ISSN: 0973-7510
E-ISSN: 2581-690X
The genus Marasmius is distributed worldwide, along with a precise identification of this fungus as a pathogen on the Imperata cylindrical plant, although its bioactive role is not well understood. The current work was designed to examine the population of Marasmius palmivorus and its antagonistic patterns. Sequencing the ITS genetic region of this fungus occurs through verifying the cladistics nature, and taxonomic validity, in addition to studying the antagonistic potential of this fungus. M. palmivorus (MG717877.1) was recorded in this plant, which represents a new finding in Iraq mycobiota. Data from the sequence alignment were used to plot a phylogenetic tree, using the Neighbor-Joining method (NJ) in molecular evolutionary genetics analysis based on Mega 7. Phylogenetic trees show a close relationship of 99% between this strain of M. palmivorus (MG717877.1) and international strains. On the other hand, the maximum antagonistic effect of M. palmivorus has been found to act against Fusarium solani and F. thapsinum, and to a lesser degree against Penicillium sp., T. harzianum, and P. cyclopium after 6 to 8 days of incubation. Results confirm the precise nomination of this taxon is M. palmivorus, which may act as a bioherbicidal and could be used as bioagent against various phytopathogenic fungi.
Marasmius palmivorus, Phylogenetic structure, Antagonism.
Marasmius is a genus of mushroom-forming fungi in the family Marasmiaceae, order: Agaricales, class: Agaricomycetes, division: Basidiomycota1. In 2005, Kirk2 listed 955 species belonging to Marasmius in the Index Fungorum. There has been recent attention in understanding worldwide biodiversity and the evolution of Marasmius3-7.
Marasmius palmivorus is most phonetically like to a complex of the Marasmiellus species, which is reportedly pathogenic in a range of economically important monocots, including bananas, oil palms, sugar cane, corn, maize, taro (Colocasia esculenta), turf grass, cocoa, and reed plants8-14. Marasmius is not closely related to the species Marasmiellus, as it is best to use its present taxon – where it was originally described15, and until more data from multiple genes and a large Marasmiaceae dataset provide the information to clarify its taxonomic position16.
Members of the genus Marasmius are saprotrophic fungi, often in forests, that play a key role in the breakdown of wood and leaf litter, nutrient cycling, and soil genesis; this is where they serve an important ecological role in the biodegradation of lignocellulosic material17. Fifty cultures, investigated for enzyme activities and antimicrobial and antioxidant properties, indicate that the Marasmius species may have potential applications in biotechnology17.
Based on our knowledge, there is no scientific report available to characterize Imperata cylindrical wilt disease, caused by the Marasmiys genus. Therefore, the current study was carried out to determine the molecular sequence of the causative agent of this disease by sequencing the ITS-5.8s-ITS2 region of rDNA and investigating the antagonistic potential of this agent against fungal pathogens; however, this is poorly documented, and may be used to develop new biocontrol agents.
Source of fungal isolates
Fusarium solani, F. thapsinum, Penicillium sp., T. harzianum and Penicillium cyclopium are procured from the Advanced Mycology Unit, Biology Department, College of Science, University of Babylon.
Isolation and identification
Disease samples from stems and rhizomes of I. cylindrical are grown close to the reed plant or in agricultural fields, and were collected from 20 agricultural fields in Babylon Province, Iraq, in November, 2017. Samples were transferred to the laboratory, with inoculation and incubation conducted as described14. Identification of fungal isolates were based on morphological and microscopic features, such as characteristics of colony and mycelium clamp connections, using 40X Microscopic Objective Lens18,19.
Growth and maintenance of fungal isolates
Due to the similarity among all isolates of Imperata Cylindrical Wilt Disease Fungus (IcWDF), only one isolate was chosen for the following experiments. The microbial cultures include IcWDF isolate, Fusarium solani, F. thapsinum, Penicillium sp., T. harzianum, and Penicillium cyclopium, which were separately inoculated in petri dishes with potato dextrose agar (PDA) (pH 7.0), followed by incubation for 5 days at 26 ± 2oC. To make the slants, 20 ml of PDA was poured into glass tubes and left until solidified. Fungal isolates were maintained at 50C in a refrigerator, followed by subculturing at regular intervals (every 30 days)20.
Preparation of fungal inoculum
The inoculum of IcWDF was prepared with 250 ml of millet seeds (Panicum miliaceum L.) in conical flasks, according to the procedure described by Dewan and Sivasithamparam (1989)21 and others14,22.
Pathogenicity
Twenty uniform I. cylindrical plants at similar age were selected from vegetable fields and from the channels of irrigation and planted as described by Tamur et al.14. I. cylindrical plants were grown for 2 months under glasshouse conditions (25±5 oC). The pots were then inoculated individually with a causal agent, by mixing the previously prepared inoculum in the pot soil at a rate of 5% each. Ten pots without inoculation served as controls. The symptoms of disease were reported at 15 days after inoculation14.
DNA extraction
The fungal isolate from IcWDF was recultured on PDA, as previously mentioned, and incubated at 28 ± 2oC for 3 days. The fungus cells were collected and then transported to a new sterilized tube for DNA extraction, which was achieved with a special purification kit (Genomic DNA purification kit, Zymogene, Orlando, FL, USA), and used according to the manufacturer’s protocol.
Detection of gene ITS by using PCR
Detection of ITS gene was conducted by using primers for amplification. A fragment 565 bp of ITS was amplified using a forward primer (ITS1 F: 5′ – TCCGTAGGTGAACCTGCGG -3′) and a reverse primer (ITS4 R:5′ TCCTCCGCTTATTGATATGC-3′) (Primer set supplied by IDT (Integrated DNA Technologies, Coralville, IA, USA). The PCR amplification was performed in a volume of 25µl, containing 1.5µl DNA, 5 µl Taq PCR Pre Mix (Intron Biotechnology, Seoul, S. Korea), 1 µl of each primer (10 pmol), at which point distilled water was added into a tube for a total volume of 25 µl. The thermal cycling conditions were as follows: denaturation at 94°C for 3 min, followed by 35 cycles of 94°C for 45 s, 52°C for 1 min, and 72°C for 1 min, with final incubation at 72°C for 7 min, using a thermal cycler (Gene Amp, PCR system 9700; Applied Biosystem, Waltham, MA, USA). The PCR products were separated by 1.5% agarose gel electrophoresis and visualized by exposure to ultraviolet light (302 nm) after red staining (Intron, Seoul, S. Korea).
Gene sequencing was done at the National Instrumentation Center for Environmental Management (nicem) online (http://nicem.snu.ac.kr/main/?en_skin=index.html), biotechnology lab, with a DNA sequencer 3730XL, Applied Biosystems, Waltham, MA, USA). A homology search was conducted using a Basic Local Alignment Search Tool (BLAST) program, available at the National Center of Biotechnology Information (NCBI) online at (http:// www.ncbi.nlm.nih.gov) and the BioEdit program. After morphological and molecular identification, these experiments were carried out:
Bioactivity of Marasmius palmivorus Temperature
The growth of M. palmivorus was measured at various temperatures (15, 20, 25, and 30oC). 20 ml PDA were added to sterile petri dishs, each plate, was inoculated by taking 0.5 cm from the edge of colonies. Plates were separated into 5 groups before incubation for 3 days (4 replicates each). Radial growth of this fungus (from two intersecting lines in the center of the dish.) was daily estimated on an Petri plate until full expansion of growth, with different temperature treatments clearly determined.
pH
Different ranges of pH (4.5, 5.5, 6.5, 7.5, and 8.5) were established for the growth of M. palmivorus, using a sterilized petri-dishes (20 ml of PDA each). The inoculation, the incubation process, and the fungal growth measurement were achieved as described in the previous paragraph (for temperature).
Growth of Marasmius palmivorus in different nutrient sources
Different nutrient sources were prepared by collecting leaves of wheat, I. cylindrical, and reed (youngest expanded). Leaf samples were dried in the shade, as well as in an oven (60÷C) for 48 h, and grinded into fine powder by an electronic blender14. All leaf powders were stored in sterilized containers until usage23.
Preparation of media was achieved by dissolving 2 g of each nutrient source, in 100 ml of distilled water with or without dextrose using a magnetic stirrer. Supplementation with dextrose and agar growth medium were carried out at the same rate as for PDA, then mixed gently and autoclaved at 15 psi for 15 min, with PDA as a control treatment. Autoclaved media were poured into petri plates for each type, with the plates inoculated after solidification with fungal tissue (0.5 cm) taken from the edge of an M. palmivorus colony (4 days old). Three replicates of each source were taken and incubated for 6 days at 28 ± 20C24. Radial growth was regularly recorded as mentioned before.
Antagonistic activity of Marasmius palmivorus: Pathogenic and nonpathogenic fungi
The antagonistic activity of M. palmivorus isolate was screened for the growth of pathogenic and nonpathogenic fungi, including Fusarium solani, F. thapsinum, Penicillium sp., T. harzianum, and Penicillium cyclopium by using the dual culture technique25. Five mm discs of tested fungus were placed individually in the center of each PDA plate half, whereas the other half was inoculated with IcWDF followed by incubation at 26 ± 2°C, in a controlled experiment, with each fungus grown on PDA. Growth inhibitory antagonistic activity of the tested fungus against phytopathogenic fungi was assessed in terms of percent inhibition in cm by using this formula26:
Inhibition % = (r1-r2/r1) ×100,
where, r1 is the diameter of phytopathogenic fungal growth without IcWDF, while r2 is represents the radial growth of the pathogenic fungus with IcWDF. Furthermore, all treatments were carried out in three replicates.
Statistical analysis
The current work were arranged in a randomized complete block design. Data were assessed by analysis of variance. The level of significance was determined by Fisher’s least significant difference (LSD) comparisons at the 5% probability level.
Growth Characters of Marasmius palmivorus
Disease symptoms
Infection of I. cylindrical by IcWDF fungus was appeared as wilt symptoms on stems and rhizomes, which produced blight symptoms on the leaves. Symptoms primarily initiated on the base of the stem’s lower section as brown to black rot lesions. Mycelium which was grown rapidly show u as white cottony growth around the plant base (Fig. 1A).
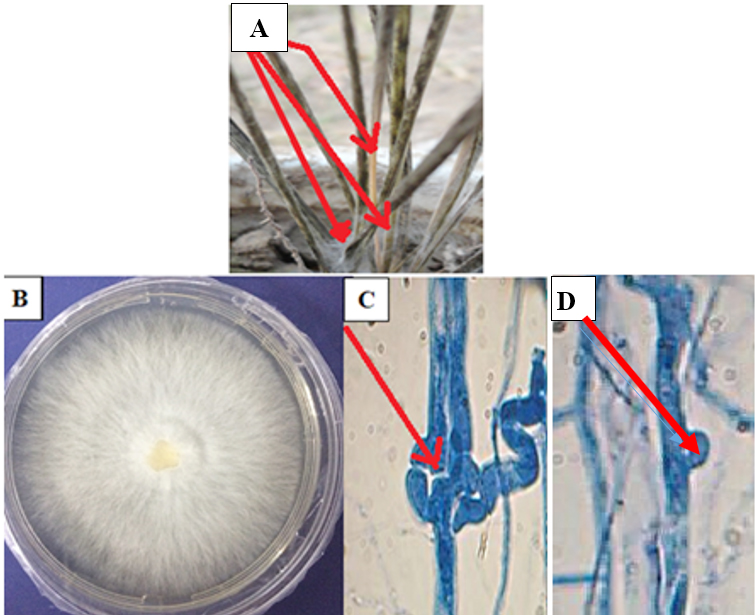 Fig. 1. Disease symptoms of wilt disease caused by M. palmivorus on I. cylindrical plants at 15 days after inoculation (A), morphological feature of M. palmivorus growing on PDA at 28 ± 2oC after 4 days of incubation (B), septal wall of fungal hyphae (C), and clamp connections (D), under a light compound microscope, 25 µm (40X-1 at 10 days after incubation)
Fig. 1. Disease symptoms of wilt disease caused by M. palmivorus on I. cylindrical plants at 15 days after inoculation (A), morphological feature of M. palmivorus growing on PDA at 28 ± 2oC after 4 days of incubation (B), septal wall of fungal hyphae (C), and clamp connections (D), under a light compound microscope, 25 µm (40X-1 at 10 days after incubation)IcWDF isolate was tentatively identified as M. palmivorus, according to colony features. Microscopic observation revealed that this fungus produced wilt colonies on petri dishes and covered the plate within 3 to 4 days at 28 ± 2oC, with hairy-branched and hyaline mycelium (Fig. 1B). Colonies were creamy after 10 days of inoculation. Septation is recognized in the fungal mycelium (Fig. 1C), with clamp connections clearly distinguished (Fig. 1D). Spores, and reproductive aspects were not observed under the light microscope examination, with a temperature range of 28 ± 2oC. The macroscopic fruiting body of M. palmivorus was not detected under lab conditions.
Pathogenicity
The signs and symptoms of M. palmivorus were confirmed according to Koch’s postulates. Different symptoms of I. cylindrical wilt disease were recorded after 2 weeks and found to be similar to symptoms observed during natural infection. In controlled experimentation, no disease symptoms appeared. Death of I. cylindrical plants tended to occur after 3 weeks, while the infection process increased in moist circumstances.
Phylogenetic analysis
In this study, the confirmation process of one isolate, M. palmivorus, was conducted by conventional PCR technique to detect the presence of a specific gene, with sequence analysis of the ITS1-5.8s-ITS2 region, and BLAST analysis from the NCBI database. After sequencing, this isolate was identified. The extracted genomic DNA of these isolates was used as a template spacer (ITS). The PCR amplification products show that M. palmivorus yielded about 565 pb. The product is shown in Fig. 2.
 Fig. 2. PCR product, the band size of 565 bp. The product was electrophoresis on 1.5% agarose at 5 volt/cm2. Also, 1x TBE buffer for 1.5 h, DNA ladder (100), lane (1 to 2) PCR product of band size 565 bp, visualized under UV light
Fig. 2. PCR product, the band size of 565 bp. The product was electrophoresis on 1.5% agarose at 5 volt/cm2. Also, 1x TBE buffer for 1.5 h, DNA ladder (100), lane (1 to 2) PCR product of band size 565 bp, visualized under UV lightPhylogenetic analysis by rDNA sequencing found a genetic variation among isolates from M. palmivorus in other countries. BLAST results were explored by searching with partial nucleotide sequences at the gene bank database (Table 1).
Table (1):
Homology sequence identified for local Marasmius palmivorus isolate.
ACCESSION |
Gene |
country |
Source |
Compatibility |
|---|---|---|---|---|
ID: MG251431.1 |
18S ribosomal RNA |
India |
Marasmius palmivorus |
99% |
ID: MF100969.1 |
18S ribosomal RNA |
USA
|
Marasmius palmivorus |
98% |
ID: KR056289.1 |
18S ribosomal RNA |
Philippines
|
Marasmius palmivorus |
98% |
ID: KJ865843.1 |
18S ribosomal RNA |
India
|
Marasmius palmivorus |
98% |
ID: JQ653445.1 |
18S ribosomal RNA |
Malaysia
|
Marasmius palmivorus |
98% |
ID: JQ653435.1 |
18S ribosomal RNA |
Malaysia
|
Marasmius palmivorus |
98% |
ID: MF100965.1 |
18S ribosomal RNA |
USA\ California
|
Marasmius palmivorus |
98% |
ID: KT273356.1 |
18S ribosomal RNA |
Kenya
|
Marasmius palmivorus |
98% |
ID: KR056290.1 |
18S ribosomal RNA |
Philippines
|
Marasmius palmivorus |
98% |
ID: JQ653446.1 |
18S ribosomal RNA |
Malaysia
|
Marasmius palmivorus |
98% |
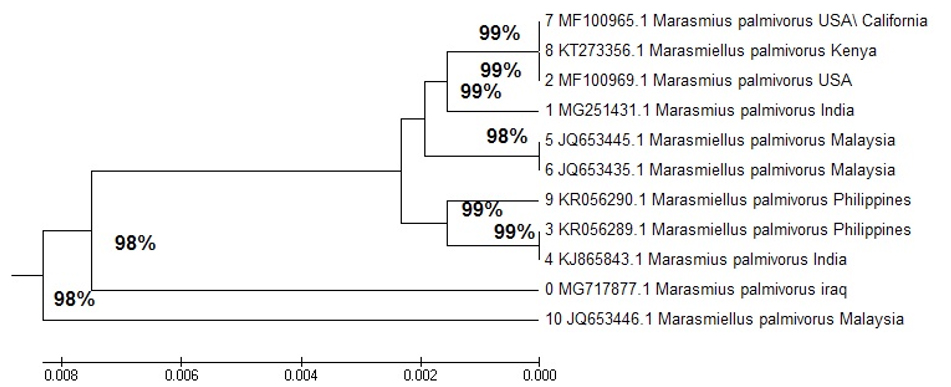
A phylogenetic isolate tree in Iraq showed a close relationship with M. palmivorus in India, identified at 99% (4 KC771224.1), in the Philippines at 98% (4 KR05 6289.1), and in Kenya at 98% (9 KT27 3356.1). M. palmivorus in Malaysia was identified at 98% (7 JQ65 3440.1), with a distant relationship to M. palmivorus (MG717877) in Iraq (Fig. 3 and Table 1).
Bioactivity of Marasmius palmivorus
Temperature
The results of this study revealed a substantial effect of temperature (Fig. 4) for the growth of M. palmivorus. The optimum growth occurred at 30°C after a 3-day incubation, followed by 25°C, then 20°C, while the lowest growth occurred at 15oC. The rising or lessening temperatures below 15°C, or higher than 30°C, caused obstruction of M. palmivorus growth.
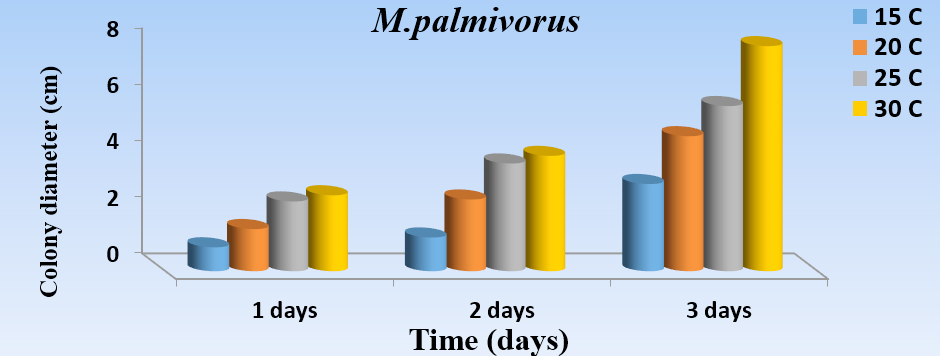 Fig. 4. Effect of different temperatures: 15oC (A), 20oC (B), 25oC (C), and 30oC (D) in the growth diameter of M. palmivorus on PDA after 3 days of incubation (LSD(0.05)=0.263)
Fig. 4. Effect of different temperatures: 15oC (A), 20oC (B), 25oC (C), and 30oC (D) in the growth diameter of M. palmivorus on PDA after 3 days of incubation (LSD(0.05)=0.263)pH
As a constant, pH (4.5, 5.5, 6.5, 7.5, and 8.5) has a significant effect on the growth of M. palmivorus with PDA, at temperatures 28 ± 2˚C. Maximum growth of M. palmivorus was obtained at pH 7.5 (8 cm), followed by pH 6.5 (6.3 cm), with the radial growth of this fungus increasing at pH 5.5 (6 cm), which was less than pH 7.5 and 6.5, and higher than pH 8.5 (5 cm). Growth of this fungus decreased to 4.5 cm at pH 4.5 at the end of the incubation (Figs. 5A and 5B).
 Fig. 5.A Effect of different pH levels (4.5, 5.5, 6.5, 7.5, and 8.5 on colony growth of M. palmivorus after 4 days of incubation)
Fig. 5.A Effect of different pH levels (4.5, 5.5, 6.5, 7.5, and 8.5 on colony growth of M. palmivorus after 4 days of incubation)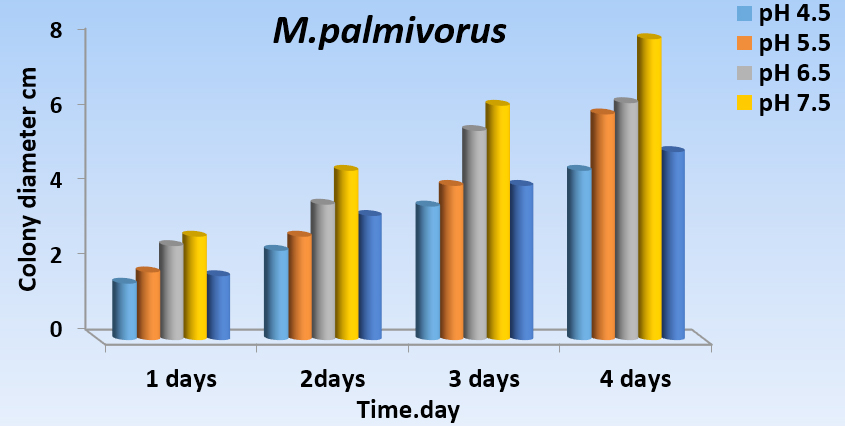 Fig. 5.B Effect of different pH levels (4.5, 5.5, 6.5, 7.5, and 8.5) on growth diameter of M. palmivorus after 4 days of incubation (LSD(0.05)=0.377)
Fig. 5.B Effect of different pH levels (4.5, 5.5, 6.5, 7.5, and 8.5) on growth diameter of M. palmivorus after 4 days of incubation (LSD(0.05)=0.377)Growth of Marasmius palmivorus in different nutrient sources
Growth of M. palmivorus varied considerably, according to the composition of growth media, which was most profound with wheat and reed leaves, supplemented with powdered material in the medium (Fig. 6). Therefore, the growth diameter of M. palmivorus was dramatically affected by the source of the added nutrient. After 4 days of incubation, the growth of the tested fungus significantly differed in wheat (6.8), reed (9), caladium (8.5) supplemented with dextrose, compared to PDA. In contrast, the average growth reached 5.5, 8.3, and 7.1 in wheat, reed, and caladium, respectively, which were not supplemented with dextrose (compared to PDA). Thus, proportional fungal growth was increased in all formulated media with and without dextrose, except in the wheat treatment, which was not supplemented with dextrose (Fig. 7).
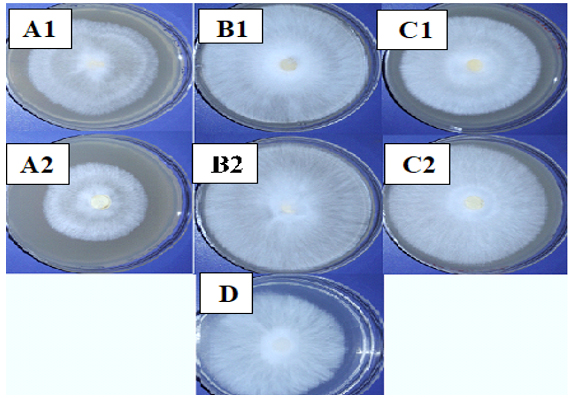 Fig. 6. Colony growth of M. palmivorus on culture media from powdered leaves of the following plants with dextrose (1) and without dextrose (2): A (wheat), B (reed), C (I. cylindrica), and D (PDA) after 4 days of incubation at 30oC
Fig. 6. Colony growth of M. palmivorus on culture media from powdered leaves of the following plants with dextrose (1) and without dextrose (2): A (wheat), B (reed), C (I. cylindrica), and D (PDA) after 4 days of incubation at 30oC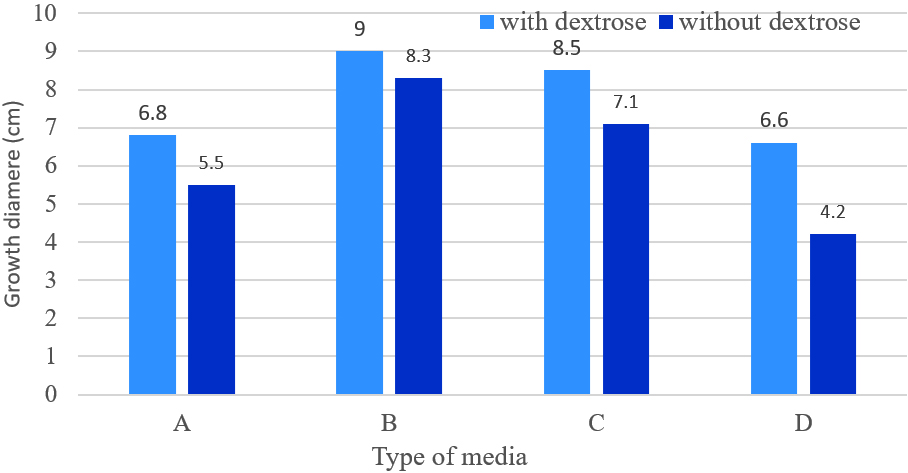 Fig. 7. Growth diameter of M. palmivorus on culture media of powdered leaves of the following plants with dextrose (1) and without dextrose (2): A (Wheat), B (Reed), C (I. cylindrica), and D (PDA) after 4 days of incubation at 30oC.(LSD(0.05)=0.631)
Fig. 7. Growth diameter of M. palmivorus on culture media of powdered leaves of the following plants with dextrose (1) and without dextrose (2): A (Wheat), B (Reed), C (I. cylindrica), and D (PDA) after 4 days of incubation at 30oC.(LSD(0.05)=0.631)Antagonism
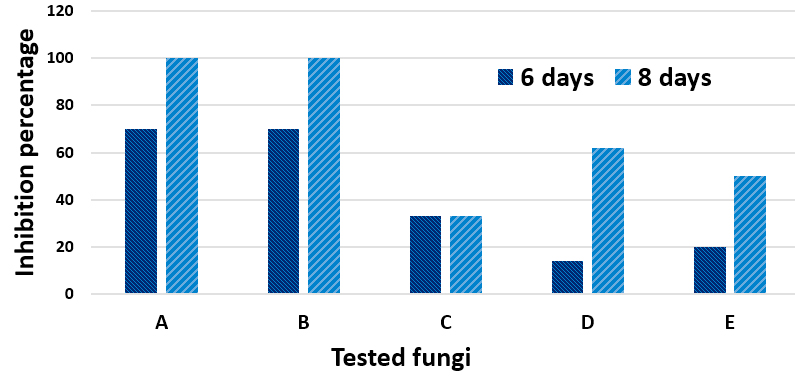
Substantial antagonistic effect of M. palmivorus was appeared against F. solani and F. thapsinum, in the range of 70-100% at 6 to 8 days after incubation, respectively. The growth inhibitory percentages of RBDF isolate were 33% (Penicillium sp.), 14-62% (T. harzianum), and 20-50% (P. cyclopium) within the 6th – 8th days after incubation, respectively. The most remarkable antagonism was observed with T. harzianum, in which Marasmiellus sp. started to grow over the already-grown tissues of T. harzianum. Another exciting results about antagonism were occurred between Penicillium sp. and M. palmivorus, where in spite of the fast growth of the Penicillium sp. covering nearly 70% of the petri area, the fungal mycelia of M. palmivorus rapidly approached Penicillium sp.; this then directly inhibited its growth upon contact (Figs. 8 & 9).
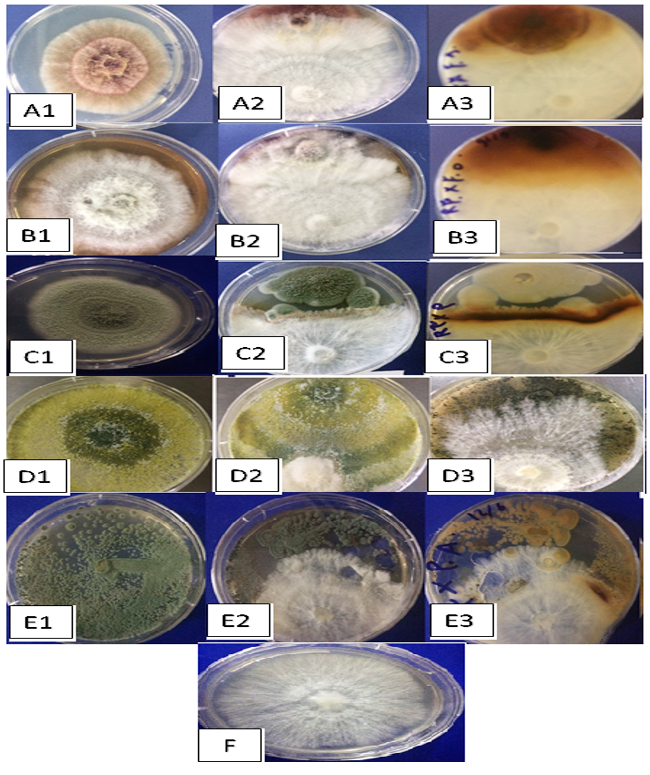 Fig. 8. Antagonistic efficiency of M. palmivorus towaed: F. solani (A1), F. solani + M. palmivorus– front face (A2), F. solani + M. palmivorus– reverse face (A3), F. thapsinum– alone (B1), F. thapsinum + M. palmivorus– upper face (B2), F. thapsinum + M. palmivorus– lower face (B3), Penicillium sp. alone (C1), Penicillium sp. + M. palmivorus– front face (C2), Penicillium sp + M. palmivorus – reverse face (C3), T. harzianum (D1), T. harzianum. + M. palmivorus-4 d.a.i. (D2), T. harzianum + M. palmivorus– 8 d.a.i. (D3), Penicillium cyclopium (E1), Penicillium cyclopium. + M. palmivorus– fron face (E2), Penicillium cyclopium + M. palmivorus– reverse face (E3), Control, M. palmivorus (F) at 28 ± 2oC
Fig. 8. Antagonistic efficiency of M. palmivorus towaed: F. solani (A1), F. solani + M. palmivorus– front face (A2), F. solani + M. palmivorus– reverse face (A3), F. thapsinum– alone (B1), F. thapsinum + M. palmivorus– upper face (B2), F. thapsinum + M. palmivorus– lower face (B3), Penicillium sp. alone (C1), Penicillium sp. + M. palmivorus– front face (C2), Penicillium sp + M. palmivorus – reverse face (C3), T. harzianum (D1), T. harzianum. + M. palmivorus-4 d.a.i. (D2), T. harzianum + M. palmivorus– 8 d.a.i. (D3), Penicillium cyclopium (E1), Penicillium cyclopium. + M. palmivorus– fron face (E2), Penicillium cyclopium + M. palmivorus– reverse face (E3), Control, M. palmivorus (F) at 28 ± 2oCImperata cylindrical plants are known to invade agriculture fields and irrigation channel systems in the majority of agriculture lands in Iraq. In the present study, the observations of disease symptoms and the distribution of disease demonstrated that the rate of wilt disease on I. cylindrical by M. palmivorus varied according to agricultural region, weed intensity, and type of soil.
Based on colony characteristics, like the appearance of clamp connections in fungal tissues (mycelium), this fungus was identified as Marasmius palmivorus. This finding is closely harmonious with the observations presented by Singer (1973)8.
The generic situation of M. palmivorus is questionable, since it’s morphology is quite similar to that in Marasmiellus than to Marasmius sensu stricto19. The original was published as M. palmivorus by Corner (1996)27. In 2005, Wilson and Desjardin7 formally proposed including this taxon as Marasmiellus palmivorus, but the proposal was not accepted, and there is no taxon available as Marasmiellus palmivorus. Some workers read the formal proposal of Wilson and Desjardin, but wrongly named their collected taxon Marasmiellus palmivorus; they published some of this work as well16.
Phylogenetic tree
The transformative history was deduced by utilizing the Neighbor-Joining method (NJ)28. The ideal tree with the entirety branch length = 0.01559401 is appeared. The tree is drawn to scale, with branch lengths in the similar units as the evolutionary distances used to conclude the phylogenetic tree. These separations were processed with the most extreme Composite Likelihood Method29, and are in units of the number of base substitutions per site. The investigation included 21 nucleotide sequences.
Codon positions included were 1st+2nd+3rd+Noncoding. All positions containing gaps and missing data were eliminated. There were a entire of 325 locations in the final dataset. Evolutionary analyses were conducted in MEGA730.
The multiple sequence alignment analysis of partial 18S ribosomal RNA gene sequence for local M. palmivorus isolate and the NCBI gene bank, using Mega 7, was a multiple alignment analysis tool. Sequence comparisons of the ITS region are broadly utilized in taxonomy and molecular phylogeny, as it is direct to enhance from little amounts of DNA (due to the high copy number of rRNA genes), including that there is a high level of variations between firmly related species
It has been demonstrated that partial 18S ITS1 5.8S ITS2, and partial 28S ribosomal RNA gene sequence data on an individual strain with the nearest neighbor, and exhibiting a similarity score of < 97%, represents a new species; however, the meaning of similarity scores > 97% is not as clear. The first step of alignment analysis for all sequences in this study with other selected references used the (Clustal W) program step in MEGA 6.0. This program demonstrated an accurate degree of identity with all world sequences, including those in this study. The results with (Clustal W) are important, because they were directly used in the phylogenetic tree construction, and agreed with previous studies using specific primers, The firmly related taxa possess similar fragment distributions, while the remotely related taxa are increasingly dissimilar, thus providing impressive phylogenetic data 31.
The multiple alignment analysis showed similarity and differences in 18S ribosomal RNA gene nucleotide sequences. This corresponds to the study by Tom ovsk et al.32, in which it was revealed that all Trametes species form a single clade. The ITS region showed the highest fungi ecology33 and was recommended to be the universal fungal barcode sequence34.
A nucleotide was used with a maximum likelihood of detecting the relationship of world and local sequences, which was the preferred method of nucleotide sequences in MEGA 6.0: it uses the NJ method, a simplified version of the Minimum Evolution (ME) method. The NJ method produces an unrooted tree, as it does not require a constant rate of evolution. Finding the root requires an out-group taxon28. Further studies with different gene sequences will resolve this in the phylogenetic analysis of fungi35.
Dutta and Acharya (2018) reported that nrITS and nrLSU sequences of M. palmivorus were 760 and 593 bp, respectively, which were deposited in the GenBank database with accession numbers MG251431 (for nrITS) and MG251441 (for nrLSU). Based on BLAST searches in the NCBI database, they had the most similarity to the taxon from Malaysia [GenBank JQ653433; identities = 717/726(99%), gaps = 3/726(0%)], Hawaii [GenBank AY639434; identities = 592/593(99%), and gaps = 0/593(0%)]16.
Bioactivity of Marasmius palmivorus
Culture media is viewed as one of the fundamental necessities for the development and improvement of fungi. M. palmivorus was found to grow a lot quicker in supplemented growth medium of powdered leaves, comprising of wheat, reed, and caladium, compared with PDA; this suggests that there are preferable growth sources with the possibility to substitute the utilization of PDA for culturing M. palmivorus.
The alteration in growth patterns was studied by Thiruchchelvan et al.36, which observed that at 4 days’ incubation of fungus on PDA, King yam and Elephant foot yam media were more preferable for fungal growth than that in filter paper, sago nutrient agar, and water agar. Another study acheived by Ravimannan et al.37 showed the utility of legume seeds (cowpea, green gram, black gram, and soya meat) for the growth of Trichoderma, Sclerotium, Fusarium, Aspergillus, and Penicillium sp.
The antagonistic effect of Basidiomycetes mushroom on the growth of plant pathogenic fungi can be used as a promising biocontrol system. The results of this study show the broad spectrum of antagonistic activity of M. palmivorus to pathogenic fungi, such as F. solany, F. thapsinum, P. cyclopium, and T. harzianum. M. palmivorus surprisingly colonizes the antibiotic effect of T. harzianum, which indicates overgrowth of M. palmivorus or production of antifungal substance(s) against the tested fungi.
However, the growth of M. palmivorus was found to be suppressed by Penicillium sp., as they reached the vicinity of the other. This might be due to the production of toxic substance(s) by Penicillium sp., which undermine the growth of M palmivorus. In the present scenario, one interesting aspect to be investigated is the possible mechanistic insight behind the biocontrol activities of such a fungus. One potential mechanism of antagonism behavior of M. palmivorus can be associated with nutrients and niche competition. In addition, the antibiosis effect of M. palmivorus against soil-borne fungi, as well as weed plants, may also be associated with the production of volatile and nonvolatile compounds. Previously, it was documented that the antagonistic effect of a living cell might be due to the release of extracellular bioactive molecules.
The present results disagree with those of Thiruchchelvan et al.36, in which they report that Trichoderma sp. causes growth inhibition of Marasmiellus sp. up to 92% under in vitro conditions. The antifungal activity of T. harzianum to fungal pathogens have been reported: Aspergillus niger, A. flavus, Phytophthora sp., Fusarium oxysporum, Rhizoctonia solani, Penicillium notatum, and Alternaria solani38.
To the best of our information, this is the first record of the presence of M. palmivorus in Iraq, and likely around the world for weeds. Using M. palmivorus as a biocontrol agent against I. cylindrical plants is a new approach. The earliest experimental studies also show the selective action of M. palmivorus against weeds, without causing any deteriorations to neighboring plant seedlings. However, further studies are required to explore its selective herbicidal activity in the near future.
Acknowledgements
Authors would like to thank the Ministry of Science and Technology – Babylon Section – for providing formal facilities to Mr. Tamur, and the Department of Biology, University of Babylon, for technical assistance. Authors are also thankful to Prof. Ali H. K. El-Bahadili for scientific advice.
Conflict of Interest
The authors declares that there is no conflict of interest.
Authors’ Contribution
All authors listed have made a substantial, direct and intellectual contribution to the work, and approved it for publication.
Funding
This work was financially supported by the Department of Biology, College of Science, University of Babylon, Iraq.
Data Availability
All datasets generated or analyzed during this study are included in the manuscript.
Ethics Statement
This article does not contain any studies with human participants or animals performed by any of the authors.
- Tan Y-S, Desjardin DE, Vikineswary S, Noorlidah A. Basidiomycota: The genus Marasmius in Peninsular Malaysia. In: Malaysian Fungal Diversity (eds. E.B.G. Jones, K.D. Hyde and S. Vikineswary). Xxxx, Malaysia, 2007, 61-73.
- Kirk PM. Species Fungorum (version 18th May 2015). In: Species 2000 & ITIS Catalogue of Life. 2015. Retrieved 2015-06-17.
- Antonםn V. Supplements to the monograph of tropical African species of Marasmius (Basidiomycota, Marasmiaceae). Cryptogamie Mycol. 2013; 34(2): 113–135.
Crossref - Antonםn V, Noordeloos ME. A monograph of marasmioid and collybioid fungi in Europe. Eching, IHW Verlag. 2010; 480.
- Wannathes N, Desjardin DE, Hyde KD, Perry BA, Lumyong S. Fungal Div. 2009; 37: 209–306.
- Desjardin DE, Ovrebo CL. New species and new records of Marasmius from Panama. Fungal Div. 2006; 21: 19-39.
- Wilson AW, Desjardin DE. Phylogenetic relationships in the gymnopoid and marasmioid fungi (Basidiomycetes, euagarics clade). Mycol. 2005; 97(3): 667–679.
Crossref - Singer, R. The genera Marasmiellus, Crepidotus and Simocybe in the neotropics. Beih. Nova Hedwigia. 1973; 44: 1–517.
- Miller GL, Desjardin DE, Tredway LP. First Report of Marasmiellus mesosporus causing Marasmiellus Blight on Seashore Paspalum. Plant Dis. 2010; 94(11): 1374.
Crossref - Almaliky BSA, Mior Ahmad MA, Kadir J, Mui W. Pathogenicity of Marasmiellus palmivorus (Sharples) Desjardin comb. Prov. on Oil Palm Elaeis guineensis. Wulfenia 2012; 19(8): 144-160.
- Pong VM, Zainal Abidin MA, Almaliky BSA, Kadir J, Wong MY. Isolation, Fruiting and Pathogenicity of Marasmiellus palmivorus (Sharples) Desjardin (comb. prov.) in Oil Palm Plantations in West Malaysia. Pertanika J. Trop. Agric. Sci. 2012; 35: 38 – 48.
- Thiruchchelvan N, Thirukkumaran G, Mikunthan G. Banana stem rot disease; causal agent (Marasmiellus sp.), host range and selection of low cost media for its cultivation. J. Nat. Prod. Plant Resour. 2013; 3(2): 97-104.
- Amoako-Attah I, Akrofi AY, Bin Hakeem R, Asamoah M, Kumi-Asare E. White thread blight disease caused by Marasmiellus scandens (Massee) Dennis & Reid on cocoa and its control in Ghana. African J. Agricultural Res. 2016; 11(50): 5064-5070.
Crossref - Tamur HA, Mohsin LY, Al-Janabi JKA, Al-Yassiry ZAN. Marasmiellus palmivorus as a new causal agent of Reed Wilt disease in Iraq. Pak. J. Biotechnol. 2018; 15(1): 29-35.
- Sharples A. Palm diseases in Malaya. Malayan Agric. J. 1928; 16(9–10): 313–360.
- Dutta, AK, Acharya K. A new host for the parasitic macrofungus Marasmius palmivorus Sharples (Marasmiaceae). Current Sci. 2018; 114 (7): 1400-1402.
Crossref - Tan Y-S, Desjardin DE, Perry BA, Vikineswary S, Noorlidah A. Marasmius sensu stricto in Peninsular Malaysia. Fungal Diver. 2009; 37: 9-100.
- Kirk PM, Cannon PF, Minter DW, Stalpers, JA. Dictionary of the Fungi, 10th ed. CABI, Wallingford, UK, 2008.
- Desjardin DE, Perry BA. The gymnopoid fungi (Basidiomycota, Agaricales) from the Republic of Sדo Tomי and Prםncipe, West Africa. Mycosph. 2017; 8(9): 1317–139.
Crossref - Obaid AJ, Al-Janabi JKA, Taj-Aldi WR. Antifungal Activity of Anise Essential Oil against Growth and Morphological Characteristics of Trichophyton Rubrum. J. Global Pharma. Technol. 2017; 7(9): 53-68.
- Dewan MM, Sivasithamparam K. Efficacy of treatment with a sterile red fungus for control of take-all in wheat. New Zealand J. Crop Horticul. Sci. 1989; 17: 333-336.
Crossref - Alnuaimy, AA, Al-Janabi JKA, Al-Obaidi LAH, Marjan AF. The expression of resistance genes in tomato induced by abiotic and biotic factors against Fusarium oxysporum f. sp. lycopersici. International J. Chem. Tech. Res. 2017; 10(6): 838-850.
- Umechuruba CF, Elenwo EN. Diagnostic Techniques in Mycology. Beek Publishers, Port Hercout. 1999, 30 – 56.
- Mohsen LY, Kadhim HJ, Al-Janabi JKA, Al-Yassiry ZAN. Alternative culture media for growth and sporulation of Trichoderma harzianum. Pak. J. Biotechnol. 2017; 14(4): 587-593.
- Cherif M, Benhamou N. Cytochemical aspects of chitin breakdown during the parasitic action of a Trichodermasp. on Fusarium oxysporum f. sp. radicis-lycopersici. Phytopathol. 1990; 80: 1406-1414.
- Kulkarni NS, Lingappa S. Compatibility of entomopathogenic fungus Nomuraea rileyi (Farlow) Samson with a fungicide carbendazim. Karnataka J. Agric. Sci. 2001; 14(3): 819-821.
- Corner EJH. The agaric genera Marasmius, Chaetocalathus, Crinipellis, Heimiomyces, Resupinatus, Xerula and Xerulina in Malesia. Beih. Nova Hedwigia 1996; 111: 1-175.
- Saitou N, Nei M. The neighbor-joining method: A new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 1987; 4(4):406-25.
- Tamura K, Nei M, Kumar S. Prospects for inferring very large phylogenies by using the neighbor-joining method. Proc. Nat.Acad. Sci. (USA) 2004; 101: 11030-11035.
Crossref - Kumar S, Stecher G, Tamura K. MEGA7: Molecular Evolutionary Genetics Analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 2015; 33(7):1870–1874.
Crossref - Frye SR, Healy M. Molecular strain typing using repetitive sequence-based PCR, In: Tang, Y.-W., Stratton, C.W. (Eds.), Advanced Techniques in Diagnostic Microbiology, 1st ed. Springer, New York, 2006, 444–471.
Crossref - Tom ovsk M, Kolaֽםk MS, Paסoutovב S, Homolka L. Molecular phylogeny of European Trametes (Basidiomycetes, Polyporales) species based on LSU and ITS (nrDNA) sequences. Nova Hedwigia 2006; 3: 269-280.
Crossref - Peay KG, Kennedy PG, Bruns TD. Fungal community ecology: a hybrid beast with a molecular master. BioSci. 2008; 58: 799–810.
Crossref - Schoch, CL, Seifert KA, Huhndorf S, Robert V, Spouge JL, Levesque CA, et al. Nuclear ribosomal internal transcribed spacer (ITS) region as a universal DNA barcode marker for Fungi. Proc. Nat. Acad. Sci. (USA) 2012; 109: 6241–6246.
- Petti CA. Detection and identification of microorganisms by gene amplification and sequencing. Clin. Infect. Dis. 2007; 44: 1108–1114.
Crossref - Thiruchchelvan N. Thirukkumaran G, Mikunthan G. In vitro Biological Control of Marasmiellus sp. The Causal of Stem Rot of Banana Grown in Jaffna Peninsula, Sri Lanka. Acad. J. Plant Sci. 2012; 5(3): 94-101.
- Ravimannan N, Arulanantham R, Pathmanathan S, Niranjan K. Alternative culture media for fungal growth using different formulation of protein sources. Ann. Biol. Res. 2014: 5: 36-39.
- Patale SS, Mukadam DS. Management of plant pathogenic fungi by using Trichoderma species. Biosci. Discov. 2011; 2(1): 36-37.
© The Author(s) 2019. Open Access. This article is distributed under the terms of the Creative Commons Attribution 4.0 International License which permits unrestricted use, sharing, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.