ISSN: 0973-7510
E-ISSN: 2581-690X
Antibiotic resistant bacteria (ARB) in aquatic environments is attracting increasing attention. However, the spread of ARB along Perfume River in Hue City is poorly understood. This study aimed to phenotypically and genotypically characterize β-lactam- and fluoroquinolone-resistant Escherichia coli isolates from this river. Water samples were collected from the urban, rural, agricultural, and less-affected areas in March 2020. E. coli susceptibility to seven commonly employed antibiotics was analyzed using the disk diffusion method, and the antibiotic resistance genes (ARGs), qnrA, qnrB, qnrS, TEM, SHV, and CTX-M, were identified using polymerase chain reaction (PCR) and DNA sequencing. The antibiotic susceptibility patterns of E. coli revealed that the rate of amoxicillin resistance was the highest (60%). PCR assays and sequencing of 12 β-lactam-resistant E. coli isolates indicated the presence of blaTEM and blaCTX-M-15 in 58.3% and 16.7% of the isolates, respectively. Only one of four fluoroquinolon -resistant E. coli isolates harbored the qnrS, while qnrA or qnrB genes were not detected. These findings suggest that this water may be an essential source of transmissible ARGs in Hue City, which may have a detrimental impact on the people living in this area.
Aquatic Environments, Escherichia coli, Antibiotic Resistance, Perfume River
Since their discovery, antibiotics have been extensively used to treat infectious diseases caused by various bacteria in humans and animals. However, one of the biggest global health concerns is the increase in antibiotic resistance (AR) in hospitals, communities, and the environment. Antibiotic-resistant illnesses cause more than 700,000 deaths annually, and this number is estimated to reach 10 million by 2050.1 The misuse and overuse of antibiotics for treating infectious diseases in humans and animals are thought to be the main contributors to AR.2 In addition, environmental antibiotic residues exert selective pressures on the evolution of antibiotic resistance genes (ARGs). It is known that these ARGs can spread between different microorganisms via mobile genetic components like plasmids, integrons, and transposons,3,4 promoting the emergence of antibiotic-resistant bacteria (ARB). The environmental presence of ARB may endanger human health through direct or indirect contact with contaminated water (for drinking and recreational use).5
Escherichia coli, one of the most prevalent indicator microorganisms of fecal pollution in the environment, acts as the main reservoir of ARGs because of its ability to collect and preserve resistance genes from other bacteria.6 E. coli strains that are resistant to antibiotics are becoming more frequently reported in various environments, including surface water,3,5,7,8 and drinking water.9 Strains of E. coli producing enzymes known as extended-spectrum lactamases (ESBLs) have threatened public health since they were first reported in 1979. They can hydrolyze lactam antibiotics, such as penicillins and cephalosporins, rendering them ineffective. This mechanism is particularly concerning because it can lead to resistance to multiple classes of antibiotics, making treatment difficult. Recently, there has been an increased occurrence of ESBL-producing E. coli in surface water, particularly in Southeast Asia.10 Plasmid-mediated quinolone resistance (PMQR) refers to resistance to quinolone antibiotics, which are commonly used to treat E. coli infections. This mechanism is mediated by plasmids, small DNA molecules that can be transferred between bacteria. PMQR has also been found in E. coli strains globally and is an increasing public health concern.11,12
Antibiotic use is high in Vietnam and Asia. Their consumption is associated with high rates of misuse and overuse.4 In 2015, 3,838 tons of antibiotics were consumed in Vietnam, with 71.7% for animal use and the remainder for human use.13 According to a study published in 2019, Vietnam uses the most antibiotics in the aquaculture industry, and antimicrobial use has increased over the past 10 years as a result of uncontrolled drug use in this country.14 In Vietnam, the discovery of E. coli strains resistant to antibiotics and residual antimicrobials in river water are causes of concern.15,16 The isolation of these antibiotic-resistant strains of E. coli from river water in Vietnam raises questions regarding the source of contamination. It is possible that these bacteria originated from untreated human or animal waste discharged into the river. The overuse of antibiotics in the agricultural sector may also have played a role, as antibiotic-resistant bacterial strains can develop in livestock treated with antibiotics.
In central Vietnam, the Perfume River is the largest river flowing through Hue City, which has a population of more than 1 million residents as of 2021. This river is a crucial water source for drinking, domestic use, irrigation, and other purposes. However, the water quality of the Perfume River has deteriorated in some locations over the past few decades because of rapid population and economic growth in Hue City. Despite increasing contamination of the Perfume River, there is limited information about the antibiotic resistance of E. coli in the surface water of Hue City. This preliminary investigation aimed to examine the antibiotic susceptibility patterns and molecular characteristics of E. coli isolates resistant to seven antibiotics, including lactams and fluoroquinolones, obtained from the Perfume River.
Sampling sites
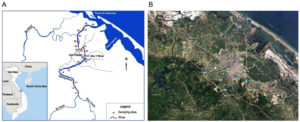
Water samples were collected along the Perfume River (length of the river channel: 104 km; catchment area: 2,830 km2) from upstream (Bang Lang Junction, Thuy Bang Commune, Hue City) to downstream (Thao Long Bridge, Phu Thanh Commune, Hue City). Figure and Table 1 show detailed maps of the locations and descriptions of all sampling locations, respectively. Twenty sampling locations were chosen based on the predominant land use in Hue City. Four distinct activities were represented by the sites: five were less-affected areas, five were urban areas, five were rural areas, and five were agricultural areas.
Figure. Location of sample collection. Water samples were collected from Perfume River (20 locations): A. A map from Department of Natural Resources and Environment, Thua Thien Hue province; B. A satellite image from Goolge Map
In March 2020, all water samples were collected between 08:00 h and 10:00 h in the morning at a depth of 50 cm below the water surface to avoid surface scum and debris. There was no rainfall for a couple of days. The samples were collected into 0.5-L glass containers previously sterilized by autoclaving at 121°C for 15 minutes. A total volume of 500 mL was collected from each location, and the samples were labeled, stored on ice for transportation to the laboratory, and processed for further analysis within 24 hours.
Table (1):
Description and coordinates for each sample site in the Perfume River.
| Human activities | Sample label | Description | Coordinates | |
|---|---|---|---|---|
| Latitude | Longitude | |||
| Less affected areas (L) | L1 | This site is under Tuan Bridge, upstream of the Perfume River, which is close to Bang Lang junction. | 16°23’59.6″N | 107°34’29.3″E |
| L2 | Suburban area with sparse residential apartments | 16°24’36.4″N | 107°34’01.2″E | |
| L3 | Sparse residential apartment, close to Hon Chen Temple | 16°25’17.0″N | 107°33’49.0″E | |
| L4 | Perfume River and Bach Yen River junction close to Long Ho Bridge | 16°26’54.9″N | 107°31’48.4″E | |
| L5 | This site is close to Thien Mu Pagoda, which is about 5 km from urban areas in Hue City | 16°27’06.1″N | 107°33’02.2″E | |
| Rural areas (R) | R1 | Rural area; the junction between Perfume River and Pho Loi River | 16°30’00.4″N | 107°35’19.0″E |
| R2 | Rural area with residential apartments | 16°29’45.6″N | 107°35’01.0″E | |
| R3 | Sampled at the junction of the canal, Dong Ba River and Perfume River; residential and business area | 16°29’40.9″N | 107°34’41.5″E | |
| R4 | Rural area with residence | 16°30’05.0″N | 107°34’35.4″E | |
| R5 | Rural and residential area | 16°30’51.1″N | 107°34’28.2″E | |
| Urban areas (U) | U1 | Residential and business area with shopping malls, Dong Ba Market and floating restaurant on Perfume River | 16°28’14.5″N | 107°35’23.3″E |
| U2 | Residential apartment area, behind a local market | 16°28’58.1″N | 107°35’30.1″E | |
| U3 | This site is below the Con Bridge, which connects Con Island to the city | 16°28’54.8″N | 107°35’41.3″E | |
| U4 | Sampled at the Stock Dam where Nhu Y River connects with the main river; tourist boat docking area | 16°28’24.2″N | 107°35’41.6″E | |
| U5 | This site is close to Cho Dinh Bridge and an aquatic processing company (small scale) | 16°29’23.6″N | 107°35’34.1″E | |
| Agricultural areas (A) | A1 | Rural and agricultural area; sampled at the junction between Bo River and the main river | 16°31’39.9″N | 107°34’24.7″E |
| A2 | Rural area with agriculture and sparse residential apartments; junction of Pho Loi River and Perfume River | 16°32’16.4″N | 107°35’13.9″E | |
| A3 | Rural, sparse residential and agricultural area | 16°32’47.4″N | 107°35’32.6″E | |
| A4 | Rural and agricultural area | 16°32’59.6″N | 107°35’56.4″E | |
| A5 | Rural and agricultural area; sampled near Thao Long Bridge, where there are several livestock farms | 16°32’49.6″N | 107°36’57.6″E | |
E. coli isolation and identification
Each water sample was filtered to collect microbiological cells using a sterile membrane filter with a 0.45-µm pore (47 mm diameter, cellulose nitrate; Sartorius, Gottingen, Germany). Membranes were placed on m-FC agar (CriterionTM; Hardy Diagnostics, Santa Maria, CA, USA) plates and cultured at 44.5°C for 24 hours. Fecal coliform bacteria were detected as blue colonies. A maximum of three blue colonies were picked from each m-FC dish and grown on CHROM agar plates (Difco, Franklin Lakes, NJ, USA) for E. coli isolation. Dark pink to reddish colonies, thought to be E. coli, were chosen and sub-cultured on nutrient agar plates (Merck, Darmstadt, Germany). BHI medium supplemented with sterile glycerol at a final concentration of 20% was stored at -80°C for further analysis. Approximately 4–5 colonies of putative E. coli isolates were grown overnight at 37°C on nutrient agar plates and suspended in tubes containing 500 µL of sterile distilled water. DNA was extracted using the phenol-chloroform method, according to the manufacturer’s instructions, using an iVApDNA Extraction Kit (Viet A Technology Corporation, Ho Chi Minh, Vietnam).
The uidA gene, which is specific to E. coli species, was examined in all putative E. coli isolates. The forward primer (52 -CCAAAAGCCAGACAGAGT-32 ) and reverse primer (52 -GCACAGCACATCAAAGAG-32 ) were used in the polymerase chain reaction (PCR) assay, and the amplicon was 623 bp. One microliter of each template DNA sample was added to 24 µL of the 1× Q5 High-Fidelity Master Mix (New England Biolabs, Ipswich, MA, USA) reaction mixture, containing each primer at a final concentration of 0.2 µM. The PCR cycling conditions were an initial denaturation at 94°C for 3 minutes; followed by 30 cycles of denaturation at 94°C for 30 seconds, annealing at 56°C for 30 seconds, and extension at 72°C for 50 seconds; and a final extension at 72°C for 7 minutes. GelRedTM Nucleic Acid Stain 10,000× (Biotium, Fremont, CA, USA) was used to stain the PCR products on a 1.5% agarose gel. Electrophoresis was performed using a 100V direct current for 30 minutes, and the bands were observed under ultraviolet light and compared using a 100 bp ladder (Invitrogen, Carlsbad, CA, USA).
Antibiotic susceptibility tests
Following the recommendations of the Clinical Laboratory Standards Institute (CLSI) recommendations, antibiotic susceptibility was evaluated using an agar diffusion assay with antibiotic-impregnated paper disks (disk diffusion method).17 The following antibiotics were used: amoxicillin (10 µg), cefotaxime (30 µg), ceftazidime (30 µg), ceftriaxone (30 µg), ciprofloxacin (5 µg), nalidixic acid (30 µg), and meropenem (10 µg).
Confirmed E. coli strains were suspended in nutritional broth and cultured at 35 ± 2°C for 5 hours. The broth was diluted in a normal saline solution to achieve the density of a 0.5 McFarland turbidity standard. The diluted broth was applied to cotton swabs on Mueller-Hinton agar plates (Merck). Antibiotic platelets were placed 30 mm apart and 10 mm from the edge of the disk after air drying. The plates were incubated under aerobic conditions at 35 ± 2°C for 16–18 h. CLSI breakpoints were used to analyze the data after measuring and recording the zones of inhibition and resistance.17 Strains that were resistant to three or more antibiotics were classified as multidrug-resistant (MDR) E. coli strains.
The presence of ESBL was identified using a combination disk test (CDT) in isolates that exhibited resistance to cephalosporins. Pairs of cephalosporin-containing disks (30 µg of cefotaxime or ceftazidime), with and without 10 µg of clavulanic acid, were put on opposite sides of a Mueller-Hinton plate (Merck) inoculated with a 0.5 McFarland suspension of the test isolate. After overnight incubation in the air at 37°C, zones of inhibition were observed. If the zone of inhibition around the combination disk was at least 5 mm, the test isolate was considered an ESBL producer.
Detection of antibiotic resistance genes
Detecting β-lactamase genes
The blaTEM, blaSHV, and blaCTX-M genes were detected by PCR amplification of isolates resistant to penicillins or cephalosporins. The primers were chosen based on previous studies. Table 2 lists the target genes, primer sequences, product sizes, and references. PCR was performed in a 25 µL final reaction volume containing 1× Q5 High-Fidelity Master Mix (New England Biolabs), 0.2 µM primer pairs for blaTEM or 0.4 µM primer pairs for blaSHV and blaCTX-M, nuclease-free water, and 1 µL of DNA. The amplification protocol was as follows: 94°C for 5 minutes; 30 cycles of 95°C for 20 seconds, 61°C for 30 seconds, and 72°C for 1 min; and finally, 72°C for 5 minutes. Agarose gels (1.5%) were used to separate and visualize all PCR products, as described previously.
Table (2):
Oligonucleotide primers for PCR detection of β-lactamase resistance and PMQR genes in E. coli.
| Target gene | Primer sequence (5’ à 3’) | Product size (bp) | Reference |
|---|---|---|---|
| blaTEM | F: AGTGCTGCCATAACCATGAGTG R: CTGACTCCCCGTCGTGTAGATA |
431 | [18] |
| blaSHV | F: GATGAACGCTTTCCCATGATG R: CGCTGTTATCGCTCATGGTAA |
214 | |
| blaCTX-M | F: ATGTGCAGYACCAGTAARGT R: TGGGTRAARTARGTSACCAGA |
593 | |
| qnrA | F: ATTTCTCACGCCAGGATTTG R: GATCGGCAAAGGTTAGGTCA |
516 | [19] |
| qnrB | F: GATCGTGAAAGCCAGAAAGG R: ACGATGCCTGGTAGTTGTCC |
469 | |
| qnrS | F: ACGACATTCGTCAACTGCAA R: TAAATTGGCACCCTGTAGGC |
417 |
Detecting fluoroquinolone resistance genes
PCR amplification was used to detect PMQR genes, including qnrA, qnrB, and qnrS, in isolates resistant to ciprofloxacin and nalidixic acid. The primers were selected from those used in previous studies. Table 2 lists the target genes, primer sequences, and sizes of the products and references. PCR was performed in a 25 µL final reaction volume containing 1× Q5 High-Fidelity Master Mix (New England Biolabs), 0.4 µM each primer, nuclease-free water, and 1 µL of DNA. The following amplification protocol was used: 94°C for 5 minutes; 32 cycles of 94°C for 45 seconds, 58°C for 45 seconds, and 72°C for 1 minute; and finally, 72°C for 5 minutes. All PCR products were separated on 2% agarose gels and electrophoresis was performed under 100V of direct current for 60 minutes.
Sequencing analysis of ESBL genes
To identify the ESBL gene types detected by the multiplex PCR assay, the DNA sequences of the amplicons were analyzed. A Clean & ConcentratorTM-5 (Zymo Research, Irvine, CA, USA) was used to purify the amplified PCR products before bidirectional sequencing was performed. The resulting sequences were analyzed using Geneious R11 software and compared with known ESBL gene sequences by multiple sequence alignment using the BLAST program.
Data analysis
The statistical analyses were performed using Excel software.
E. coli antibiotic resistance pattern
Twenty different bacterial strains of E. coli were detected in the surface water samples and evaluated for their resistance to seven antibiotics. All isolates demonstrated susceptibility to meropenem but showed different levels of resistance to the six other antibiotics. The AR rate of E. coli was highest for amoxicillin (Table 3).
Table (3):
Antibiotic susceptibility profiles of E. coli strains (N=20).
Name of antibiotics |
Resistant n (%) |
Intermediate n (%) |
Susceptible n (%) |
|---|---|---|---|
Amoxicillin |
12 (60) |
0 |
8 (40) |
Nalidixic acid |
4 (20) |
2 (10) |
14 (70) |
Ciprofloxacin |
3 (15) |
10 (50) |
7 (35) |
Cefotaxime |
3 (15) |
1 (5) |
16 (80) |
Ceftriaxone |
3 (15) |
0 |
17 (85) |
Ceftazidime |
2 (10) |
1 (5) |
17 (85) |
Meropenem |
0 |
1 (5) |
19 (95) |
Table 4 displays the number of E. coli isolates that exhibit antibiotic resistance at various sampling locations along the Perfume River, which are impacted by diverse human activities in Hue City. In general, urban areas showed the largest concentration of antibiotic-resistant E. coli.
Three of 20 E. coli isolates tested positive for cephalosporin resistance and underwent confirmatory testing (CDT). They were discovered to be ESBL producers, and all were MDR.
Table (4):
The number of environmental isolates resistant to six antibiotics based on their sample locations (N=20).
| Location | E. coli isolates | The number of antibiotic-resistant E. coli isolates | |||||
|---|---|---|---|---|---|---|---|
| AX | CTX | CAZ | CRO | CIP | NA | ||
| Less affected area | 5 | 2 | 0 | 0 | 0 | 0 | 0 |
| Urban area | 5 | 4 | 3 | 2 | 3 | 2 | 2 |
| Rural area | 5 | 3 | 0 | 0 | 0 | 0 | 1 |
| Agricultural area | 5 | 3 | 0 | 0 | 0 | 1 | 1 |
| Total | 20 | 12 | 3 | 2 | 3 | 3 | 4 |
AX: amoxicillin, CTX: cefotaxime, CAZ: ceftazidime, CRO: ceftriaxone, CIP: ciprofloxacin, NA: nalidixic acid.
Identification of genes in E. coli causing β-lactam and fluoroquinolone resistance β-lactam
All 12 penicillin- or cephalosporin-resistant isolates were selected and screened for the presence of b-lactamase genes, including blaTEM, blaSHV, and blaCTX-M. blaTEM and blaCTX-M-15 were present in 58.3% and 16.7%) of the E. coli isolates, respectively, whereas blaTEM and blaCTX-M-65 were present in 8.3% of the isolates. No isolates contained blaSHV genes (Table 5).
Table (5):
Distribution of the resistance genotypes in β-lactam- and fluoroquinolone-resistant isolates.
Site |
Resistance pattern |
Presence of ARGs |
|---|---|---|
L2 |
AX |
– |
L5 |
AX |
blaTEM |
U1 |
CIP/NA |
– |
U2 |
AX |
blaTEM |
U3 |
AX/CTX/CAZ/CRO |
blaCTX-M15 |
U4 |
AX/CTX/CAZ/CRO |
blaCTX-M15 |
U5 |
AX/CTX/CRO/CIP/NA |
blaTEM, blaCTX-M65 |
R1 |
AX |
– |
R2 |
AX/NA |
blaTEM |
R3 |
AX |
blaTEM |
A1 |
AX |
blaTEM |
A2 |
CIP/NA |
qnrS |
A4 |
AX |
blaTEM |
A5 |
AX |
blaTEM |
AX: amoxicillin, CTX: cefotaxime, CAZ: ceftazidime, CRO: ceftriaxone, CIP: ciprofloxacin, NA: nalidixic acid.
Fluoroquinolone
PMQR genes qnrA, qnrB, and qnrS were examined in four fluoroquinolone-resistant strains. The qnrS gene was present in only one sample, but qnrA and qnrB genes were absent in all isolates (Table 5).
Antibiotic resistance pattern of environmental E. coli
Aquatic environments, such as rivers, are ideal reservoirs for the spread of AR. Many studies have demonstrated that ARB or genes confer resistance in aquatic settings.8,12,20,21 Interestingly, this has also been observed in countries where antibiotic use is strictly regulated. For example, ARGs have been discovered in Swedish waters.20 This study assessed the antibiotic susceptibilities of 20 different E. coli strains recovered from the Perfume River. The highest prevalence of AR in E. coli was observed for amoxicillin (60%), followed by nalidixic acid (20%). The antibiotic resistance patterns of E. coli isolates observed in our study are common in other aquatic environments.8,12,21. This may be because these antibiotics are the most widely manufactured and prescribed in human and veterinary medicine in Vietnam. In addition, β-lactams and fluoroquinolones were prescribed for humans at eight defined daily doses per 1,000 individuals in 2015.13 Therefore, the discovery of E. coli resistant to these antibiotics in environments is not surprising. As the final resort among β-lactams, meropenem is exclusively used for patients with particularly challenging infections that are unresponsive to other β-lactam antibiotics. Fortunately, no meropenem-resistant E. coli isolates were detected.
The number of E. coli isolates at the sampling sites along the Perfume River that were affected by anthropogenic activities in Hue City is shown in Table 4. In general, the highest number of antibiotic-resistant E. coli strains was observed in metropolitan areas, followed by agricultural locations. It is important to note that, although the majority of the municipal wastewater in Hue City is appropriately collected and treated at a wastewater treatment plant, untreated wastewater is discharged into the canals before entering the Perfume River. As a result, water samples collected from urban areas are at risk of being polluted by untreated wastewater from a variety of sources, including households, urban runoff, and commercial activities. Agricultural areas were also affected by contamination with antibiotic-resistant E. coli. This can be attributed to the overuse of chemical insecticides and fertilizers for growth-promotion purposes in farming practices. In addition, agricultural waste and untreated wastewater may be released into the environment without surveillance. Our results were similar to those of previous studies.4,12 The results reflect how human activities along the river contributed to an increase in contamination with antibiotic-resistant E. coli from upstream to downstream, suggesting a potential risk of AR spreading in the aquatic environment.
ESBL-producing E. coli strains are becoming more common worldwide. They are not only present in clinical settings, but also found in environmental niches, such as livestock; wildlife; and most notably, water.22 The spread of these isolates, especially MDR isolates, is a source of concern in both developing and developed countries, as they constitute a potential public health risk.21 In this study, 15% (n = 3) of isolates were ESBL-producing E. coli isolates, all of which were isolated in metropolitan settings. It is of particular concern that all of the ESBL producers identified in this study displayed an MDR phenotype. It is highly likely that the Perfume River has been contaminated by sewage discharge, such as municipal sewage and wastewater from animal farms, which are located in or close to the sampling sites. Hon et al. reported that most ESBL-producing E. coli isolated from wild and farmed fish in the Mekong Delta of Vietnam were MDR.23 This is not surprising, given that ESBL producers are routinely resistant to several antibiotics, particularly fluoroquinolones. This is because blaESBL genes are frequently found in conjugative plasmids, which usually contain genes conferring resistance to other antibiotics.24
Distribution of the genes for b-lactamase and PMQR
The most commonly administered antibacterial medicines are b-lactams, and the most prevalent and major mechanism by which bacteria resist these drugs is by the production of b-lactamase.25 In our study, b-lactamase gene screening was performed on E. coli isolates. The percentages of E. coli harboring blaTEM and blaCTX-M-15 were 58.3% and 16.7%, respectively. blaTEM appeared at the highest frequency, similar to the results reported in China.8,25 In addition, one isolate carried blaTEM and blaCTX-M-65. Several drug-resistance genes have been identified in bacterial strains isolated from aquatic environments in previous reports.12,25 This may be attributed to the coexistence of blaTEM and blaCTX-M genes in the plasmids.12 However, the isolated E. coli strains did not possess blaSHV genes, as blaSHV is typically found in Klebsiella spp.12
Among the strains with ESBL phenotypes, blaCTX-M-15 was the most common ESBL gene, which is in agreement with data from India and Turkey.3,12 Since the beginning of the 21st century, blaCTX-M-producing E. coli isolates have spread and they are now major contributors to urinary tract and bloodstream infections in humans, occurring in both nosocomial settings and in the general population. This was not surprising because these enzymes constitute the majority of ESBLs in E. coli isolates recovered from surface water.
Fluoroquinolone resistance was assumed to be caused by mutations in gyrase and topoisomerase IV genes. However, it is now clear that it is also related to plasmid-associated resistance factors that may transmit antibiotic resistance in the environment. In our study, qnrA, qnrB, and qnrS were chosen to investigate PMQR. Although four E. coli strains were ciprofloxacin- or nalidixic-acid-resistant, only one strain possessed the qnrS gene. It has been reported that E. coli strains isolated from rivers contain the qnrS gene,11,12 but not fluoroquinolone resistance determinants, such as qnrA and qnrB. The lack of qnrA and qnrB may be explained by the fact that these genes are frequently included in complex sul1-type integrons.26 Resistance to fluoroquinolones in one E. coli strain was attributed to a plasmid-derived qnrS gene. The remaining isolates may be a result of point mutations in the gyrase and topoisomerase subunit genes, gyrA, gyrB, parC, or parE on the chromosome,27 but this was not been investigated in this study.
However, our study has some limitations. The study was conducted for only 1 month; therefore, the impact of seasonal changes on the antibiotic-resistant rate of E. coli remains unclear. Furthermore, there may have been a correlation between water quality parameters and the prevalence of antibiotic-resistant E. coli in this study. Further research is required to investigate these possibilities.
The discovery of antibiotic-resistant E. coli in water samples from the Perfume River, Vietnam, suggests that this body of water was contaminated with feces. Our findings demonstrated that environmental isolates of E. coli have a significant prevalence of AR, particularly amoxicillin and nalidixic acid resistance. The proportion of antibiotic-resistant E. coli strains was higher in metropolitan regions, where large amounts of waste from human activities enter the river without treatment. ESBL-producing E. coli strains were also observed, and these showed an MDR phenotype for all antibiotic classes. In addition, the results of this study provide a clear genetic description of E. coli isolates resistant to β-lactams and fluoroquinolones from the Perfume River.
The rising prevalence of antibiotic resistance in aquatic environments should create concern for public health, as the Perfume River is utilized for irrigation, livestock, and domestic uses. Therefore, wastewater treatment systems should be improved to ensure the effective removal of bacteria and other contaminants from water sources. Antibiotics should only be used when they are truly necessary, and efforts should be made to reduce their overuse in agriculture. Furthermore, the development of monitoring programs to track the occurrence of ARB and ARGs in the Perfume River and other major water bodies around the country is recommended. By taking these steps, it may be possible to reduce the prevalence of ARB in water sources and prevent the spread of infections that are difficult to treat using standard antibiotics.
ACKNOWLEDGMENTS
The authors would like to thank all faculty members, residents and technical staff members of Department of Microbiology and Faculty of Public Health, University of Medicine and Pharmacy, Hue University and Department of Microbiology, University of Sassari, Italy for their contribution and support.
CONFLICT OF INTEREST
The authors declare that there is no conflict of interest.
AUTHORS’ CONTRIBUTION
NVQT, NHB, PTTT contributed in conceptualization, study design, literature review, data collection, writing original draft and editing the manuscript. NHB wrote, reviewed and edited the manuscript. NVQT, NHB, PTTT, NTDK, NTT, PTHC, NTDH, TTCT, BP contributed in data analysis, interpretation, read and approved the final manuscript for publication.
FUNDING
None.
DATA AVAILABILITY
All datasets generated or analyzed during this study are included in the manuscript.
ETHICS STATEMENT
This study was approved by the Institutional Ethics Committee, Hue University of Medicine and Pharmacy, Hue University, Hue City, Vietnam, with reference number H2020/019.
- Jim O. Antimicrobial Resistance: Tackling a crisis for the health and wealth of nations. Review on Antimicrobial Resistance: London, UK. 2014.
- Martin MJ, Thottathil SE, Newman TB. Antibiotics overuse in animal agriculture: A call to action for health care providers. Am J Public Health. 2015;105(12):2409-2410.
Crossref - Kurekci C, Aydin M, Yipel M, Katouli M, Gundogdu A. Characterization of extended spectrum β-lactamase (ESBL)-producing Escherichia coli in Asi (Orontes) River in Turkey. J Water Health. 2017;15(5):788-798.
Crossref - Truong T, Hoang TL, Tran LT, Pham TPT, Le TH. Prevalence of Antibiotic Resistance Genes in the Saigon River Impacted by Anthropogenic Activities. Water. 2021;13(16):2234.
Crossref - dela Pena LBRO, Nacario MAG, Bolo NR, Rivera WL. Multiple Antibiotic Resistance in Escherichia coli Isolates from Fecal and Water Sources in Laguna Lake, Philippines. Water. 2022;14(9):1517.
Crossref - Talukdar PK, Rahman M, Rahman M, et al. Antimicrobial resistance, virulence factors and genetic diversity of Escherichia coli isolates from household water supply in Dhaka, Bangladesh. PloS One. 2013;8(4):e61090.
Crossref - Liu H, Zhou H, Li Q, Peng Q, Zhao Q, Wang J, et al. Molecular characteristics of extended-spectrum β-lactamase-producing Escherichia coli isolated from the rivers and lakes in Northwest China. BMC Microbiol. 2018;18:125.
Crossref - Zhang SH, Xiaoyang LV, Han B, et al. Prevalence of antibiotic resistance genes in antibiotic-resistant Escherichia coli isolates in surface water of Taihu Lake Basin, China. Environ Sci Pollut Res Int. 2015;22(15):11412-21.
Crossref - Chen Z, Yu D, He S, et al. Prevalence of antibiotic-resistant Escherichia coli in drinking water sources in Hangzhou City. Front Microbiol. 2017;8:1133.
Crossref - Chong Y, Shimoda S, Shimono N. Current epidemiology, genetic evolution and clinical impact of extended-spectrum β-lactamase-producing Escherichia coli and Klebsiella pneumoniae. Infect Genet Evol. 2018;61:185-188.
Crossref - Rutgersson C, Fick J, Kristiansson E, et al. Fluoroquinolones and qnr genes in sediment, water, soil, and human fecal flora in an environment polluted by manufacturing discharges. Environ Sci Technol. 2014;48(14):7825-732.
Crossref - Dhawde R, Macaden R, Saranath D, Nilgiriwala K, Ghadge A, Birdi T. Antibiotic Resistance Characterization of Environmental E. coli Isolated from River Mula-Mutha, Pune District, India. Int J Environ Res Public Health. 2018;15(6):1247.
Crossref - Carrique-Mas JJ, Choisy M, Nguyen VC, Thwaites G, Baker S. An estimation of total antimicrobial usage in humans and animals in Vietnam. Antimicrob Resist Infect Control. 2020;9(1):16.
Crossref - Lulijwa R, Rupia EJ, Alfaro A. Antibiotic use in aquaculture, policies and regulation, health and environmental risks: a review of the top 15 major producers. Reviews in Aquaculture. 2019;12(2):640-663.
Crossref - Yamasaki S, Le TD, Vien MQ, Dang CV, Yamamoto Y. Prevalence of extended-spectrum β-lactamase-producing Escherichia coli and residual antimicrobials in the environment in Vietnam. Anim Health Rev. 2017;18(2):128-35.
Crossref - Le TH, Truong T, Hoang TL, Pham TTV, Pham TPT, Tran LT. Influences of anthropogenic activities on water quality in the Saigon River, Ho Chi Minh City. J Water Health. 2022;20(3):491-504.
Crossref - CLSI. Performance Standards for Antimicrobial Susceptibility Testing 30th Edition. Clinical and Laboratory Standard Institute. 2020.
- Hijazi SM, Fawzi MA, Ali FM, Abd El Galil KH. Prevalence and characterization of extended-spectrum beta-lactamases producing Enterobacteriaceae in healthy children and associated risk factors. Ann Clin Microbiol Antimicrob. 2016;15:3.
Crossref - Robicsek A, Strahilecitz J, Sahm DF, Jacoby GA, Hooper DC. qnr prevalence in ceftazidime-resistant Enterobacteriaceae isolates from the United States. Antimicrob Agents Chemother. 2006;50(8):2872-2874.
Crossref - Khan FA, Söderquist B, Jass J. Prevalence and Diversity of Antibiotic Resistance Genes in Swedish Aquatic Environments Impacted by Household and Hospital Wastewater. Front Microbiol. 2019;10:688.
Crossref - Li S, Zhu ZC, Wang L, Zhou YF, Tang YJ, Miao ZM. Prevalence and characterization of extended-spectrum beta-lactamase-producing Enterobacteriaceae in spring waters. Lett Appl Microbiol. 2015;61(6):544-548.
Crossref - Kamruzzaman M, Shoma S, Naymul Bari SM, et al. Genetic diversity and antibiotic resistance in Escherichia coli from environmental surface water in Dhaka City, Bangladesh. Diagn Microbiol Infect Dis. 2013;76(2):222-226.
Crossref - Nguyen TNH, Tran TTH, Nguyen QT, Hinenoya A. Spread of antibiotic and antimicrobial susceptibility of ESBL-producing Escherichia coli isolated from wild and cultured fish in the Mekong Delta, Vietnam. Fish Pathology. 2016;51(Special issue): S75-S82.
Crossref - Wang J, Stephan R, Kaczmarczyk M, Yan Q, Hachler H, Fanning S. Molecular characterization of blaESBL-harboring conjugative plasmids identified in multi-drug resistant Escherichia coli isolated from food-producing animals and healthy humans. Front Microbiol. 2013;4:188.
Crossref - Zou LK, Li LW, Pan X, et al. Molecular characterization of β-lactam-resistant Escherichia coli isolated from Fu River, China. orld J Microbiol Biotechnol. 2012;28(5):1891-1899.
Crossref - Garnier F, Raked N, Gassama A, Denis F, Ploy MC. Genetic environment of quinolone resistance gene qnrB2 in a complex sull-type integron in the newly described Salmonella enterica serovar Keur Massar. Antimicrob Agents Chemother. 2006;50(9):3200-3202.
Crossref - Hopkins KL, Davies RH, Threlfall EJ. Mechanisms of quinolone resistance in Escherichia coli and Salmonella: Recent developments. Int J Antimicrob Agents. 2005;25(5):358-373.
Crossref
© The Author(s) 2023. Open Access. This article is distributed under the terms of the Creative Commons Attribution 4.0 International License which permits unrestricted use, sharing, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.