan AB, Aissa RB.Genotypic and phenotypic profiles of enterotoxigenic Escherichia coli associated with acute diarrhea in Tunis, Tunisia. CurrMicrobiol,2007; 55(1): 47–55.
Crossref - Mosquito S, Ruiz J, Pons JM, Durand D,Barletta F, Ochoaa JT. Molecular mechanisms of antibiotic resistance in diarrhoeagenic Escherichia coli isolated from children. Int J Antimicrob Agents,2012; 40(6): 544–548.
Crossref - Ochoa TJ, Ecker L, Barletta F, Mispireta ML, Valle del J L, Martha Vargas O, Gil I A, Ecker L, Barletta F, Hall R E, Cleary GT, Lanata FC.High frequency of antimicrobial resistance of diarrheagenic E. coli in Peruvian infants. Am J Trop Med Hyg, 2009; 81(2): 296–301.
Crossref
© The Author(s) 2020. Open Access. This article is distributed under the terms of the Creative Commons Attribution 4.0 International License which permits unrestricted use, sharing, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.