ISSN: 0973-7510
E-ISSN: 2581-690X
Groundwater is the main water source for many areas in Saudi Arabia and the only source of water in some areas. Many local studies have reported that high nitrate concentrations in some wells of groundwater. To provide safe drinking water, the excess amounts of nitrate have to be removed by bio-denitrification process. This study aims to develop a denitrifying biological filter using denitrifying bacteria immobilized on microbial cellulose for the removal of nitrates from water contaminated with nitrate. Denitrifying bacteria that can form biofilter on microbial cellulose were isolated from different regions in Saudi Arabia and were characterized by molecular techniques. They were evaluated for their ability to analyze nitrates and to develop biofilter to remove nitrates from contaminated water. In the results of this project, an optimal microbial cellulose production was achieved by Gluconacetobacter xylinus ATCC 23768 in the lab, which had facilitated the use of biofilter with the immobilized nitrate-reducing bacteria Pseudomonas aeruginosa. The reduction rate of nitrate was reached 1.9mg/L from the starting concentration of 100 mg/L after 18h. Promising results of nitrate removal rate on MC immobilized with Pseudomonas aeruginosa on biofilter at optimized lab conditions of pH, and proper carbon source were achieved. The results suggest that water contaminated with nitrate can be removed by the bio-denitrification process effectively.
Nitrate-reducing bacteria, Saudi Arabia, Biofiltration, bio-denitrification, Groundwater
Geographically, Saudi Arabia constitutes 2.2 million km2. The rainfall level is considered scarce between 50-100 mm. Therefore, Saudi Arabia facing water shortage, which could affect its water demands by the year 2025. In Saudi Arabia, the primary water supply is the conventional source, which is groundwater, and non-conventional source, which is desalination. In Saudi Arabia, there are thirty-five desalination plants capable of producing potable water from seawater and fresh groundwater. Currently, Saudi Arabia is one of the biggest countries that produce desalinized water in the world. The groundwater is used in many regions of the country and sometimes it is the only source of water in some areas. Due to the high number of people and lifestyle, the yearly water needs were enlarged in 1980 from 2,352 MCM (million cubic meters) to 27,239 MCM in 1990. In 1992 the needs increased to 30,000 MCM1,2. The decline in groundwater is due to many misused in industrial, agricultural activities humans in Saudi Arabia3.
Nitrate is considered the most dangerous compound in wastewater and rain because of its toxicity and its ability to contaminate the groundwater. Nitrate is regarded as one of the drinking water contaminants that come mainly from wells and groundwater. Studies reported that nitrate concentrations in some wells of groundwater exceed the permitted limit. Well water in Saudi Arabia contains a different level of nitrate ranging from 1.1 to 884 mg/l Nitrate, and its derivatives cause several medical problems such as stomach and bladder cancer and blue-baby syndrome. The excess amounts of nitrate have to be removed in order to provide safe drinking water. Different approaches such as reverse osmosis, ion exchange, chemical reduction, in addition to biological denitrification were employed to eliminate or reduce the nitrate in drinking water. These treatments removed some of the nitrates, but with varying degrees of efficiencies, much of which depended on the presence of other substances found in the water. Despite their lab efficacy, many of these methods are still in the process of validation for commercial uses.
The health risk of groundwater depends on the use of humans, animals, crops, or industrial processes. The toxicity level of nitrate to humans is caused by the total accumulation of nitrate to nitrite inside the human body. One of the outcomes of nitrite accumulation is the formation of Methemoglobinemia in which hemoglobin is oxidized to methemoglobin, which affects the oxygen level significantly. Therefore, in the US, the current maximum contaminant nitrate limit in safe water is 10 mg/L, that is concordant with the WHO guidelines limits4.
Denitrification is a biological treatment considered effective in removing nitrates. It is a process by which highly oxidized forms of nitrogen, such as nitrate (NO3-) and nitrite (NO2-), are reduced to nitrogen intermediates, nitric oxide (NO), nitrous oxide (N2O), and ultimately molecular nitrogen (N2) to be available for consumption by various groups of the organism. The remediation of groundwater contaminated with nitrate biologically may provide an efficient and cost-effective methodology for water denitrification5. The biological denitrification was studied largely by employing different methods in order to provide a safe drinking water. It was shown that the denitrifying bacteria can assimilate nitrate to produce a harmless nitrogen gas and nitrous oxide could provide highly specific, efficient, and cost-effective technology6-8.
Fixed biofilm reactors were also used for nitrate removal by employing immobilized denitrification bacteria on diverse materials. Many different bacterial strains have been considered for immobilization and biofilm formation including Paracoccus denitrificans, Nitrosomonas europe and Paracoccus denitrificans. In a separate study for nitrate removal using pumice as immobilized supporting material, Paracoccus denitrificans had shown a high removal rate of 97.69% when methyl alcohol was used as a primary carbon source9. Biofilms is formed when microbial cells are attached to a solid surface (a substratum). Biofilms are made up of microbial cells and exopolysaccharides (EPS). The vast majority of bacterial EPS are made up of specific and nonspecific polysaccharides. Biofilms form readily on substances such as wood chips, frittered glass particles, polyester foams, and various plastics10. In this study, we aimed to develop a denitrifying biological filter using denitrifying bacteria immobilized on microbial cellulose for denitrification process from contaminated water.
Sampling Area and Sample Collection
Medina region: Medina is a city in the Al-Hejaz region of western Saudi Arabia at an altitude of 26.57 (26° 34’ 0 N) and a longitude of 50.07 (50°4’ 0 E). The soil of the Medina consists of basalt primarily. While the south of the city is made of volcanic ash. Jazan region: Jazan is situated in the south-west of Saudi Arabia on the coast of the Red Sea at a latitude of 16° 53′ 21″ N and a longitude of 42° 33′ 40″ E and serves a vast agricultural region. Riyadh region (Hanifa Valley): Riyadh is the capital city of Saudi Arabia. Historically, it belongs Najd and Al-Yamama regions. It is located in the middle of the Arabian Peninsula at an altitude of 24° 38′ 0″ N and a longitude of 46° 43′ 0″ E 11. Samples were collected from different soil’s trees and soil layers from the rhizosphere of Morus alba (TO), Aloe perryi, Lactuca sativa, Citrus latifolia (LM), Lycopersicon esculentum (TM) and Mentha pennyroyal (MI). Three samples of soil were collected from 0-10 cm depth at intervals of 200 meters from each other. Jazan region: Four samples of agricultural soil (P) were collected from 0-10 cm depth of at intervals of 200 meter from each site were collected. Four other samples were collected from 0-10 cm depth of at intervals of 200 meters from each other. Riyadh region (Hanifa Valley): Six samples were collected from Hanifa Valley (WP, W) at a depth of 10 cm with 200 meters intervals from each site11.
Bacterial isolation and DNA extraction
Each 10 gram of the soil sample was suspended in 90 ml of ddH2O and agitated for 2 min. The soil suspensions were serially diluted in sterile ddH2O. The different dilutions from 10-1 to 10-6 were plated on nutrient agar and Luria-Bertani Agar media12. All plates were incubated for 48 hours at 30°C13. The grown bacterial colonies on the agar were studied by colors, elevation, edge, and form. Pure bacterial colonies were subcultured on new nutrient agar and LB agar media single and pure colonies. DNA extraction or lysate of bacterial isolates was used for bacterial identification (16S rRNA) and nitrite reductase gene detection (Nir) by the molecular method. About 1.5 ml of pure culture was transferred into an Eppendorf tube and centrifuged for 13000 g at 10 mins. The pellet was suspended again and washed with 100μl of PBS buffer, followed by an additional centrifugation step at 13000 g for 10 min. The pellet was resuspended in 40μl of sterile PBS buffer and was boiled at 100°C in a water bath for 10 min. It was then immediately cooled on ice and centrifuged at 13000g x for 10s. The supernatant was collected in TE buffer and frozen at -20°C in fresh Eppendorf tubes.
Amplification and sequencing of Nitrate gene and 16S rRNA gene
PCR reactions were performed to amplify the 16S rRNA gene for microbial identification and to amplify the Nir gene from bacterial isolates13. The final volume of the PCR reaction mixture was 25μl. The reaction mixture containing beads (beads contain buffer salts, nucleotides and Taq polymerase in a lyophilized powder), 3μl of DNA or lysate, 10 pmol of forward primer for Cd1-nir F1acd (TAY CAC CCS GAR CCG C), and 10 poml of reverse primer Cd1-nir R4cd (CCG ACC GGT AAG TTC AAC GTG). Degenerate letters denote D (AGT), R (AG), M (AC), S (GC), B (GCT), N (AGCT), V (AGC), Y (CT), H (ACT), W (AT) and ddH2O to a final volume of 25μl. The primers used for 16s rRNA amplification were as followed forward primer 8FLP (5’GCG GAT CCG CGG CTG CAG AGT TTG ATC CTG GCT CAG3’) and reverse primer 806R (5’GCG GAT CCG CGG CCG CGG ACT ACC AGG GTA TCT AAT3’)14,15. The PCR was performed with the following cycling conditions to amplify the 16S rRNA gene or Nir gene; 94°C for 5 min, then denaturation at 94°C for 30 s, annealing at 57°C for 30 s and extension at 73°C for 1 min for 35 cycles followed by final extension at 73°C for 10 min. The PCR amplification was analyzed by 1% agarose gel electrophoresis in 1X TBE buffer after it was stained with Ethidium Bromide. The gel image was generated by UV light at 254 nm in a UV transilluminator. The data obtained by PCR was confirmed further by sequencing using the BigDye sequencing kit according to the manufacturer’s instructions (Thermo Fisher Scientific) and forward primers of each gene under investigation in a sequencer 3730XL DNA analyzer (Thermo Fisher Scientific).
Detection of Decomposition of nitrates
The detection was performed on multiple media including liquid nutrient medium (Beef extract 3.0 g/l, NaCl 5.0 g/l, Peptone 5.0 g/l, KNO3 0.5 g, ddH2O to 1000 ml, pH 7-7.5) and Mineral medium (K2HPO4 7.0 g, KH2PO4 3.0 g, sodium citrate.2H2O 0.5 g, MgSO4 7H2O 0.1 g, FeSO4 7H2O 0.05 g, KNO3 0.5 g, Glucose 1%)16.
Denitrification tests
All pure bacterial isolates were subcultured on liquid LB medium for 7 days under static culture conditions at 30°C. The nitrate and nitrite reduction tests were completed by different biochemical assays. The denitrifying bacteria were selected based on the pH indicator and the results of the reduction tests17. Alternatively, semi-quantitative nitrate strips assay Quantofix (MERCK) was used to detect nitrate reduction by nitrite strips. The strips detect nitrite decrease in the media. These semi-quantitative test strips are rapid and reliable in the determination of Nitrate or Nitrite in any solutions, including bacterial media, were used according to manufacturer’s instructions. It gives you an easy and reliable result within 1 minute. The color reaction ranges from white to red-violet, and the numerical ranges from 0. 15. 10. 20. 40. 80 mg/L NO2 or color from red to yellow ranging from 0 · 100 · 300 · 600 · 1000 · 2000 · 3000 mg/l NO2– for larger scale. For nitrate detection, the following ranges were used 0· 10· 25· 50· 100· 250· 500 mg/l NO3– with color reactions from white to red-violet. A direct nitrate assessment technology was also used to measure the level of nitrate in a bacterial solution using HACH ISENO318103 probe (according to manufacturer’s instructions). To read nitrate concentration in the media, the Hach Hq440d Benchtop dial input multipara meter was used. Nitrate Ionic Strength Adjustor Powder Pillows was used to ionize nitrate before reading the resolute by the probe.
Optimization of nitrate denitrification
The Genus Pseudomonas was one of the isolates obtained from the field collections and have given promising denitrification results. The strain was grown in 100 ml Mineral medium broth in 250 ml conical flasks. Optimization of denitrification was performed under several condition, three different carbon sources were used (glucose, sucrose and cellulose) with different concentration (0.1%, 0.3%, 0.5%, 0.7% and 0.9%). The concentration of 100 mg/L of KNO3 was used. Five different incubation times (2 h, 4 h, 8 h, 16 h, and 24 h) also experimented. Finally, five different inoculations (200 ml, 400 ml, 600 ml, 800 ml, and 1000 ml) and five different pH levels (5, 6, 7, 8, and 9) were investigated.
Microbial cellulose production for biological denitrification
Microbial cellulose production from Gluconacetobacter xylinus ATCC 23768, Nitrosospira multiformis ATCC 25196, Nitrosococcus oceani ATCC 19707, Achromobacter denitrificans ATCC 13138, Pseudomonas stutzeri ATCC 53819. Acetobacter xylinum ATCC 23768 was attempted to establish the biological denitrification assay18. All bacterial strains were purchased directly from ATCC (the American Type Culture Collection). The optimal Microbial cellulose production was achieved by Gluconacetobacter xylinus ATCC 2376. The process of MC production is better achieved under static culturing conditions. The process depends primarily on the air level supplied to culture media. However, the yield of the MC (Microbial Cellulose) correlates positively with the source of the carbon available and the growth time. The bacterial isolates were grown under static conditions at 30°C in the SH (Schenk and Hildebrandt) medium which consists of 0.5% peptone, 0.5% (W/V) yeast extract, 2% (W/V) glucose, 0.115% (W/V) citric acid and 0.27% (W/V) Na2HPO4 was used to cultivate the microbial cellulose bacteria under static conditions at 30°C. The SH medium consists of 0.5% (W/V) yeast extract, 0.5% peptone, 2% (W/V) glucose, 0.115% (W/V) citric acid and 0.27% (W/V) Na2HPO4 (Merck KGaA). One pure single colony was used in the inoculum on the SH agar media before inoculating the 50 ml of SH liquid media. The culture was kept for 2, 3, 4, and 5 days and the bacterial optical density was measured throughout the course of the experiment to determine the best MC production conditions. The MC or (cellulose sheets) was removed, washed with ddH2O, 4 % NaOH, and 2% SDS (sodium dodecyl sulfate) at a temperature of 80°C in a water bath. The MC sheets were dried and cut into a circle bioreactor column size for immobilization.
In-house biofilter Design
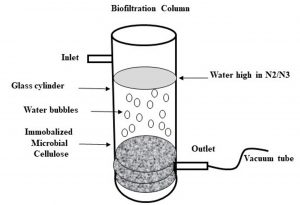
The biofilter for testing denitrification consists of a plexiglass cylinder with dimensions 90 cmx8 cm. The bioreactor column was built in-house to meet our specifications (Fig. 1). The bioreactor was packed with Pseudomonas aeruginosa immobilized on microbial cellulose up to 60 cm synthesized by Gluconacetobacter xylinus ATCC 23768 and the top of the bioreactor was sealed with a cork and the distant side was closed with an insulating material made from fibers of glass. Inlet and outlet are adjusted at 2 centimeter in the column. The biofiltration process in the reactor is initiated by running the discharges18.
Preparation of Column Biofilter
We used bacterial MC sheets to immobilize denitrifying bacteria on the column biofilter. When glucose is used as the primary carbon source, the Pseudomonas aeruginosa (OD600= 0.5) was co-incubated with the previously cut MC sheets to reach a stationary phase after 24 hrs. at maximum speed (120 rpm) on a rotary shaker. The MC sheets were loaded up into the column biofilter in stacks format and were let to settle down for 60-120 mints before they were used for water denitrification processes.
Microbial isolation by location
Medina region: Medina is situated in western of Saudi Arabia at an altitude of 26.57 (26° 34’ 0 N) and a longitude of 50.07 (50°4’ 0 E). The soil nearby Medina areas composed of basalt, while in the south especially hills areas are mainly volcanic ash that dates to Paleozoic era geologically. Jazan region: Jazan is situated in the south west of Saudi Arabia. Jazan lies on the shore of the Red Sea at a latitude of 16° 53′ 21″ N and a longitude of 42° 33′ 40″ E and serves a large agricultural region. Riyadh region (Hanifa Valley): Riyadh is located in the center of Saudi Arabia at an altitude of 24° 38′ 0″ N and a longitude of 46° 43′ 0″ E.
Six samples of soil were collected from the rhizosphere of Morus alba (TO), Aloe perryi, Lactuca sativa, Citrus latifolia (LM), Lycopersicon esculentum (TM) and mentha pennyroyal (MI). Three samples of soil were collected from 0-10 cm depth at intervals of 200 meters from each other. Jazan region: Four samples of agricultural soil (P) were collected from 0-10 cm depth of at intervals of 200 meter from each site were collected. Four other samples were collected from 0-10 cm depth of at intervals of 200 meters from each other. Riyadh region (Hanifa Valley): Three samples were collected from 0-10 cm depth at intervals of 200 meters from each site of Hanifa Valley (WP). Three samples of Hanifa Valley soil (W) were collected from 0-10 cm depth at intervals of 200 meters from each other.
The total samples used for the study were Ninety-three isolates (n= 93) that were isolated from the rhizosphere. They are divided as follows: Sixteen isolates were isolated from the rhizosphere of Morus alba (TO). Twenty-two isolates were isolated from the rhizosphere of Aloe perryi. Eleven isolates were isolated from the rhizosphere of Lactuca sativa. Twenty isolates were isolated from the rhizosphere zone of Citrus latifolia (LM). Fourteen isolates were isolated from the rhizosphere of Lycopersicon esculent um (TM). Ten isolates were isolated from the rhizosphere of Mentha pennyroyal (MI). Twenty-five isolates were isolated from different Soil Farms from Jazan regions (P). Six strains isolates were isolated from valley soil of Hanifa Valley in Riyadh (W).
Fig. 1. Biofilter column was built in-house and consists of glass cylinder with a dimension of 90 cmx8 cm with an inlet and outlet points. The bottom of the cylinder is packed with MC sheets that were previously co-incubated with Pseudomonas aeruginosa to be immobalized on the MC sheets.
Detection of Denitrification
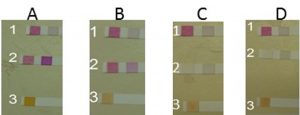
Using semi-quantitative analysis, our isolates (Pseudomonas aeruginosa, Pseudomonas putida, Pseudomonas monteilii, and Bacillus sp.) and our control (Pseudomonas stutzeri) were tested for their denitrification abilities. Tests were completed by strips Quantofix. Each strip tests for a change in color from white to red-violet of Nitrate or Nitrite in solutions such as bacterial media (Macherey-Nagel, GmbH, Duren, Germany). The color reaction is from white to red-violet ranging from 0,1, 5, 10, 15, 25, 50, 80 mg/L NO2 or color from red to yellow ranging from 0, 100, 300, 600, 1000, 200, 3000 mg/L NO2 for larger scale. In this result, the following ranges were used 0, 10, 25, 50, 100, 250, 500 mg/L NO2– with color reactions from white to red-violet. It was shown that P. stutzeri and P. aeruginosa were able to denitrify No3 to NO2 on Nutrient and Mineral media. We also observed that our isolates (P. aeruginosa) was able to do the denitrification process at a higher rate when compared to control isolate (Fig. 2) Some factors may affect the rate of denitrification processes such as the component of the media such as carbon source, pH, temperature, and the OD of inoculum.
Fig. 2 A. Strip (1) control for nutrient media for detection NO3/NO2. Strip (2) Moderate reduction by P. stutzeri for NO3/NO2. (3) P. stutzeri strep detected NO2. B (1) Control for Mineral media strep for detection of NO3/NO2. (2) Moderate reduction by P.stutzeri strep for NO3/NO2. (3) P.stutzeri strep for detection of NO2. C. (1) Control for nutrient media strep for detection of NO3/NO2. (2) Full reduction by aeruginosa strep for NO3/NO2. (3) P. aeruginosa strep for detection of NO2. D. Control for Mineral media strep for detection of NO3/NO2. (2) Full reduction by aeruginosa strep for NO3/NO2. (3) P. aeruginosa strep for detection of NO2.
Molecular Detection of Nitrate reductase gene
All bacterial isolates, including control ATCC strains we subjected to PCR amplification to detect the gene responsible for the denitrification process (Nir). The primers were designed by Hallin utilizing consensus regions in sequences for the structural genes encoding Nir, which were retrieved from the GenBank 13. The PCR amplicons of 800bp of the Nir gene were obtained (Fig. 3), which were also confirmed by Sanger sequencing.
Fig. 3. Identifying the Nir gene by PCR amplification from our isolates collection. The PCR expected size is 800 bp and was obtained. 1.100bp Marker, 2. +ve control, 3.Rals, 4.BAA, 5. 27853, 6. 9721, 7. TO114, 8. TM105, 9. LS106, 10. W1003, 11. –ve control.
Microbial cellulose production
A thick sheet of the MC membrane was obtained after extended incubation time under static aerobic culture conditions. The production of MC was achieved by the model bacterium Gluconacetobacter xylinu ATCC 23768, which is a gram-negative microorganism that has the capacity of synthesizing a substantial amount of MC (Fig. 4).
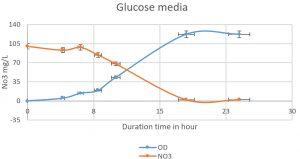
Fig. 4. The nitrate was reduced on mineral media with glucose by Pseudomonas aeruginosa. The higher reduction of nitrate was reached at 18 h (1.9mg/L). An inverse relationship was shown in duration time from 0 to 18 h, Nitrate reduction in log phase (the more bacterial growth, the higher the denitrification).
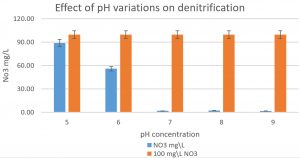
Fig. 5. The effect of pH variations on the denitrification process is shown in the graph. At least 100mg/L nitrate was used as a fixed starting point. It was shown that pH7, 8, and 9 were optimum for denitrification processes by Pseudomonas aeruginosa on glucose media.
The denitrification process by Pseudomonas aeruginosa
Pseudomonas aeruginosa (isolated from our sample collection) was able to denitrify the nitrate on mineral media with different carbon sources. The maximum level of denitrification was achieved after 18 hours with glucose as a carbon source (Fig. 5). At least 100mg/L of nitrate was used as a fixed starting point. The maximum denitrification rate was achieved after 18h when the glucose media was inoculated with 0.8 % and 1% inoculum of OD600=1. The pH effect variables on the denitrification processes was also investigated. It was shown that pH 7, 8, and 9 were suitable for the denitrification process by Pseudomonas aeruginosa on glucose media (Fig. 6). When employing the proper experimental conditions on synthesized MC and glucose media using P. aeruginosa in the biofiltration column, the nitrate was significantly reduced from 70 mg/L to 21mg/L after 6 hr. (Fig. 7).
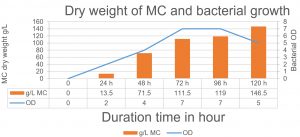
Fig. 6. The relationship between the production of MC and bacterial growth. The maximum MC production has been achieved after 120 h when the bacteria started in the death phase.
Ground Aquatic contamination has great importance in many countries around the globe, including Saudi Arabia, due to the scarce of groundwater levels in this region. Therefore, controlling wastewater contamination with ammonia and nitrate will result in the safe use of groundwater. The efficiency of the biological treatment of wastewater by biofiltration can minimize wastewater pollution to an undetectable level. This method is characterized by its low cost compared to the chemical and physical treatment approach19. The biofiltration process involved a separation between the microbes and the wastewater. The bacteria are immobilized on the biofilter (microbial cellulose), and the wastewater flows through the biofilter. The wastewater adsorbed to the biofilter, and the biological waste degradation begins. The waste biodegradation activity increases with a high concentration of biomass. The increased activity is linked to the high nutrients available for the attached microbial biofilm20.
The microbial production of cellulose is mainly dependent on carbon sources. It has been shown that the process by which the bacteria produce the cellulose is during their active growth phase. Unlike plant cellulose, bacterial cellulose consists of linear β-1-4-glucan chains, which is a unique form found of cellulose found only in bacteria. After the polymerization process, the secretion is achieved by the pores on the outer membrane of the bacteria. Interestingly, the MC is characterized by its nano-structure that permits a larger surface area for molecules interaction21.
The microbial bioreactor model has shown promising results of denitrification level with immobilized P. aeruginosa cell isolate. The key benefit of this biological system is the use of microbial cellulose biofilm as an adsorbent filter. Increasing the efficiency of denitrification in the microbial bioreactor model is due to the use of microbial cellulose, where the bacterial surface contact with oxygen, resulting in a lack of oxygen in the media and with the lack of oxygen flowing conditions ideal for the denitrification in a faster time22. Although we did not characterize the synthesized microbial cellulose, it seems to be working with high efficiency. It appears that the source of carbon has an important role in the denitrification procedure by the bacteria23. Interestingly, the pH level and the source of carbon may accelerate the nitrate removal and improve the denitrification process. In this experiment, it was observed that the pH 7, 8, 9 were the most efficient in the denitrification. This is because the pH of major denitrifying bacteria is between 7 and 8. The pH values lesser than 6 or upper than 9 might cause substantial decline or even halt the denitrification progression for various denitrification systems24. It was also noted that the denitrification was better achieved when using glucose as a source of carbon. Although cheap carbon source like ethanol may be employed, compared to glucose or succinate, which may be relatively more expensive.
In this current study, Pseudomonas aeruginosa was immobilized on synthesized MC in a biofiltration column. It was shown that Pseudomonas aeruginosa was able to remove nitrate of an initial nitrate concentration of 70mg/ml under constant flow and optimized experimental conditions. The production and treatment of MC in the lab have facilitated the process of denitrification and were a cost-effective method and were easy to prepare and to deal with in the lab, and It was not expensive to produce. Different bacteria were used to form microbial cellulose. Yet, Gluconacetobacter xylinus provided the highest level of microbial cellulose. It was shown that Pseudomonas aeruginosa was able to denitrify nitrate effectively on immobilized microbial cellulose on natural pH and on cultivating microbial media. The key utility of the MC system was its ease of use and its efficiency. The work of this study is suggesting that immobilizing Pseudomonas aeruginosa on microbial cellulose may offer an effective and alternative groundwater denitrification treatment. However, to achieve a superior efficiency at a large-scale level, it may be beneficial to immobilize more denitrifying bacteria on the same microbial cellulose sheet to accelerate the bio-filtration activity of nitrate from contaminated water.
ACKNOWLEDGMENTS
The authors also would like to thank those who contributed to this project Dr. Hatem M. Elzefzaf, and Mr. Baraa J. Alyamani for their scientific inputs and contributions.
CONFLICT OF INTEREST
The authors declare that there is no conflict of interest.
AUTHORS’ CONTRIBUTION
All authors designed the experiments. EA and RB performed the experiments. EA, RB, AB, SA analyzed and interpreted the data. EA, RB gathered information from the literature and drafted the manuscript. AB, SA reviewed the final manuscript. All authors have made a substantial, and intellectual contribution to the work in this manuscript, and approved it for publication.
FUNDING
This paper contains the results and funding of a research project that is funded by King Abdulaziz City for Science and Technology (KACST) Grant No. ARP.30.34.
ETHICS STATEMENT
This study does not contain human subjects or animals performed by the authors
AVAILABILITY OF DATA
This article does not contain any studies with human participants or animals performed by any of the authors.
- Dabbagh AE, Abderrahman WA. Management of groundwater resources under various irrigation water use scenarios in Saudi Arabia. Arab J Sci Eng. 1997;22(1):47-64.
- Al-Omran AM, Mousa MA, AlHarbi MM, Nadeem MEA. Hydrogeochemical characterization and groundwater quality assessment in Al-Hasa, Saudi Arabia. Arabian Journal of Geosciences. 2018;11(4):79.
Crossref - Alabdula’aly Ai. Nitrate Concentrations In Riyadh, Saudi Arabia Drinking Water Supplies. Environmental Monitoring and Assessment. 1997;47(3):315-324.
Crossref - Canter LW. Nitrates in groundwater. Routledge. 2019. doi: 10.1201/9780203745793
- Koren D, Gould W, Bedard P. Biological removal of ammonia and nitrate from simulated mine and mill effluents. Hydrometallurgy. 2000;56(2):127-144.
Crossref - Mateju V, Cizinska S, Krejci J, Janoch T. Biological water denitrification-a review. Enzyme and Microbial Technology. 1992;14(3):170-183.
Crossref - Soares M. Biological denitrification of groundwater. Water, Air, and Soil Pollution. 2000;123(1-4):183-193.
Crossref - dos Santos VAPM, Bruijnse M, Tramper J, Wijffels RH. The magic-bead concept: an integrated approach to nitrogen removal with co-immobilized micro-organisms. Appl Microbiol Biotechnol. 1996;45(4):447-453.
Crossref - Karagozoglu B, Sarioglu M, Peker I. Nitrate removal in a fixed-film column reactor using Paracoccus denitrificans affected by different carbon sources. Fresenius Environ Bull. 2002;11(10):927-932.
- Kumar CG, Anand S. Significance of microbial biofilms in food industry: a review. Int J Food Microbiol. 1998;42(1):9-27.
Crossref - El Maghraby MMS. Hydrogeochemical characterization of groundwater aquifer in Al-Madinah Al-Munawarah City, Saudi Arabia. Arabian Journal of Geosciences. 2015;8(6):4191-4206.
Crossref - Kross BC, Hallberg GR, Bruner DR, Cherryholmes K, Johnson JK. The nitrate contamination of private well water in Iowa. Am J Public Health. 1993;83(2):270-272.
Crossref - Hallin S, Lindgren P-E. PCR Detection of Genes Encoding Nitrite Reductase in Denitrifying Bacteria. Appl Environ Microbiol. 1999;65(4):1652-1657.
Crossref - Braker G, Fesefeldt A, Witzel K-P. Development of PCR primer systems for amplification of nitrite reductase genes (nirK and nirS) to detect denitrifying bacteria in environmental samples. Appl Environ Microbiol. 1998;64(10):3769-3775.
Crossref - Weisburg WG, Barns SM, Pelletier DA, Lane DJ. 16S ribosomal DNA amplification for phylogenetic study. Journal of Bacteriology. 1991;173(2):697-703.
Crossref - Blaszczyk M. Effect of Medium Composition on the Denitrification of Nitrate by Paracoccus denitrificans. Appl Environ Microbiol. 1993;59(11):3951-3953.
Crossref - Mazoch J, Kucera I. Detection, with a pH indicator, of bacterial mutants unable to denitrify. J Microbiol Methods. 2002;51(1):105-109.
Crossref - McAdam E, Judd S. A review of membrane bioreactor potential for nitrate removal from drinking water. Desalination. 2006;196(1):135-148.
Crossref - Cohen Y. Biofiltration – the treatment of fluids by microorganisms immobilized into the filter bedding material: a review. Bioresource Technology. 2001;77(3):257-274.
Crossref - Madigan MT, Martinko JM, Parker J. Brock biology of microorganisms. Vol 11: Prentice hall Upper Saddle River, NJ. 1997.
- Lynd LR, Weimer PJ, van Zyl WH, Pretorius IS. Microbial Cellulose Utilization: Fundamentals and Biotechnology. Microbiol Mol Biol Rev. 2002;66(3):506-577.
Crossref - Rezaee A, Godini H, Dehestani S, Yazdanbakhsh AR, Mosavi G, Kazemnejad A. Biological denitrification by Pseudomonas stutzeri immobilized on microbial cellulose. World J Microbiol Biotechnol. 2008;24(11):2397-2402.
Crossref - Peng YZ, Ma Y, Wang SY. Denitrification potential enhancement by addition of external carbon sources in a pre-denitrification process. J Environ Sci (China). 2007;19(3):284-289.
Crossref - Oh S-E, Kim K-S, Choi H-C, Cho J, Kim IS. Kinetics and physiological characteristics of autotrophic dentrification by denitrifying sulfur bacteria. Water Science and Technology. 2000;42(3-4):59-68.
Crossref
© The Author(s) 2020. Open Access. This article is distributed under the terms of the Creative Commons Attribution 4.0 International License which permits unrestricted use, sharing, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.