ISSN: 0973-7510
E-ISSN: 2581-690X
Heterotrophic bacteria are organic and inorganic decomposer functioning to inhibit pathogenic bacteria in the waters. This research is aimed at understanding the heterotrophic population in Siak River estuary and the antagonism of heterotrophic bacteria against pathogenic bacteria (Vibrio sp, Aeromonas sp dan Pseudomonas sp), identified based on their genotype and phenotype. The research was conducted from July to October 2017 in the estuary of Siak River using a survey method. Estuary water sample was used to classify the heterotrophic bacteria. The antagonism test of 25 isolated bacteria resulted in 10 isolates potential in inhibiting the growth of pathogenic bacteria (Vibrio sp, Pseudomonas sp and Aeromonas sp). The mean of the biggest transparent zone of Vibrio sp. bacterium was 1P11 isolate by 20.3 mm and the smallest one was 1P19 by 5 mm, and for the Aeromonas sp bacterium. Its largest transparent zone diameter was 2P20 isolate for 9.3 mm and 1P12 (3.3 mm) for the smallest one. Further, the Pseudomonas sp biggest transparent zone was 1P12 isolate (17mm) and the smallest ones were 1P11 and 2P22 (8.3 mm). The result of molecular identification with 16S rRNA and BLAST analysis showed that there was a relationship between 10 heterotrophic isolated bacteria and that of Bacillus genus; Bacillus cereus (1P11, 1P12, 1P14, 1P15, 1P19, 2P20, 2P21, 2P23), Bacillus toyonensis (2P22) and Virgibacillus salarius (1P18).
Heterotrophic bacteria, Isolation, Antagonism test, Identification, PCR
Siak River estuary is one of the biggest river systems in Riau that has a vital role in supporting the society’s activity as well as industries. Numerous activities around Siak watershed generate various organic and inorganic polluting agents (waste), either directly or indirectly pass into the river and is accumulated in the river mouth.
Bacteria are the centre of carbon (C), nitrogen (N) and phosphorous (P) cycles in an ecosystem (Godwin and Cotner, 2015). Heterotrophic bacteria are critical decomposers of pollutant (Kong and Ye, 2014). The nutrient concentration, pH, and temperature have important role in creating freshwater bacterial community (Rubin and Leff, 2007; Kuznetsov et al., 1979). Moreover, even with the insufficient nutrition in the water, microorganism can proliferate and evolve using any available components around like an oligotroph. Heterotrophic bacteria have a crucial role in the food chain. They resist in the marine environment by means of taking food from the degrading and diluted organic materials (Allada and Kondalarao, 2016).
The excessive heterotrophic bacteria is intensely related to the nutrition supply and distribution within an ecosystem. The distance of waterways from the mainland will influence the number of heterotrophic bacteria (Varquez-Dominguez et al., 2008). Microbial components are as well counted to have a focal part in duction pro biomassa and carbon biogeochemical cycle and such other nourishments (Weinbauer, 2004).
Heterotrophic bacteria prevent the occurence of organic nitrogen accumulation which could decrease the quality of water on its cultivation media (Avimelech, 1999; Jurelevicius et al., 2013; Thyssesn et al., 2005; McGraw, 2002; Lima-Bittencourt et al., 2014). To be precise, heterotrophic bacteria have a significant and effective contribution for degradation of pollutant in the sea.
The heterotropic bacteria in the marine ecosystem decompose organic materials into essential mineral elements. The result of that mineralization is a source of nutrition for marine organism according to the tropics level in the marine ecosystems.
One of the processes of converting nitrogen in an aquaculture system is the bacterial heterotrophic process which directly transform ammonia to bacterial biomass. The heterotrophic bacteria that grow and evolve inside the media can be used by fish as a source of fodder. For its importance in biogeochemical cycles and its impact towards the quality of water, many research projects have scrutinized and assessed the composition of this bacterial community.
A detection method which is currently developed is the molecular detection mode through an analysis of genomic DNA; encoding the whole genetic information necessary for the bacteria cells functioning. A rapid microorganism DNA isolation is important for DNA analysis, such as PCR, Genocloning, Sequencing, dan Fingerprinting (Cheng and Jiang, 2006).
Due to its significance as an antagonism towards pathogenic bacteria (Vibrio sp, Aeromonas sp and Pseudomonas sp)-which are harmful for aquaculture animals (fish and shrimp), it is necessary to conduct an in-depth study.
The research was conducted from July to September 2017. The samples were taken in Siak River estuary, Riau province (Fig. 1). The analysis was accomplished in the Marine Microbiology Laboratory, Department of Marine Science and Technology, Riau University; whereas the DNA analysis was completed at the Genetics Laboratory, Biology Department, Riau University.
Antagonism test towards pathogen
Isolated bacteria were identified based on their morphological and biochemical attributes. The assembled data from the observation were used as supporting data in identifying the obtained bacteria species.
The antagonistic activity was done by employing an agar diffusion method and using a paper disc dripped with 0.5 µl amoxan as a positive control, NB (Nutrient Broth) 0.5 µl for negative control (-) and bacterial isolates repeated for three times. Afterwards, it was incubated at 28°C temperature for 24 hours. Furthermore, the formed transparent zone was observed and its diameter was gauged.
If the diameter of the inhibition zone is more than 20 mm, the bacteria growth inhibition response will be very powerful; but if the diameter was roughly between 10-20 mm, it will elicit a strong response; a 5-10 mm inhibition zone yields a moderate response; and when it is less than 5 mm, the response is categorized as weak (Greenwood, 1995).
DNA analysis was based on Feliatra et al (2016). DNA isolation cultures of heterotrophic bacteria were grown in 4 mL of liquid Luria-Bertani medium (LB) at 37oC for 24 hours. The cultures were then moved to 1.5 mL microtubes and centrifuged at a speed of 13,000 rpm (revolutions per minute) at 4oC for 2 minutes. The supernatant was discarded, and the pellet dried. Dissolved in 40 µL of of Tris-EDTA (TE) buffer solution with 8 pH and 20 µL of lysozyme added to the solution. The solution was inverted 3-5 times, and incubated for 10 minutes at 37oC. After incubator 50 µL of 10%, sodium dodecyl sulfate (SDS) was added and mixture was gently rotated (20 rpm) for 5-10 minutes on a rotamix rotator. Added 550 V of Phenol: Chloroform: Isoamyl alcohol (25:24:1) was added to the mixture and the mixture further rotated gently (20 rpm) for 10 minutes. The mixture was then centrifuged at 7.000 rpm for 10 minutes. The upper aqueos phase was transferred to a new microtube, and an equal volume of Chloroform:Isoamyl alcohol (24:1) was added to the tube. The mixture was gently rotated (20 rpm) on a rotamix rotator for 10 minutes. The mixture was then centrifuged at 7.000 rpm for 10 minutes. The upper aqueos phase was transferred to a new microtube and 3 M sodium acetate was added (40% volume) to the aqueos phase. Two times volume of 99% cold alcohol was then added to this solution to precipitate the DNA, and the solution chilled at -20oC for 30 minutes. The precipitated DNA was centrifuged at 13.000 rpm, 4°C for 10 minutes and the supernatant discarded. The DNA pellet was washed in 1 mL 70% cold ethanol centrifuged for another 10 minutes at 13.000, 4°C. The supernatant discarded, and resulting DNA pellet was then air dried. The pellet was dissolved in 40 µLTE-buffer (pH 8) and 2.5 µL of RNAse solution were added and homogenized using vortex mixer and incubated at 37°C for 1 hour. Finally, the DNA of probiotic candidate bacteria was ready to be used, or stored -20°C.
Polymerase Chain Reaction (PCR)
The PCR reaction was done by using 8 µL DNA of each probiotic candidate bacteria as template, and adding 23 µL of H2O, 4 µL of 10x Taq polyrmerase, 0.5 µL of 12mM dNTPmix, 2 µL for each of universal primers consisting of forward primer 24F (5’AGAGTTTGATCCTGGCT-3′) and reverse primer 1541R (5’AAGGAGGTGATCCAGCCGCA-3′) and 0.5 µL Taq polymerase. PCR conditions were: hot start for 2 minutes at 94oC, followed by 30 cycles of denaturation for 1 minute at 94oC, annealing for 40 seconds at 50oC, and strand extention for 1 minute at 72oC.
Purification of DNA PCR fragments
After PCR, the DNA obtained was separated on a 1% agarose gel by electrophoresis. PCR DNA bands were purified from the agarose gels using the Qiagen High Pure PCR Purification Kit (Japan), following instructions by the manufacturer.
Sequencing, BLAST and phylogenetic analysis
Purified PCR products were sequenced with ABI 3010 Genetic Analyzer XL Applied Biosystems. Sequencing was done at the Institute for Biotechnology Serpong, Tangerang, Banten. BLAST analysis was done using the tools and data of GenBank accessed at http://www.nebi.nih.gov/blast. DNA sequences were aligned using CLUSTALX 2.1. (Larkin et al 2007). Phylogenetic trees were generated using the tools provided by CLUSTAL X 2.1., and trees were visualized using TreeView X version 0.5.0.
Data Analysis
Data gained from the isolated bacteria identification and antagonism test towards pathogenic bacteria is presented in the form of table and picture. At the DNA examination, the outcomes were analysed using BLAST analysis. The data were then used for phylogenetic analysis to recognize the tag or name of the obtained species.
Phenotypic Characterization
Phenotypic characterization was completed to categorize bacteria from their morphological as well as physiological properties to particular reaction. The results of the classification can be seen in Table 1.
Table (1):
Morphological and biochemical observation.
| No | Isolate | Colony colours | Catalase | Gram | Motility | Indol | H2S | TSIA test | Citrate | MR | |||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| G | L | S | |||||||||||
| 1 | 1P11 | Yellowish white | + | + | + | – | + | – | – | – | + | – | |
| 2 | 1P12 | Yellowish white | + | + | + | – | – | + | – | – | + | – | |
| 3 | 1P14 | Yellowish white | + | + | + | – | – | – | – | – | + | – | |
| 4 | 1P15 | Yellowish white | + | + | + | – | + | + | + | + | + | – | |
| 5 | 1P18 | Milky-white | + | + | + | – | – | + | + | + | + | – | |
| 6 | 1P19 | Milky-white | + | + | + | – | – | – | – | – | + | – | |
| 7 | 2P20 | Milky-white | + | – | + | – | – | – | – | – | – | – | |
| 8 | 2P21 | White | + | + | + | – | – | – | + | + | – | + | |
| 9 | 2P22 | White | + | + | + | – | – | – | + | + | – | – | |
| 10 | 2P23 | Yellowish white | + | + | + | – | – | – | – | – | – | – | |
Note: G: glucose, L: lactose, S: sucrose, MR: methyl red
Antagonism Test of Heterotrophic Bacteria towards Pathogenic Bacteria
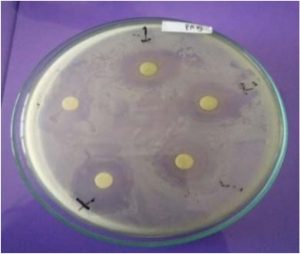
The antagonism observation result of 25 isolated bacteria towards 3 pathogenic bacteria indicates a fact that not all isolated bacteria are capable of inhibiting the growth of those three pathogenic bacteria.
The antagonistic activity of 25 isolates is demonstrated by the existence of transparent zone around the disc (Fig. 2). The transparent zone was then measured and the mean was obtained.
Based on the result of the mean of the established transparent zone diameter, 10 isolated bacteria having a capability to inhibit the growth of the three pathogenic bacteria (Vibrio sp, Aeromonas sp and Pseudomonas sp) were selected. The antagonism test result of the 10 isolated bacteria is displayed in Table 2.
Table (2):
Heterotrophic bacteria antagonism test observation result.
| Isolates | Inhibition Zone Diameter (mm) | ||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Vibrio sp | Aeromonas sp | Pseudomonas sp | |||||||||||||
| (+) | U1 | U2 | U3 | R | (+) | U1 | U2 | U3 | R | (+) | U1 | U2 | U3 | R | |
| 1P11 | 9 | 18 | 23 | 20 | 20,3 | 8,5 | 8 | 4,6 | 3,1 | 5,2 | 6 | 5 | 8 | 12 | 8,3 |
| 1P12 | 4 | 15 | 9 | 9,5 | 11,1 | 14 | 3,3 | 2,4 | 4,1 | 3,3 | 8 | 20 | 15 | 16 | 17 |
| 1P14 | 5,5 | 13 | 12 | 14 | 13 | 2,8 | 7,5 | 7,8 | 5,5 | 6,9 | 6 | 12 | 9 | 15 | 12 |
| 1P15 | 6 | 11,5 | 18 | 15,5 | 15 | 5 | 7,1 | 6,9 | 8,6 | 7,5 | 5 | 11 | 13 | 22 | 15,3 |
| 1P18 | 4 | 9 | 11,5 | 8 | 9,5 | 2,2 | 6,1 | 7,8 | 6,6 | 6,8 | 4 | 7 | 6 | 16 | 9,6 |
| 1P19 | 6 | 6 | 5 | 4 | 5 | 13,4 | 11,7 | 6 | 2,3 | 6,7 | 6 | 16 | 16 | 13 | 15 |
| 2P20 | 18 | 14 | 10 | 13 | 12,3 | 9 | 7,4 | 8,2 | 12,3 | 9,3 | 4 | 15 | 8 | 5 | 9,3 |
| 2P21 | 6 | 15 | 15,5 | 17 | 15,9 | 6 | 8,4 | 6,6 | 6,5 | 7,2 | 5 | 16 | 14 | 15 | 15 |
| 2P22 | 6 | 10 | 7 | 8,5 | 8,5 | 4 | 5 | 6 | 9 | 6,7 | 6 | 9 | 7 | 9 | 8,3 |
| 2P23 | 6 | 17 | 11 | 17 | 15 | 4 | 6,3 | 6 | 5,2 | 5,8 | 6 | 13,5 | 12,5 | 9 | 11,7 |
Note : (+): Positive control (Amoxan) , U1: 1-replication, (-): Negative control, (Nutrient Broth), U2: 2- replication, U3: 3- replication, R: Transparent zone mean.
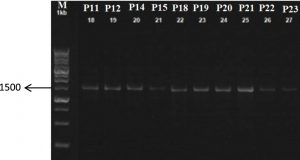
Electrophoresis of PCR product results
The PCR results dimension
Based on the result of the electrophoresis, DNA bands were on a scale of 1500. This scale was in accordance with the expected measure of bacterial 16S rDNA genes, that is 1500-1600 bp.
Genotypic Characterization
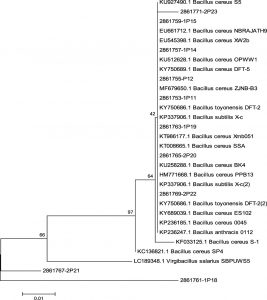
Sequencing was carried out using primer 24F and 1541R. The sequencing results were subsequently analyzed using BLAST and also phylogenetic to understand the species from the isolated bacteria. The BLAST investigation result from each isolate is presented in Table 3, whereas the phylogenetic tree analysis upshot is illustrated through Fig. 4.
Table (3):
The result of isolated bacteria 16S rRNA sequence with BLAST system.
Isolates |
Species |
Strain |
Access code |
Query Coverage |
Homology |
|---|---|---|---|---|---|
1P11 |
Bacillus cereus |
ZJNB-B3 |
MF679650.1 |
98% |
98% |
1P12 |
Bacillus cereus |
DFT-5 |
KY750689.1 |
100% |
99% |
1P14 |
Bacillus cereus |
XW2b |
EU545398.1 |
98% |
99% |
1P15 |
Bacillus cereus |
S5 |
KU927490.1 |
98% |
97% |
1P18 |
New spesies 1 |
SBPUWS5 |
LC189348.1 |
100% |
85% |
1P19 |
Bacillus cereus |
Xmb051 |
KT986177.1 |
100% |
99% |
2P20 |
Bacillus cereus |
BK4 |
KU258288.1 |
98% |
96% |
2P21 |
New spesies 2 |
SP4 |
KC136821.1 |
100% |
91% |
2P22 |
Bacillus toyonensis |
DFT-2 |
KY750686.1 |
98% |
99% |
2P23 |
Bacillus cereus |
NBRAJATH9 |
EU661712.1 |
98% |
99% |
Source: BLAST
On BLAST analysis result (Table 3), seven isolated bacteria are known to have similarity level to those bacteria from Bacillus genus with homology level is approximately between 96-99%. An isolate approaches the Virgibacillus salarius. Additionally, there were 2 isolates believed to originate from Indonesia for their homologous level was merely between 85% and 91% from the heterotrophic bacteria exist in Gen Bank. Table 3 depicting a morphological observation designates that generally the colour of bacteria colony graded from white to yellowish-white.
The results are in line with a study conducted by Montanari et al (2004) revealing two types of colony encountered on the media. The first colony colour was cream with shining surface while the second one was light yellow with rough surface (Schoeni and Wong, 2005). Meanwhile, the biochemical test result disclosed isolates belonging to gram positive bacteria, catalase positive and motile. In fact, some isolates were also able to ferment glucose, lactose, and sucrose.
Bacillus is a gram positive bacterium characterized by motility with perictric flagellum, oval endospores, and is sometimes cylinder or circular in shape, and very resistant to unfavorable conditions. The colony was milky white to yellow with wavy edges. This bacterium is aerobic, catalase positive, negative indole, and capable of fermenting glucose, lactose, and sucrose. In addition, they are widely distributed in various habitats and few of them were pathogenic. The optimum growth temperature was 28- 35oC (Holt, 1994). Bacillus is grouped to gram positive and positive catalase bacteria, which can grow at pH 6-8, no strains can produce indole and cannot flourish in anaerobic circumstance. Citrate and propionate are not usable carbon source (Costanzo et al., 2005).
The Bacillus genus comprises of members showing broad variety of physiological and ecological relation from DNA sequences and gene regulations. The most appealing species is predispose to be known for its pathogenicity and is closely associated genetically (Barbosa et al., 2005). Besides, on the ground of antagonism test conducted, heterotrophic isolated bacteria have found to be able to impede the development of the three pathogenic bacteria.
Antibiotics were intensively used as a medication and a growth promotor in agriculture industry. However, these chemical compounds lacks of attention as a pollutant in aquatic environment (Abriouel et al., 2010).
Bacillus has an anti-microbial resistant force and at the same time can produce antimicrobial so that this bacterium could survive within digestive tract (Stein, 2005 ; Moore et al., 2013; Ash et al., 1991). Another bacterium such as Bacillus spp is capable of synthesizing antagonistic molecules and secreting large amount of antibiotics out of cell. Members of Bacillus genus are known to generate extensive arsenal, antimicrobial subtances, including peptide and lipopeptide antibiotics, and bacteriocin.
A few Bacillus strains synthesize bacteriocin, which is only effective in resisting the same species of bacteria and negative gram bacteria, and some other strains have an extensive antibiotic activity spectrum (including antifungal and antiprotozoan). Bacteria from the Bacillus genus have enormous potential as probiotic cultures for their abundant existence. The bacteria are a producer of more than 200 antibiotics.
The genotypic identification result designates that isolated bacteria are actually part of Bacillus cereus (1P11, 1P12, 1P14, 1P15, 2P20, 2P23), Virgibacillus salarius (1P18) and Bacillus toyonensis (2P22). The 1P19 and 2P21 are the native species from Indonesia. Bacillus cereus is gram positive, aerobic-facultative, and spore widely disseminated in the ecosystem, and has intimate phenotypic and genetic (16S rRNA) relationship with several other species, especially B. an-thracis (Ash et al., 1991).
The research result discloses that the 10 heterotrophic isolate bacteria isolated from the Siak River estuary can inhibit the growth of tested pathogenic bacteria (Vibrio sp, Pseudomonas sp dan Aeromonas sp). Based on the phenotypic analysis result, 7 isolated bacteria have an association with the Bacillus genus. One isolate belongs to Virgibacillus salarius and the remaining two are assumed to be native Indonesian heterotrophic bacteria.
Acknowledgements
Grateful thankgoes to the Ministry of Technology,Research and Higher Education ofIndonesia who has support this research through the Competence Grant No. 304/UN.19.5.1.3/PP/2018.
Conflict of Interest
The author(s) declare that there is no conflict of interest.
- Abriouel, H., C.M. A. P. franz, N. B. Omar and A. Gálvez. Diversity and applications of Bacillus bacteriocins. FEMS Microbiol Rev. 2010; 35 (2011): 201-232. 10.1111/j.1574-6976.2010.00244.x.
- Allada, V and B. Kondalarao. Distribution of heterotrophic bacteria in the near shore waters and intertidal sediments of Visakkhapatnam, east coast of India. Res. J. Marine. Sci. 2016; 4(6): 1-12. http://www.isca.in/MARINE-SCI/Archive/v4/i6/1.ISCA-RJMarineS-2016-004.php [Accessed 22 February 2018].E-ISSN 2321-1296.
- Ash, C., J. A. Farrow, M. Dorsch, E. Stackenbrandt, and M. D. Collins. Comparative analysis of Bacillus anthracis, Bacillus cereus, and related species on the basis of reverse transriptase of 16S rRNA. Int. J. Syst. Bacteriol. 1991; 41: 343-346.
- Avimelech, Y. Carbon nitrogen ratio as a control element in aquaculture system. Aquaculture. 1999; 176: 227-235.
- Barbosa, M.T., et al. Applied and Environmental Microbiologi: Screening for Bacillus Isolates in The Broiler Gastrointestinal Tract. 2005; Vol 71.No. 2. American Society for Mikrobiologi. Amerika.
- Cheng, HR and N. Jiang. Extremely rapid extraction of DNA from bacteria and yeasts. Biotechnol. Let. 2006; 28: 55-59.
- Costanzo, S. D., J. Murby, and J, Hates. Ecosystem Response to anticiotic entering the aquatic environment. Marine pollution Bulletin. 2005; 31: 218-223.
- Feliatra, F., I. Lukistyowati, D. Yoswaty, H. Rerian, D. Melina,W. Hasyim, T.T. Nugroho, A.R. Fauzi and R. Yolanda. Phylogenetic analysis to compare populations of acid tolerant bacteria isolated from the gastrointestinal tract of two different prawn species Macrobrachium rosenbergii and Penaeus monodon. AACL Bioflux. 2016; 9(2):360-368.
- Godwin, C.M. and Cotner, J.B. Aquatic heterotrophic bacteria have highly flexible phosphorus content and biomassa stoichiometry. The ISME. J. 2015; 9: 2324-2327. 10.1038/ismej.2015.34.
- Greenwood, D. Antibiotics, susceptibility (sensitivity) test antimicrobial and chemotherapy. USA: Mc. 1995; Graw Hill Company. P. 47.
- Holt, J.G. Bergey’s Manual of Determinative Bacteriology. Ninth Ed.A Wolters Kluwer Company. 1994; Philadelphia. Page: 562-570.
- Jurelevicius D., V. M. Alvarez and J. M Marques. Bacterial Community Response to Petroleum Hydrocarbon Amendments in Freshwater, Marine, and Hypersaline Water-containing Microcosms. Appl Environ Microbiol. 2013; 79: 5927–35.
- Kong X, P and S. H. Ye, The impact of water temperature on water quality indexes in north of Liaodong Bay.Mar Pollut Bull. 2014; 80: 49–245.
- Kuznetsov, S., G. Dubinina, and N. Lapteva. Biology of oligotrophic bacteria. Annu. Rev. microbial. 1979; 33: 377-387. http://www.annualreviews.org/doi.abs/10.1146/annurev.mi.33.100179.002113.
- Lima-Bittencourt, C. I., P. S. Cousta, M. P. Reis, A. B. Santos, F. A. R. Barbosa, J. L. Valentin, F. L. Thompson, E. Chartone-Souza and A. M. A. Nascimento. A survey on cultivable heterotrophic bacteria inhabiting a thermally unstratified water column in an Atlantic Rainforest Lake. Peer J. 2014; 2: e478. 10.7717/peerj.478.
- McGraw, W. J. Utilization Of Heterotrophic and Autotrophic Bacteria In Aquaculture. 2002; Global Aquacultere Advocate.82-83 Pp.
- Montanari, G., A. Borsari, C. Chiavari, G. Ferri, C. Zabonelli and L. Grazia, Morphological and phenotypical characterization of Bacillus sporotherma-durans. App. Mic. J. 2004; 97: 802-809. 10.1111/j.1365-2672.2004.02371.x
- Moore, T., L. Globa, J. Barbaree, V. Vodyanoy and I. Sorokulova, Antagonistic activity of Bacillus bacteria against Food-Borne Pathogens. J. Prob. Health. 2013; 1:3. http://dx.doi.org/10.4172/2329-8901.1000110.
- Rubin, M. A and L. G. Leff. Nutrients and other abiotic factors affecting bacterial communities in an Ohio River (USA). Microbia l Ecology. 2007; 54: 374-383. 10.1007/s00248-007-9209-2.
- Schoeni, J. L., and Wong, A. C. L. Bacillus cereus food poisoning and its toxins. J. Food. Protection. 2005; 68(3): 636-648.
- Stein, T, Bacillus subtilis antibiotics: structures, syntheses and specific functions. Mol Microbiol. 2005; 56: 845-857.
- Thyssesn, M., D. Lefevre, G. Caniaux, J. Ras, C.I. Fernandez, and M. Denis. Spatial distribution of heterotrophic bacteria in the northeast Atlantic (POMME stud area) during spring 2001. J. Geophys.Res., 2005; 110: 1–16.
- Varquez-Dominguez.E., C.M. Duarte, S. Agusti, K. Jurgen, D. Vangue and J. M. Gasol, Microbial plankton abundance and heterotrophic activityacross the central Atlantic Ocean. J. Prog. In Ocean. 2008; 79(1): 83-94.
- Weinbauer MG. Ecology of prokaryotic viruses. FEMS MicrobiolRev 2004; 28: 127–81.
© The Author(s) 2018. Open Access. This article is distributed under the terms of the Creative Commons Attribution 4.0 International License which permits unrestricted use, sharing, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.