ISSN: 0973-7510
E-ISSN: 2581-690X
Mercury (Hg) being one the most toxic element on the earth and its capability to result in neurotoxic disorder has become a serious problem for human health and environmental issues. Methyl mercury is the organic form of the mercury which easily accumulates within the aquatic animals and led to biomagnification in other higher organisms. Hg in the environment is resulted from unregulated discharge of mercury within the environment via numerous industries. Hence, it is crucial to bio-convert the toxic form of mercury to non-toxic form with the useful resource of microbes. One of the pronounced site of mercury contamination reported in India (Panipat) containing 147ppm Hg content, was selected to construct metagenomic library of mer operon using E.coli as host. The clone showed maximum tolerance towards mercury (90ppm) accompanied through effective volatilization (91.89% to 41.23%) for Hg (10-90ppm). The clone was able to efficiently bio-convert Hg in actual contaminated site as well. It was also encapsulated in sodium alginate beads and polyacrylamide gel in order to test its reusability for conversion of Hg.
Mercury, mer operon, CV-AAS, sodium alginate bead, polyacrylamide gel entrapment.
The outflow of industrial effluents has been always the cause of environmental concern as they pollute rivers and lands by adding up the toxic element and harmful metals. Among various heavy metals, mercury is known as a lethal metal which is positioned on 6th number in toxicity1. The unregulated discharge of mercury from industries and other human interventions has led to widespread of various neurological diseases2. The continuous and prolonged exposure to mercury may be source of fatigue, neurasthenia, numbness in toes and fingers, clumsiness, inability to cognizance, hearing to and visual disabilities, acute bronchitis, intestinal pneumonitis3. The worst case is followed by coma and death. Biomagnification of mercury in each trophic level has become a major cause of health risks at each level as it has entered the food chain through bio-accumulation. The level of mercury has been found to increase by 20-100 or even more folds than its permissible limit as the result of the unregulated and uncontrolled discharge of mercury4.
Increase in mercury pollution attracted the attention of some scientist to encounter the problem and they came up with the most widely used technique of bioremediation which uses microbes to convert toxic metals to less toxic form. This inquisitiveness of scientist helped them to discover microbes that can effectively grow well in the presence of mercury. Scientist discovered the sets of the genes which were responsible for providing resistant to such a toxic metal and called it as mercury (mer) operon4. Further researches confirm the presence of this operon on transposons element, conjugative plasmid and chromosomes which confers the bacteria with ability to survive mercury contamination and also confers the antibiotic resistance towards some antibiotics5,6.
The indigenous researches on mer operon has discovered two major genes which are mainly responsible for mercury conversion in bacteria are merA and merB genes. Where merB genes encodes methylmercury lyase which convert organic form of mercury (CH3Hg2+) to Hg2+ and merA gene code for mercury reductase which converts inorganic form of mercury (Hg2+) to non-toxic risky form (Hgo). Other genes of mer operon have been also found which are equally important for transportation of Hg within the system7.
This study was undertaken to clone the mer operon derived through metagenomics. The metagenome from the sample was isolated and the desired fragment (mer operon) was cloned in E.coli. The clone was checked for its ability to convert toxic form of Hg to non-toxic form in Hg contaminated site (Panipat Soil Sample). The clone was also immobilized to examine its reusability for the same.
Sample collection and mer operon library construction
Panipat industrial area was selected for soil sample collection because it is reported to have much higher concentration of mercury than the permissible limit. The soil was taken 10 cm under the ground level by digging it and was kept in sterile container at 4oC for further investigation.
In order to determine the concentration of mercury present in the soil sample, 10 gm of sample was weighed to check Hg concentration present in the soil using CV-AAS.
MOBIO kit was used to isolate metagenomic DNA from the soil sample following the protocol provided by the manufacturer. Before constructing the library the DNA was quantified using UV-absorption at 260nm at dilution fold of 500. After the quantification the partial restriction digestion was performed to get the desired fragment size of 5 to 10Kb in order to make clones.
For restriction digestion reaction, sterile micro-centrifuge tube was used to set up the reaction of 150µl which consist of 100ml of DNA, 15µl of 10X enzyme buffer, 1.5ml of Sau 3A1 and 33.5ml autoclave distilled water. The reaction was setup in the ice bucket and immediately transferred to 16oC for 3h. The reaction was terminated by heating the mixture at 70oC in water bath. The digested metagenomic DNA was run on 1% Agarose using gel electrophoresis assembly (BIORAD) and the fragments of the DNA in range of 5-10Kb were gel eluted using HiYield Gel/PCR DNA Mini Kit (Real Biotech Corporation, Taiwan). Ligation reaction was setup using digested DNA and dephosphorylated pUC 19 vector digested with Bam HI along with T4 DNA ligase enzyme. The mixture was incubated at 4oC for 16-18h and reaction was terminated by heat inactivation of enzyme at 70oC. Electroporation at 200W, 25mF and 2.5kV was done by introducing recombinant vector into host bacteria (E.coli DH10B) with aid of Micropulser II (BIO RAD). Immediately, the tube was poured with 1ml of Luria Broth (LB) and kept in shaker incubator at 180rpm for 1h. Transformed cells (100µl) were spread on the Luria Bertani Agar (LBA) plates supplemented with ampicillin, X-gal and IPTG and incubated at 37oC for 16h.
Screening and selection of mercury resistant clones
The white colonies were picked and streaked on the LBA plates which were supplemented with ampicillin and 5ppm of Mercury Chloride (HgCl2), incubated at 37oC for 24-48h. The clones which were able to grow in the presence of 5ppm of HgCl2 were transferred to fresh LB supplemented with 10ppm of HgCl2. The clones showing visible growth at 10ppm were further transferred to 20ppm, 30ppm and so on up to 100pm for acclimatization of clones and examined by taking OD at 600nm. The clone which showed maximum growth after acclimatization at highest HgCl2 concentration was selected for further studies. To check the presence of merB gene, the clone was grown in LB with 5ppm and 10ppm of Methyl Mercury Chloride (CH3HgCl2), incubated at 37oC for 18h and OD was taken at 600nm.
Antibiotics resistance assay
Antibiotic resistance of the selected clone was checked through disk diffusion method against various antibiotic listed in Table 1. Freshly cultured clone was spread on the LBA plates and antibiotic discs were placed onto it, incubated at 37oC for 18h. Later, zone of inhibition was measured.
Table (1):
Antibiotics used for antibiotic resistance activity.
Sl.No |
Antibiotic |
Concentration (mcg) |
|---|---|---|
1 |
Streptomycin |
25 |
2 |
Norflaxin |
10 |
3 |
Tetracyclin |
30 |
4 |
Vancomycin |
30 |
5 |
Gentamycin |
30 |
6 |
Kanamycin |
30 |
Investigation of Hg volatilization by CV-AAS
The clone was inoculated in LB media containing 10 to 100ppm of HgCl2 and incubated at 37oC;180rpm. The culture was drawn periodically on 2nd, 4th and 6th day under sterile conditions in tubes and centrifuged at 8000rpm for 8 mins. The supernatant was used for CV-AAS analysis (Analytical Jena make (Vario-6) cold vapor Hg-AAS).
In-vivo efficacy of the clone for Hg Volatilization
The Panipat soil (non-sterile) was taken and mixed with the sterile soil to make 5 kg of mixture of around 10ppm Hg concentration (3:2) wt/wt). Overnight grown culture (500ml);(1OD) was sprayed onto the soil mixture. The soil sample was periodically taken out on 2nd, 4th and 6th day. The samples were used for CV-AAS in order to check the ability of clone to transform the toxic Hg to non-toxic form on the contaminated site.
Entrapment of clone to check its repeatability
Immobilization was done to check the efficiency of the clone to convert the Hg from toxic form to non-toxic form of mercury by repeated use. Two methods were used for immobilization listed below (3 cycles).
Sodium alginate Entrapment method
Pre-grown culture (16h) was mixed with sodium alginate (3%w/v) in the ratio of 1:4 and mixed well to make slurry under sterile conditions. The slurry was added to 0.2M of Calcium Chloride (CaCl2) drop-wise using sterile tips at room temperature in laminar air flow. Then the beads were stored at 4oC for 1h. After cooling the beads were washed 2-3 times with autoclaved distilled water and inoculated in conical flask containing 30ml of LB supplemented with 10, 20 and 30ppm of HgCl2 respectively. The flasks were incubated at 37oC; 180rpm and beads were drawn at the time interval of 24h (for 3 days). The beads were washed and re-inoculated to LB having same concentration of HgCl2. This procedure was repeated thrice to check reusability of entrapped clone. CV-AAS of drawn samples at different time intervals were done.
Polyacrylamide gel entrapment method
Cell suspension of pre-grown culture (16h) was prepared by adding 2ml of culture into 10ml of chilled autoclaved distilled water and kept aside. 10 ml of the Potassium Phosphate Buffer (pH 7.0, 0.2M) was prepared, Acrylamide (2.85g), Bis-acrylamide (0.15g), Ammonium persulphate (10mg) and TEMED (1ml) were added to the buffer. The chilled cell suspension was mixed with polyacrylamide gel and poured into a sterile petri plate for solidification. After solidification, the cubes (5 mm x 5mm x 5mm) were cut using sterile scalpel and kept in Sodium Phosphate Buffer (pH 7.0, 0.2M) for 1h at 4oC. The cubes were washed 3-4 times and put into conical flask containing LB (30ml) supplemented with 10, 20 and 30ppm of HgCl2. The flasks were incubated at 180rpm, 37oC and cubes were drawn each day after 24h (for 3 days). The cubes were washed and re-inoculated to LB having same concentration of HgCl2. The drawn samples were used for CV-AAS analysis.
Metagenomic library construction
Panipat soil sample was estimated for its Hg content by CV-AAS which was observed to be around 147ppm that’s 14700 folds higher than the permissible limit i.e. 0.01mg/ml for industrial effluents8.
The DNA isolated from Panipat industrial soil had the concentration of 500ng/µl at the absorbance of 260nm (Fig. 1). Metagenome was digested using Sau3A1. The desired DNA fragment in the range of 5-10Kb was cut and gel eluted. The DNA obtained after elution was ligated to dephosphorylated pUC 19 vector predigested with Bam HI.

Fig. 1. Percentage of gram-positive, gram-negative and fungal organisms isolates recovered from the culture
Approximately 200 clones were streaked on LBA plates supplemented with 5ppm of HgCl2, only 120 were able to grow among them.
Selection of the mercury resistant clone
The clones which were able to grow on 5ppm were transferred to 10ppm Hg concen-tration, and only 74 clones showed visible growth. Further with increase in concentration of Hg to 50ppm, steep decrease was seen among the selected clones (Table 2) and only 5 clones (P02, P15, P27, P58 and P98) were able to grow.
Table (2):
Numbers of clones shown growth at different Hg concentration.
Sl.No |
Concentration of HgCl2 |
Number of tolerant clones |
|---|---|---|
1 |
05 |
120 |
2 |
10 |
74 |
3 |
20 |
33 |
4 |
30 |
14 |
5 |
40 |
5 |
6 |
50 |
5 |
7 |
60 |
– |
8 |
70 |
– |
The clones which were able to grow at 50ppm of Hg had been chosen for acclimatization, by growing and transferring them to higher concentration of Hg (10ppm to 100ppm). Among five, chosen clones only one (P02) was able to grow at 90ppm and beyond it no growth was seen (Table 3). Hence P02 clone was selected for further studies.
Table (3):
Acclimatization of clones.
| Sl.No | Mercury Chloride Concentration (ppm) | |||||
|---|---|---|---|---|---|---|
| P02 | P15 | P27 | P58 | P98 | ||
| 1 | 10 | + | + | + | + | + |
| 2 | 20 | + | + | + | + | + |
| 3 | 30 | + | + | + | + | + |
| 4 | 40 | + | + | + | + | + |
| 5 | 50 | + | + | + | + | + |
| 6 | 60 | + | – | – | + | – |
| 7 | 70 | + | – | – | – | – |
| 8 | 80 | + | – | – | – | – |
| 9 | 90 | + | – | – | – | – |
| 10 | 100 | – | – | – | – | – |
Antibiotic resistant activity
Upon research of P02 for antibiotic resistance activity using disc diffusion method the zone of inhibition was measured (mm) (Fig. 2). P02 has shown resistant towards Tetracyclin (30mcg) and Vancomycin (30 mcg), whereas showed the susceptibility towards Norflaxin (10mcg) and Gentamycin (30 mcg). The microbes found to be mercury resistant are often antibiotic resistant[5]which is result of horizontal gene transfer7.
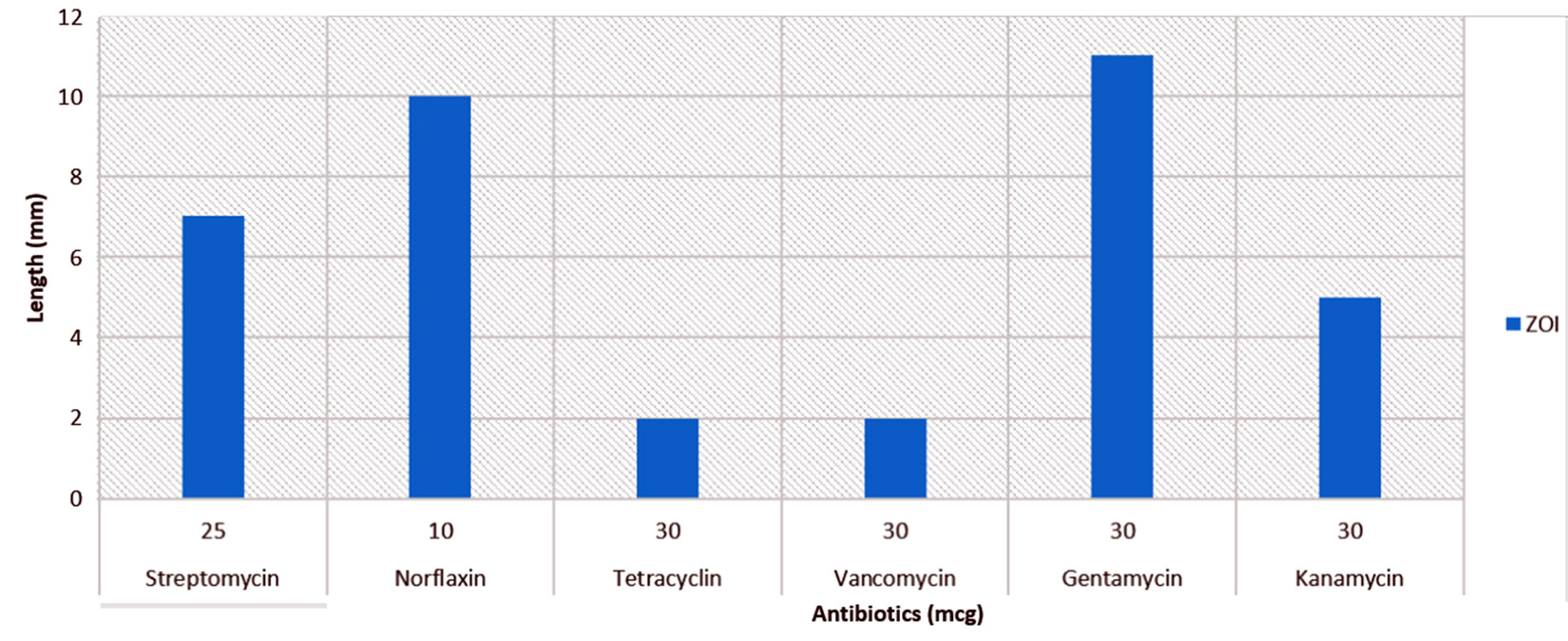
Fig. 2. Percentage of gram-positive, gram-negative and fungal organisms isolates recovered from the culture
CV-AAS of Hg volatilization
Volatilization analysis was done to quantify the amount of Hg converted to non-toxic form in media. The volatilization was found to be maximum on 6th day at 10ppm (91.89%), however it was 89% on 4th day and 87.37% on 2nd day. Gradual increase in volatilization was seen on 2nd day (37.05%), 4th day (39.46%) and 6th day (41.23%) at 90ppm (Fig. 3). The volatilization efficiency of P02 is more as compared to the volatilization by isolates as reported9,10.
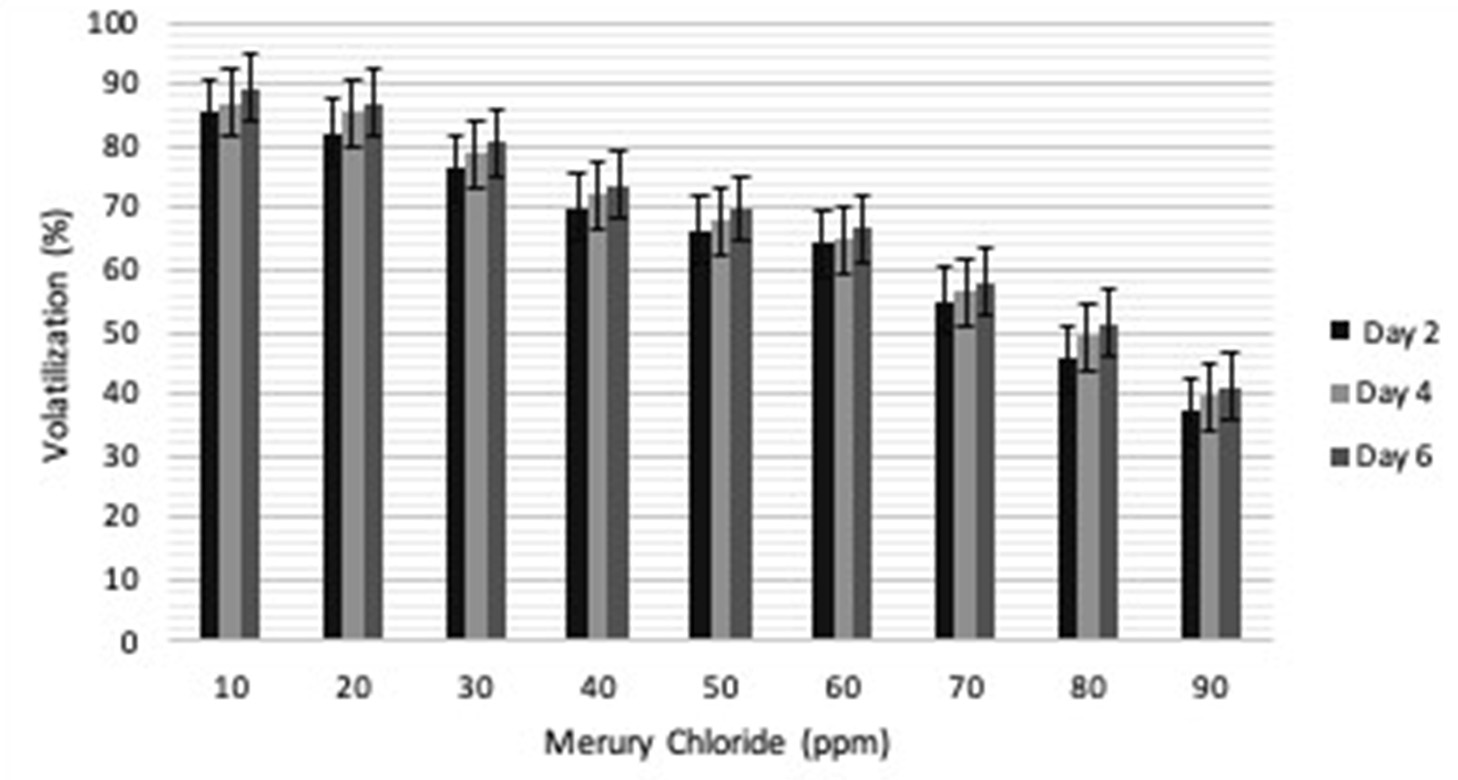
Fig. 3. Percentage of gram-positive, gram-negative and fungal organisms isolates recovered from the culture
Efficiency of volatilization by clone on soil
In order to check the functionality of P02 on mercury contaminated soil this study was undertaken and the results showed that P02 was efficient in conversion of Hg2+ to Hgo. 10ppm of concentration was chosen for soil as it showed maximum volatilization in media. The volatilization was checked on 2nd, 4th and 6th day and maximum was found to be on 6th day which was about 91.87% as compared to 4th day and 2nd day, which was calculated to be around 85.97% and 80.37% respectively (Fig. 4). The volatilization of Hg was more on mercury contaminated soil as compared to only media (LB+Hg) may be due to autochthonic bacteria which might have increased the volatilization.
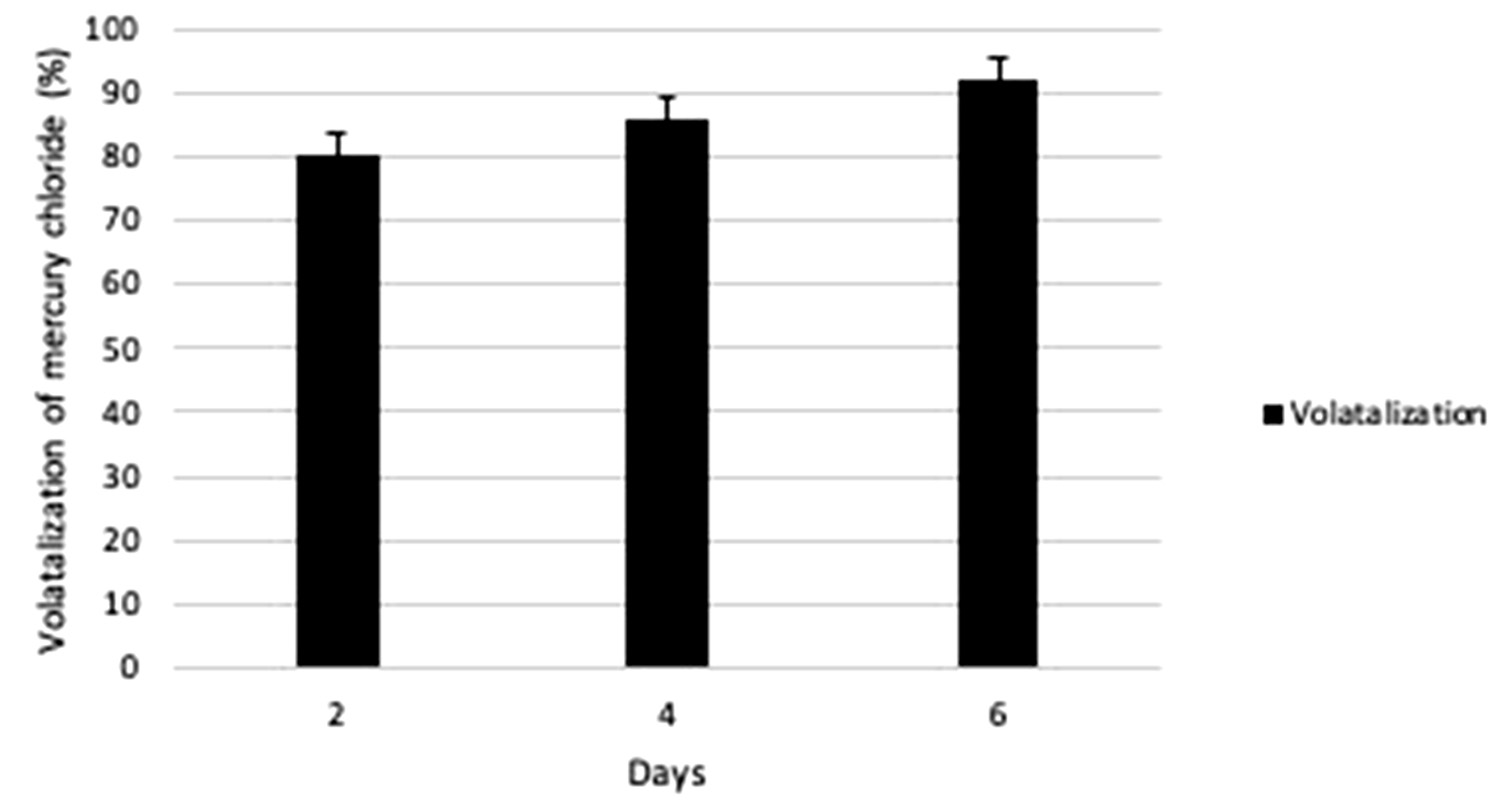
Fig. 4. Percentage of gram-positive, gram-negative and fungal organisms isolates recovered from the culture
Entrapment of clone and its reusability
Reusability of clone P02 was checked by its immobilization by two methods i.e. sodium alginate and polyacrylamide. CV-AAS analysis of supernatant showed more volatilization at low level of Hg concentration in both the cases.
The results also revealed volatilization of Hg (10-30ppm) was more with clone immobilized in sodium alginate (cycle 1: 84.88 to 49.53%; cycle 2: 37.5 to 31.76%; cycle 3: 21.82 to 18.5%) as compared to immobilization in polyacrylamide (cycle 1: 60.83 to 40.41%; cycle 2: 42.5 to 37%; cycle 3: 37.1 to 29.1%).
Though the volatilization in cycle 1 was more in case of sodium alginate but the reusability of clone was found to be more in polyacrylamide entrapment (Fig. 5a & 5b).
Numerous uses of mercury have brought about the enormous human health problem as it is one of the most toxic heavy metal found in the environment13. In order to triumph over the toxicity, some microbes have developed the mechanism to transform the toxic compound to non-toxic compound which gave perception of bioremediation. Many studies supported that microbes can convert toxic mercury to non-toxic form14.
Most of the microbes cannot be culture in lab but lives in the environment and contribute toward the bioremediation of mercury if present in their habitat3. In our study P02 was found to convert both form of Hg i.e. CH3HgCl2 (organic) to HgCl2 (inorganic) and finally to its non-toxic volatile form (Hgo), hence the clone have the broad spectrum mechanism of Hg tolerance. It has been found that antibiotic resistant genes are commonly found on the plasmid or the transposons carrying Hgr loci11 which may be transferred through horizontal gene transfer on the conjugative plasmid. P02 have shown antibiotic resistant property toward tetracyclin (30mcg) and vancomycin (30 mcg) along with the resistance to mercury. The mechanism of mercury resistance for both the form (organic and inorganic) was primarily studied on Staphy-lococcus aureus a clinical isolate which was also found to be resistance to antibiotic called penicillin4. It was observed that the maximum volatilization of Hg (10-90ppm) in media (LB+ Hg) happened on day 2 (87.37%-37.05%), whereas almost complete conversion happened on day 6 (91.89%-41.23%) which was the added volati-lization % of the residual Hg after day 2. The application study results indicated that the volatilization of P02 was much more effective in Hg contaminated soil (80.34%, 85.97% and 91.87% on day 2, 4 & 6 respectively) at 10ppm, which may be due to the activity of autochthonic microbes which has escalate the process of volatilization. We have also observed that polyacrylamide gel entrapped P02 clones were more efficient for reusability than the sodium acrylamide and can be effectively used for Hg bioremediation, as supported by other studies12.
In this study clone P02 has confirmed maximum tolerance (90ppm) of Hg with the efficiency of volatilizing it to vapor form in LB media. The volatilization turned more visible in actual mercury-contaminated soil than the LB media. Clones may be immobilized in poly-acrylamide gel and used time and again for Hg conversion efficiently15-17.
We are thankful to Department of Science and Technology, Government of India, for providing the funds to carry out the study and also thankful to Amity University, Sector-125, Noida, UP, for providing infrastructure and Dr. Rakesh Sharma, IGIB, New Delhi, Dr. Nahar Singh, NPL, Delhi for their help and support.
On behalf of all authors, the corresponding author states that there is no conflict of interest.
- Barkay T, Miller S M, Summers A O. Bacterial mercury resistance from atoms to eco-systems. FEMS microbiol. Rev., 2003; 27(2-3): 355-384.
- Woods J S,Heyer N J, Russo J E, Martin M D,Farin F M. Genetic polymorphisms affecting suscep-tibility to mercury neurotoxicity in children: Summary findings from the Casa Pia Children’s Amalgam. Clinical Trial Neurotoxicology, 2014; 44: 288-302.
- Mathema V B,Thakuri B C,&Sillanpהה M. Bacterial mer operon-mediated detoxification of mercurial compounds: a short review. ArchMicrobiol, 2011; 193(12): 837-844.
- Jaiswal G, Porwal S. Role of mercury resistance (mer) operon in bioremediation of mercury contamination. J. BioChem. Res., 2014; 2: 986-997.
- Porwal S, Singh R. Cloning of merA Gene from MethyloteneraMobilis for Mercury Bio-transformation. Indian J. Microbiol, 2016; 56(4): 504-507.
- McArthur J V,Tuckfield R C. Spatial patterns in antibiotic resistance among stream bacteria: effects of industrial pollution. Appl Environ Microbiol., 2000; 66(9): 3722-6.
- Sant’ana YX,Chartone-Souza E, Ferreira MD. Drug resistance and colicinogeny of Salmonella typhimurium strains isolated from sevrage-contamined surface water and humans in Belo Horizonte, Brazil.Revista Microbiol., 1989; 20(1): 41-9.
- Nascimento A M,Chartone-Souza E. Operon mer: bacterial resistance to mercury and potential for bioremediation of contaminated environments. Genetics Mol. Res., 2003; 2(1): 92-101.
- Mercury pollution of India, Centre for Science and Environment. http://www.cseindia.org/node/439. Accessed 20 September 2017
- MahbubK R, Krishnan K, Naidu R,Megharaj M. Mercury resistance and volatilization by Pseudoxanthomonas sp. SE1 isolated from soil. Environ. Technol. Inno., 2016; 6: 94-104.
- Sathiyanarayanan G,Saibaba G, Kiran GS, Yang YH, Selvin J. Marine sponge-associated bacteria as a potential source for polyhydroxyalkanoates. Crit Rev Microbiol., 2017; 43(3): 294-312.
- Wireman J, Liebert CA, Smith T, Summers AO. Association of mercury resistance with antibiotic resistance in the gram-negative fecal bacteria of primates. Appl Environ Microbiol., 1997; 63(11): 4494-503.
- Ashraf W. Levels of selected heavy metals in tuna fish. Arabia J. Sci. Eng., 2006; 31(1A): 89.
- Bilal M,Asgher M,& MN Iqbal H. Polyacrylamide gel-entrapped fungal manganese peroxidase from Ganoderma lucidum IBL-05 with enhanced catalytic, stability, and reusability charac-teristics. Protein and peptide letters, 2016; 23(9): 812-818.
- Wang X, He Z, Luo H, Zhang M, Zhang D, Pan X,& Gadd GM. Multiple-pathway remediation of mercury contamination by a versatile selenite-reducing bacterium. SciTotal Environ., 2018; 615: 615-623.
- Kane AL, Al-Shayeb B,Holec PV,Rajan S, Le Mieux NE,Heinsch SC,Psarska S,Aukema KG, Sarkar CA,Nater EA,Gralnick JA. Toward bioremediation of methylmercury using silica encapsulated Escherichia coli harboring the mer operon. PloS one, 2016; 11(1): e0147036.
- Sinha A,Khare S K. Mercury bioremediation by mercury accumulating Enterobacter sp. cells and its alginate immobilized application. Bio-degradation, 2012; 23(1): 25-34.
© The Author(s) 2019. Open Access. This article is distributed under the terms of the Creative Commons Attribution 4.0 International License which permits unrestricted use, sharing, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.



