ISSN: 0973-7510
E-ISSN: 2581-690X
Yeasts are distributed in all environments and have been reported as potential biocontrol agents against various phytopathogenic fungi. To investigate their enzymatic and biological activities, 32 yeasts were isolated from 15 date vinegar samples. Evaluation of the antagonistic activities of isolated yeasts against the plant pathogens Fusarium oxysporium, Sclerotinia sclerotiorum, and Macrophomina phaseolina indicated that there are two yeasts had the highest inhibitory effect against plant pathogens, these yeasts identified as Kluyveromyces marxianus and Torulaspora delbrueckii using traditional and molecular methods. These yeast isolates were tested for fungal cell wall degrading enzymes (in vitro), and results indicated that the yeasts had strong protease and amylase enzyme activity and moderate chitinase and cellulase enzyme activity. The antagonistic activities of each yeast were evaluated using a dual culture technique. The results showed that K. marxianus inhibited the mycelial growth of F. oxysporium, S. sclerotiorum, and M. phaseolina by 70.5, 57.5, and 75.5%, respectively, whereas T. delbrueckii inhibited mycelial growth of F. oxysporum, S. sclerotiorum, and M. phaseolina by 55.3%, 66.2%, and 31.11%, respectively. The biofilm production assay indicated that the tested yeast could form biofilms as a mechanism of antagonistic activity against phytopathogenic fungi.
Biocontrol, Torulaspora delbrueckii, Kluyveromyces marxianus, Phytopathogenic fungi, Date Vinegar
Yeasts are widespread organisms in nature and are often isolated from sugar-rich food; yeasts can be found on fruit surfaces (berries, grapes, apples, and peaches), in plant secretions such as Aloe vera, in the soil, as many of them are associated with different insects, and are also found in pickles1,2. Yeasts distributed in different environments have been identified as potent antagonists with inhibitory activity against various microbial pathogens infecting plants. To prevent and control plant fungal diseases, chemical fungicides are widely used, although prolonged use can lead to the development of resistance to fungicides by the fungal pathogens3. Yeasts have been studied as biocides or biocontrol agents, primarily controlling and inhibiting the growth of molds that cause fruit rot in the post-harvest period, as they are excellent competitors for nutrients and space for growth4,5. Several yeast strains, including Debaryomyces, Pichia, Candida, Metschnikowia, , Saccharomyces, Kluyveromyces, Rhodotorula, Torulaspora, and Rhodosporidium6,7 have been identified and characterized for controlling different phytopathogenic molds. The biological control of plant fungal diseases by yeasts has been an important area of study due to the several antagonist characteristics displayed by yeasts towards fruit and beverage spoilage microorganisms, with which they compete for nutrients and space8,9. Yeasts secrete enzymes such as β-glucosidase, cellulase, amylase, protease, and chitinase that degrade the cellular components of harmful microorganisms, a common feature in several types of host-pathogen interactions and regularly reported in antagonistic yeasts and implicated in their biocontrol activity10-12. Some yeasts display host resistance to fungal pathogens13,14 and the production of volatile secondary metabolites (e.g., ethyl acetate, ethanol, and CO2)15. Other antagonistic mechanisms can be attributed to decreasing germ tube elongation, preventing mycelial growth, and suppressing conidial germination by rapidly colonizing the plant tissue16. Species of Torulaspora delbrueckii are non-Saccharomyces yeasts, often found in plants, soils, wine, and fermented food17. T. delbrueckii is possibly the most appropriate yeast for winemaking because it has ideal fermentation characteristics compared with other winemaking non-Saccharomyces yeasts18. Controlled inoculation with T. delbrueckii is widely recommended to develop the complication and increase some characteristics of wines19. The antagonistic activity and production of lethal toxins by T. delbrueckii isolated from spontaneously fermenting grape musts and fruits against some fungal pathogens have been reported in previous studies20-23. Kluyveromyces marxianus, obtained from beer wort and grapes, was first classified as an yeast belonging to the genus Saccharomyces and was named Saccharomyces marxianus, but it has since been transferred to the genus Kluyveromyces. Since then, approximately 45 species have been classified into this genus. K. marxianus is well known because it is used in traditional dairy products, such as fermented cheese, yogurt kefir, and milk24,25. The biocontrol efficacy of K. marxianus against Penicillium digitatum, causing green mold of citrus fruit, has been reported. The antagonistic activity of K. marxianus was enhanced by adding 2% sodium bicarbonate. K. marxianus has been reported to produce some lethal toxins against plant pathogens26. Unfortunately, there are few studies on the potential antifungal activity of K. marxianus against plant diseases affecting fruits and vegetables27. In a previous study, K. marxianus was identified on the surface of papaya fruit28. The present study aimed to evaluate the antagonistic activity of K. marxianus and T. delbrueckii isolated from Iraqi date vinegar against some fungal pathogens.
Culturing and fungal isolation
Fifteen samples of Iraqi date vinegar were collected from local markets in Baghdad, Iraq, from October 1 to November 1, 2018, and the yeasts were isolated from these samples by direct isolation using a series of dilution techniques, according to29. A volume of 0.1 mL of each dilution was cultured on GPY medium (20 g/L glucose, 10 g/L yeast extract, and 20 g/L peptone). The pH was adjusted to 6.0 using 1 M HCl and sterilized via autoclaving. Solid medium was prepared by adding 15 g/L agar. A pure culture of yeast isolates was obtained by spreading the growth using a sterile cotton swab, and then all the Petri dishes were incubated at 25°C for 72 h. The pure isolates were maintained on solid culture medium GPY agar slants and stored at 4°C for further processing.
Identification of yeast isolates
Macroscopic and microscopic identification
Microscopic identification of growing yeast isolates was primarily based on colony and morphological characteristics. The isolates were examined under a microscope to observe their form, ensuring that the growing colonies were yeasts and not any other microorganisms. The antagonistic activity test was performed for all isolated and purified yeasts against fungal pathogens to identify species with the most inhibitory and antagonistic activity using molecular techniques.
Molecular Identification
Molecular identification of the yeasts was based on the sequencing of the internal transcribed spacer (ITS1–ITS4) region by the conserved 5.8S rRNA gene30.
Isolation of genomic DNA from yeast
DNA was extracted from two pure yeast isolates, displaying antagonist activity when cultured on YEPD medium, using a Wizard Genomic DNA Purification kit (Promega Madison, WI, USA) according to the manufacturer’s instructions. The yeast isolates were cultured on a solid YEPD medium for 20 h at a temperature of 28°C, and 1 mL of YEPD liquid media was inoculated with yeast cells in sterile centrifuge tubes and centrifuged at 16,000 × g for 2 min.
Polymerase chain reaction (PCR)
After DNA extraction from yeast cells, species identification was conducted by amplifying the 5.8S region using the following primers according to31.
ITS1 (5′TCCGTAGGTGAACCTGCGG′3), ITS4 (5′TCCTCCGCT TATTGATATGC′3)
The PCR process was performed with a 25µL reaction mixture consisting of 12.5µL of a Green Master Mix (Promega), 1 µL of 10 pmol/µL of each primer, and 2 µL of DNA template. Then, the volume was completed to 25 µL using nuclease-free water.
The thermocycling conditions are shown in Table 1.
Table (1):
Thermal circulation conditions of the polymerase chain reaction technique.
Time |
Temperature |
Stage |
|---|---|---|
4 min |
95 °C |
Denaturation |
30 sec |
55 °C |
Primers annealing |
10 min |
57 °C |
Extension |
The final Elongation cycle is at the same temperature for 10 min. Then the program will run at 4°C.
Preparation of agarose gel and electrophoresis
The agarose gel was prepared by dissolving 1 g of agar in 100 mL of TAE solution and placing it in an electric oven at 100°C, after which it was left to cool at 55-65°C. Subsequently, 1 µL of 10 mg/mL, ethidium bromide dye was added to an electrophoresis tank, the comb was placed gently and left for cooling and solidification for 30 min. Then, the comb was carefully pulled, and the tank was the attached to electrophoresis device, where a buffer was added until the gel in the tank was covered. Aliquots of five microliters of the PCR product and the 100-bp DNA Ladder (100-1500-bp) (Promega-USA) were loaded onto 1.5% agarose gel, the run with 1x (TAE) Tris-acetate-EDTA buffer. The device was then covered, turned on, and ran for 90 min at 100 volt/50 mAmp, and the gel was subsequently photographed using UV transilluminator system.
DNA Sequencing
After DNA purity was confirmed and a sufficient quantity obtained, the product was sent to (Macrogene Company, South Korea) for ITS1–ITS4 sequencing (reading the nitrogenous base sequence making up the DNA) using a 3730xl DNA analyzer (Thermo Fisher Scientific, Waltham, MA, USA). The nitrogenous base sequence obtained was aligned using existing sequences of isolates and strains of yeast that have been incorporated into the Global Gene Bank using the National Center for Biotechnology Information (NCBI-BLAST) to identify the date vinegar yeast isolates and determine their similarity with existing isolates in the World Bank for Genetic Information. Mega 10 neighbor-joining method was used for constructing the evolutionary tree of the two yeast isolates for comparison with those in the NCBI database.
Fungal Isolates
The antagonistic effect of the isolated yeasts towards some fungal pathogens was determined for Fusarium oxysporium, Sclerotinia sclerotiorum, and Macrophomina phaseolina, identified and classified at the microbiology laboratories of the University of Baghdad, College of Science for Women, Department of Biology.
Determination of extracellular enzymatic activities
The ability of K. marxianus and T. delbrueckii to produce extracellular hydrolytic enzymes, essential for biocontrol activity, was determined by agar plate experiments. In all cases, 10 µL of yeast suspension was inoculated onto the surface of the center of agar plates containing media supplemented with specific substrates for enzymatic activity. A non-inoculated plate was used as a negative control. All the inoculated plates were incubated at 25°C, and after 7-10 days of incubation, the expression of extracellular enzymes was determined.
Cellulase
A specific medium consisting of 5.0 g carboxymethylcellulose sodium salt (CMC), 10 g yeast extract, 20 g peptone, and 15 g agar was used to determine cellulase production. The mixture was dissolved in 1 L of distilled water at pH 5 and sterilized for 20 min at a temperature of 121°C using a centrifuge system. The yeast was cultured at the center of the Petri dish, and after the incubation period, it was treated with 0.2% (w/v) Congo red solution. The dye was removed using distilled water, and the plates were decolorized by several treatments with 1 M NaCl. The inhibition zone around the yeast colony in the middle of the Petri dish indicates the yeast’s ability to produce cellulose32.
Amylase activity
To investigate the yeasts’ ability to degrade starch, a culture medium (starch agar) was prepared from the following materials: 2 g yeast extract, 5 g peptone, 10 g starch, 0.5 g NaCl, 0.5 g MgSO4, 0.15 g CaCl2, and 20 g agar. The mixture was dissolved in 1 L of distilled water and sterilized as described above. After culturing the yeast in the middle of the Petri dish and incubating it for two days, the Petri dish was covered with Lugol’s solution (iodine-potassium iodide solution). The appearance of an inhibition zone around the yeast colony indicated a positive result33.
Protease activity
The culture medium for the protease activity assay was prepared using 1 g of yeast extract, 10 g of skimmed milk, 4 g of peptone, and 10 g of agar, which were dissolved in 1 L distilled water and sterilized as described above. The appearance of an inhibition zone around the growing yeast colony after two days indicated a positive result34.
Chitinase activity
A test medium was prepared using 2 g of NaNO3, 1 g of K2HPO4, 5.0 g of MgSO4, 0.5 g of KCL, 2 g of peptone, 10 g of colloidal chitin, and 15 g of agar), which were dissolved in 1 L of distilled water and sterilized as described above. The plates were incubated for 48 h, and then the plates were covered with Lugol’s solution for three minutes. The purple area that formed around the colony indicated a positive result35.
Biocontrol potential of K. marxianus and T. delbrueckii against some phytopathogenic fungi
In this experiment, K. marxianus and T. delbrueckii were assessed as biological control agents in vitro through the dual culture method, according to36. The dual culture method included the inoculation of the solid PDA medium with K. marxianus and T. delbrueckii on one side of a 9-cm Petri dish by streaking the yeast. The Petri dishes were left for 48 h in an incubator at 25°C. Five-millimeter discs of pathogenic fungal colonies of F. oxysporium, M. phaseolina, and S. sclerotiorum were placed on the opposite side of the Petri dish. The Petri dishes were incubated at 25°C for a period ranging from 7-10 days. The control treatment consisted of placing a disk of each type of fungus used in the study in a Petri dish containing sterile solid PDA medium and incubated under the conditions described above. The control treatment was used for comparison purposes. After incubation, the diameter of fungal growth was measured for both treatments, this experiment was conducted in triplicate, and the rate of growth inhibition was calculated using the following equation:
Percent growth inhibition (PGI) = KR – RI/KR x 100 Percentage of inhibition
Where KR represents the distance from the point of inoculation to the fungal colony margin in the control treatment, RI represents the distance of fungal colony diameter from the point of inoculation to the yeast colony margin on the treated Petri dishes.
Biofilm formation assay
To determine the tested yeasts’ ability to form biofilms, the method of Vero et al. 37 was adopted, with some modifications. K. marxianus and T. delbrueckii were grown for 24 h at 25°C in YEPD medium. Biofilm formation was observed in pre-sterilized polystyrene, flat-bottom 96-well microtiter plates (Nunclon, Roskilde, Denmark). All wells, containing 900 µL YEPD medium, were inoculated with 100 µL of the grown yeast (108 cells mL-1). Afterward, the well plates were incubated at 25°C for 48 h with agitation at 100 rpm. A line of wells was used as the control treatment and prepared without the yeast inoculum. Each well was then washed three times with 250 µL of sterile normal saline (0.9%). The biofilm formed was fixed by adding 200 µL of methanol for 20 min. The plates were dried at room temperature. A crystal violet solution (1% w/v) was used to stain the adherent biofilm layer at room temperature for 20 min, and the dye was removed from each well using 95% ethanol. The absorbance was measured at 620 nm. Formation was considered positive when the absorbance mean was higher than that of the control treatment (negative treatment). The biofilm formation assay was conducted in six wells and repeated in triplicate.
Statistical analysis
The data were analyzed using one-way analysis of variance (ANOVA) in SPSS statistics software version 22 for windows. For experiments that had paired comparisons, the unpaired t-test was used. Significance was set at P < 0.0.5, and results were expressed as mean ±SD of triplicate measurements.
In the present study, 32 yeast isolates were obtained from the date vinegar samples. The antagonistic activity of all isolates was tested against selected phytopathogenic fungi. The most effective antagonistic isolates inhibiting phytopathogenic fungi growth were selected for molecular identification by sequencing the 5.8S region (ITS1–ITS4). Two yeast isolates, K. marxianus and T. delbrueckii, were selected among the initial 32 isolates using molecular techniques. After alignment between the ITS1–ITS4 region of the isolates with those in the NCBI database, the first isolate had 99% similarity with K. marxianus. The ITS1–ITS4 sequence for this isolate was deposited in the NCBI database under accession number MW131636. Fig. 1A shows the genetic evolutionary tree of these isolates and similar isolates of the same genus and species registered in the NCBI database. The second isolate, identified as T. delbrueckii, had 99% similarity with the yeast isolates recorded on the same NCBI position. The ITS1–ITS4 sequence for this isolate was deposited in the NCBI database under accession number MW131635. Fig. 1B shows the genetic evolutionary tree of this yeast and similar yeasts registered within this position.
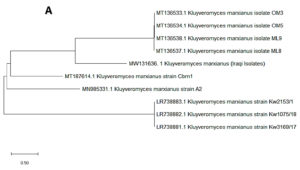
Fig. 1. Phylogenetic tree showing the position of A- K. marxianus among related species of K. marxianus B- T. delbrueckii among related species of T. delbrueckii from NCBI, as represented by the bootstrap consensus tree inferred with the neighbor-joining method from 1000 replicates derived from the analysis of 26S rDNA by sequencing the region ITS1-ITS4. The trees are drawn in scale, with branch length in the same units as those of evolutionary distance used to infer the phylogenetic tree using MEGA 10.
Enzymatic Activity Analysis of K. marxianus and T. delbrueckii
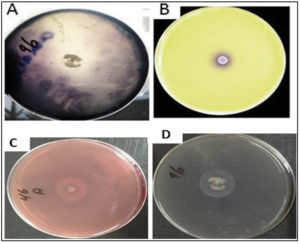
This experiment was conducted to determine the ability of K. marxianus and T. delbrueckii to produce enzymes that degrade the substances (cellulose, starch, protein, and chitin), as well as their ability to produce enzymes such as amylase, chitinase, protease, and cellulose. Results indicated the ability of K. marxianus and T. delbrueckii to produce these enzymes on solid media containing the substance material, as shown in Table 2. The yeast isolates demonstrated different levels of enzymatic activity. The K. marxianus and T. delbrueckii isolates displayed the ability to produce chitinase enzymes after adding the selected isolates to the culture medium, indicated by a color change of the medium from yellow to purple (Fig. 1). The color change is a consequence of ketone catabolism to N-acetyl glucosamine and the concomitant change in pH. In addition, lysis of the milk protein was observed within the yeast colony’s inhibition zone, indicating the yeasts’ ability to produce protease enzymes. During the experiment, there was an inhibition zone around the colony growing in the middle of the Petri dish containing CMCase. The biological control mechanisms of yeasts (e.g., oomycetes) against phytopathogenic fungi take advantage of the fungi’s cell wall constituents such as chitin, glucan, and glycoproteins as well as cellulosic materials, which, according to several studies, they degrade using enzymes such as cellulase38,39. Fig. 2 shows the yeast production of enzymes that degrade components of phytopathogenic fungi cell walls. These enzymes are important for the biological control characteristic of yeasts.
Table (2):
The enzymatic activity produced by the yeast of K. marxianus and T. delbrueckii on solid media containing the substrate for each enzyme.
| The enzymatic activity of produced enzymes by yeasts | ||||
|---|---|---|---|---|
| Protease | Chitinase | Cellulase | Amylase | Yeasts |
| +++ | + | + | ++ | K. marxianus |
| ++ | + | + | + | T. delbrueckii |
Fig. 2. Illustrates the ability of T. delbrueckii yeast to produce the enzymes that considered biological control. This was done by culturing the yeast on the growth medium that contains the substrate that stimulates the production of the enzyme, which is
A- Production of amylase enzyme by culturing yeast on a medium containing starch as a substance
B- Production of chitinase enzyme by culturing yeast on a medium containing colloidal chitin as the substrate
C- Production of cellulase enzyme by culturing yeast on a medium containing CMC as a substrate.
D- Protease enzyme production by culturing yeast on growth media containing casein.
The inhibitory activity of yeasts differs from one type to another depending on the environmental conditions in which these yeasts are present. Yeasts have various mechanisms that enable them to inhibit similar types of pathogenic yeasts and fungi, causing disease in humans or plants, including capturing iron particles and important nutrients in growth media, production of lethal toxins, and lytic enzyme secretion. These mechanisms constitute the yeast’s defense mechanisms and eliminate fungal pathogens; therefore, yeasts are considered biological control agents38.
The antagonistic activity of K. marxianus and T. delbrueckii against some pathogenic plant fungi
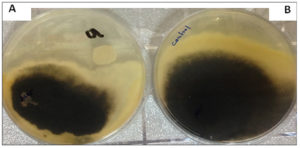
The present study was conducted to determine yeasts’ antagonistic inhibitory effects against three phytopathogenic fungi, F. oxysporum, M. phaseolina, and S. sclerotiorum using the dual culture method on solid PDA medium. It is clear from the outcome that K. marxianus and T. delbrueckii significantly inhibited the growth diameter of phytopathogenic fungi. Table 3 shows the inhibitory effect of K. marxianus on the diameter of fungal growth; the levels of inhibition were significantly higher than the control (P < 0.05), with percentage of inhibition rates of 70.5, 57.5, and 75.5% for F. oxysporum, M. phaseolina, and S. sclerotiorum, respectively. Fig. 3 shows the antagonistic activity of K. marxianus against M. phaseolina on the solid PDA medium as percentage of inhibition of the diameter growth of the fungus.
Table (3):
The diameters of the pathogenic fungal colonies and control treatment, as well as the percentage of inhibition (%) for using K. marxianus yeast as a biological control agent.
Pathological fungi |
The diameter of the pathogenic fungi colony in the control treatment / mm |
The diameter of a pathogenic fungi colony in the presence of yeast / mm |
The percentage of inhibition %±SD |
|---|---|---|---|
F. oxysporum |
85 |
60 |
70.5±0.04a |
M. phaseolina |
80 |
46 |
57.5±0.024a |
S. sclerotiorum |
90 |
68 |
75.5±0.07b |
Fig. 3. A- The diameter growth of M. Phaseolina with the presence of K. marxianus yeast as a biological control agent. B- Shows the diameter growth of M. phaseolina without the presence of yeast (control treatment). The Petri dishes were grown and incubated at 25±2°C using the PDA culture medium for seven days.
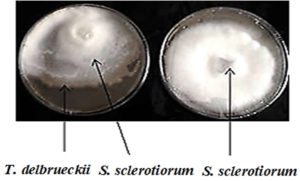
For T. delbrueckii, a decent antagonistic inhibitory effect against phytopathogenic fungi was also observed. The rates of inhibition of the growth diameter of phytopathogenic fungi were significantly higher than those of the control treatment (P < 0.05), reaching 31.11%, 66.2%, and 55.3% for F. oxysporum, M. phaseolina, and S. sclerotiorum, respectively (Table 4). Fig. 4 shows the inhibitory action of T. delbrueckii on the growth diameter of S. sclerotiorum at the center of the solid PDA medium.
Table (4):
The diameters of the pathogenic fungal colonies and control treatment, as well as the percentage of inhibition (%) of using T. delbrueckii yeast as a biological control agent.
Pathological fungi |
The diameter of the pathogenic fungi colony in the control treatment/mm |
The diameter of a pathogenic fungi colony in the presence of yeast/mm |
The percentage of inhibition% |
|---|---|---|---|
F. oxysporum |
85 |
47 |
55.3±0.25b |
M. phaseolina |
80 |
53 |
66.2±0.61a |
S. sclerotiorum |
90 |
62 |
31.11±0.9ab |
Fig. 4. A- Shows the diameter growth of S. sclerotiorum in the presence of T. delbrueckii yeast as a biocontrol control agent B- Shows the diameter growth of S. sclerotiorum without the presence of yeast (control treatment). The Petri dishes were grown and incubated at 25±2°C using the PDA culture medium for seven days.
K. marxianus and T. delbrueckii showed biofilm production (Table 5). The results obtained showed an increase in absorbance after alcohol elution, indicating that a biofilm was formed by the action of K. marxianus and T. delbrueckii and not the control. These results demonstrate that the yeast species isolated from the Iraqi date vinegar can form biofilms and survive in adverse environments.
Table (5):
Biofilm formation on microtiter plates for K. marxianus and T. delbrueckii.
Treatment |
A620 (Means ± SD) |
|---|---|
K. marxianus |
0.56±0.072b |
T. delbrueckii |
0.45±0.034a |
Control |
0.029±0.004a |
The biological control of plant fungal pathogens using antagonistic yeasts such as Debaryomyces, Hanseniaspora, Pichia, Kluyveromyces, Williopsis, and Zygosaccharomyces obtained from different habitats has been reported and is among the best alternative strategies for fungicide production7,40-43. The antagonistic activities of yeasts vary according to environmental conditions and antagonist type and are based on different principles. In some yeast species, the antagonistic effects have been attributed to hydrolytic enzyme secretion or production of volatile organic compounds. Lytic enzymes were produced by yeasts isolated from plants such as Rhodotorula mucilaginosa and C. famata and used in the post-harvest control of anthracnose in papaya fruit caused by Colletotrichum gloeosporioides44. The cell wall-degrading enzymes including proteases, chitinases, and glucanases are the main lytic enzymes produced by biocontrol yeasts45. These exo-enzymes attack and lyse the cell wall of phytopathogenic fungi, leading to death. Additionally, mycoparasitic fungi have been assumed to involve the secretion of exo-enzymes that play a significant role in degrading the fungal cell wall. The antagonistic mechanism is based on hydrolytic enzyme production, together with competition for nutrients46. Zhang et al.47,48 reported the antifungal activities of exo-1,3-β-glucanase produced by the antagonistic yeast P. guilliermondii and alkaline serine protease secreted by the biocontrol agent Aureobasidium pullulans PL5. It has been established that enzyme systems for fungal cell lysis are usually composed of mixtures of different enzymes, including one or more proteases, chitinases, 1,3-β- and 1,6-β-glucanase, and mannanases, all of which act synergistically to attack and lyse the cell wall of fungal cells49.
T. delbrueckii was applied by Author et al.21 as a biocontrol agent against the P. expansum, alone and in combination with methyl jasmonate. It was revealed that T. delbrueckii plays a significant role in reducing the growth of pathogens parasitizing apples better in combination with methyl jasmonate. The ability of T. delbrueckii to produce lethal toxins was studied by23 who reported that T. delbrueckii produced a novel lethal toxin (TdKT) with potential biocontrol efficiency against some yeasts involved in wine spoilage. T. delbrueckii produces volatile organic compounds (VOCs) whose efficacy increases at low pH. These VOCs have antagonistic potential against several toxigenic and phytopathogenic fungi, including Aspergillus, Penicillium, and Fusarium50.
There are a few studies on K. marxianus as a biological control agent against phytopathogenic fungi. K. marxianus has been shown to reduce the incidence of green mold in citrus fruits caused by P. digitatum28 and its biocontrol efficiency was increased by supplementing the growth medium with 2% sodium bicarbonate. Although K. marxianus has been demonstrated to be able to secrete some lytic and hydrolase enzymes, attempts have been made to exert direct antagonistic activities against phytopathogenic fungi12.
In addition to the biocontrol mechanisms described above, yeast biofilm production could be considered an antagonistic mechanism to manage and control phytopathogenic fungi. In the competition for growth space, yeasts typically produce a polysaccharide layer known as a capsule that can play a significant role in adhesion to fruit surfaces, covering the entire wound area51. The production of biofilms can contribute to the biocontrol activity of yeasts52. In addition, biofilm production may increase oxidative stress resistance, considered an important attribute required by yeasts to survive and maintain biocontrol activity53.
In conclusion, the yeasts isolated from Iraqi date vinegar, K. marxianus, and T. delbrueckii showed high protease and amylase activities. Moreover, they can produce biofilms, which play a significant role in the biocontrol of phytopathogenic fungi by inhibiting mycelial growth (in vitro). In particular, K. marxianus and T. delbrueckii showed inhibitory effects against selected plant pathogens using dual culture methods. This work significantly enhances our knowledge of the biocontrol of plant fungal diseases using novel yeast isolates obtained from food sources.
ACKNOWLEDGMENTS
None.
CONFLICT OF INTEREST
The authors declare that there is no conflict of interest.
AUTHORS’ CONTRIBUTION
All listed authors have made a substantial, direct, and intellectual contribution to the work and approve it for publication.
FUNDING
None.
ETHICS STATEMENT
This article does not contain any studies with human participants or animal performed by any of the authors.
AVAILABILITY OF DATA
All data obtained and generated analyzed in this work are including in the manuscript.
- Sung-Oui S, McHugh JV, Pollock DD, Blackwell M. The beetle gut: a hyperdiverse source of novel yeasts. Mycol Res. 2005;109(3):261-265.
Crossref - Slavikova E, Vadkertiova R. The diversity of yeasts in the agricultural soil. J Basic Microbiol. 2003;43(5):430-436.
Crossref - Ab Rahman SFS, Singh E, Pieterse CM, Schenk PM. Emerging microbial biocontrol strategies for plant pathogens. Plant Sci. 2018;267:102-111.
Crossref - El-Tarabily KA, Sivasithamparam K. Potential of yeasts as biocontrol agents of soil-borne fungal plant pathogens and as plant growth promoters. Mycoscience. 2006;47(1):25-35.
Crossref - Ferraz P, Cassio F, Lucas C. Potential of yeasts as biocontrol agents of the phytopathogen causing cacao Witches’ Broom Disease: is microbial warfare a solution? Front Microbiol. 2019;10:1766.
Crossref - Errampalli D. Penicillium expansum (blue mold). Postharvest Decay: Elsevier. 2014:189-231.
Crossref - Al-Qaysi SA, Al-Haideri H, Thabit ZA, Al-Kubaisy WHAA, Ibrahim JAA-R. Production, characterization, and antimicrobial activity of mycocin produced by Debaryomyces hansenii DSMZ70238. Int J Microbiol. 2017;2017:2605382.
Crossref - Liu J, Sui Y, Wisniewski M, Droby S, Liu Y. Utilization of antagonistic yeasts to manage postharvest fungal diseases of fruit. Int J Food Microbiol. 2013;167(2):153-160.
Crossref - Bencheqroun SK, Bajji M, Massart S, Labhilili M, El Jaafari S, Jijakli MH. In vitro and in situ study of postharvest apple blue mold biocontrol by Aureobasidium pullulans: evidence for the involvement of competition for nutrients. Postharvest Biol Technol. 2007;46(2):128-135.
Crossref - Fan Q, Tian S, Liu H, Xu Y. Production of β-1, 3-glucanase and chitinase of two biocontrol agents and their possible modes of action. Chinese Science Bulletin. 2002;47(4):292.
Crossref - Grevesse C, Lepoivre P, Jijakli MH. Characterization of the exoglucanase-encoding gene PaEXG2 and study of its role in the biocontrol activity of Pichia anomala strain K. Phytopathology. 2003;93(9):1145-1152.
Crossref - Pretscher J, Fischkal T, Branscheidt S, et al. Yeasts from different habitats and their potential as biocontrol agents. Fermentation. 2018;4(2):31.
Crossref - Droby S, Vinokur V, Weiss B, et al. Induction of resistance to Penicillium digitatum in grapefruit by the yeast biocontrol agent Candida oleophila. Phytopathology. 2002;92(4):393-399.
Crossref - Spadaro D, Vola R, Piano S, Gullino ML. Mechanisms of action and efficacy of four isolates of the yeast Metschnikowia pulcherrima active against postharvest pathogens on apples. Postharvest Biol Tech. 2002;24(2):123-134.
Crossref - Contarino R, Brighina S, Fallico B, Cirvilleri G, Parafati L, Restuccia C. Volatile organic compounds (VOCs) produced by biocontrol yeasts. Food Microbiol. 2019;82:70-74.
Crossref - Zheng XD, Zhang HY, Sun P. Biological control of postharvest green mold decay of oranges by Rhodotorula glutinis. Eur Food Res Technol. 2005;220(3-4):353-357.
Crossref - Canonico L, Agarbati A, Comitini F, Ciani M. Torulaspora delbrueckii in the brewing process: A new approach to enhance bioflavour and to reduce ethanol content. Food Microbiol. 2016;56:45-51.
Crossref - Benito S. The impact of Torulaspora delbrueckii yeast in winemaking. Appl Microbiol Biotechnol. 2018;102(7):3081-3094.
Crossref - Azzolini M, Tosi E, Lorenzini M, Finato F, Zapparoli G. Contribution to the aroma of white wines by controlled Torulaspora delbrueckii cultures in association with Saccharomyces cerevisiae. World J Microbiol Biotechnol. 2015;31(2):277-293.
Crossref - Ebrahimi L, Aminian H, Etebarian HR, Sahebani N. Control of apple blue mould disease with Torulaspora delbrueckii in combination with Silicon. Archives of Phytopathology and Plant Protection. 2012;45(17):2057-2065.
Crossref - Ebrahimi L, Etebarian HR, Aminian H, Sahebani N. Enhancement of biocontrol activity of Torulaspora delbrueckii with methyl jasmonate against apple blue mould disease. Archives of Phytopathology and Plant Protection. 2012;45(19):2355-2363.
Crossref - Sangorrin MP, Lopes CA, Giraudo MR, Caballero AC. Diversity and killer behaviour of indigenous yeasts isolated from the fermentation vat surfaces in four Patagonian wineries. Int J Food Microbiol. 2007;119(3):351-357.
Crossref - Villalba ML, Saez JS, del Monaco S, Lopes CA, Sangorrin MP. TdKT, a new killer toxin produced by Torulaspora delbrueckii effective against wine spoilage yeasts. Int J Food Microbiol. 2016;217:94-100.
Crossref - Karim A, Gerliani N, Aider M. Kluyveromyces marxianus: An emerging yeast cell factory for applications in food and biotechnology. Int J Food Microbiol. 2020:333:108818.
Crossref - Coloretti F, Chiavari C, Luise D, et al. Detection and identification of yeasts in natural whey starter for Parmigiano Reggiano cheese-making. Int Dairy J. 2017;66:13-17.
Crossref - Walker GM, Mcleod AH, Hodgson VJ. Interactions between killer yeasts and pathogenic fungi. FEMS Microbiol Lett. 1995;127(3):213-222.
Crossref - Geng P, Chen S, Hu M, et al. Combination of Kluyveromyces marxianus and sodium bicarbonate for controlling green mold of citrus fruit. Int J Food Microbiol. 2011;151(2):190-194.
Crossref - Geng P, Zhang Y, Hu M, Chen S, Qu F. Screening, identification of antagonistic yeast and its biocontrol efficacy against green mold caused by Penicillium digitatum of citrus fruit. Journal of Northwest A & F University-Natural Science Edition. 2011;39(6):191-196.
- Zott K, Miot-Sertier C, Claisse O, Lonvaud-Funel A, Masneuf-Pomarede I. Dynamics and diversity of non-Saccharomyces yeasts during the early stages in winemaking. Int J Food M icrobiol. 2008;125(2):197-203.
Crossref - Jespersen L. Occurrence and taxonomic characteristics of strains of Saccharomyces cerevisiae predominant in African indigenous fermented foods and beverages. FEMS Yeast Research. 2003;3(2):191-200.
Crossref - White T, Bruns T, Lee S, et al. PCR protocols: a guide to methods and applications. 1990.
- Carrasco M, Rozas JM, Barahona S, Alcaino J, Cifuentes V, Baeza M. Diversity and extracellular enzymatic activities of yeasts isolated from King George Island, the sub-Antarctic region. BMC Microbiol. 2012;12(1):251.
Crossref - Burhan A, Nisa U, Gokhan C, Omer C, Ashabil A, Osman G. Enzymatic properties of a novel thermostable, thermophilic, alkaline and chelator resistant amylase from an alkaliphilic Bacillus sp. isolate ANT-6. Process Biochemistry. 2003;38(10):1397-1403.
Crossref - Kumar CG, Takagi H. Microbial alkaline proteases: from a bioindustrial viewpoint. Biotechnol Adv. 1999;17(7):561-594.
Crossref - Hankin L, Anagnostakis S. The use of solid media for detection of enzyme production by fungi. Mycologia. 1975;67(3):597-607.
Crossref - Dennis C, Webster J. Antagonistic properties of species-groups of Trichoderma: I. Production of non-volatile antibiotics. Transactions of the British Mycological Society. 1971;57(1):25-29.
Crossref - Vero S, Garmendia G, Gonzalez MB, Bentancur O, Wisniewski M. Evaluation of yeasts obtained from Antarctic soil samples as biocontrol agents for the management of postharvest diseases of apple (Malus× domestica). FEMS Yeast Research. 2013;13(2):189-199.
Crossref - Chen P-H, Chen R-Y, Chou J-Y. Screening and evaluation of yeast antagonists for biological control of Botrytis cinerea on strawberry fruits. Mycobiology. 2018;46(1):33-46.
Crossref - Cordero-Bueso G, Mangieri N, Maghradze D, et al. Wild grape-associated yeasts as promising biocontrol agents against. Vitis vinifera.. Frontiers in Microbiology. 2017; 8: 1-15
- Hameed AR, Al-Qaysi SA, Hameed ST. Killer Activity of Hanseniaspora uvarum Isolated from Dates Vinegar: Partially Purification and Characterization of Killer Toxin. Baghdad Science Journal. 2019;16(Suppl. 1):140-150.
Crossref - El-Neshawy S, Shetaia YM. Biocontrol capability of Candida spp. against Botrytis rot of strawberries with respect to fruit quality. Paper presented at: International Conference on Quality in Chains. An Integrated View on Fruit and Vegetable Quality 6042003. 1. 727-733.
Crossref - Liu SQ, Tsao M. Biocontrol of spoilage yeasts and moulds by Williopsis saturnus var. saturnus in yoghurt. Nutrition & Food Science. 2010:40:166-175.
Crossref - Weiler F, Rehfeldt K, Bautz F, Schmitt MJ. The Zygosaccharomyces bailii antifungal virus toxin zygocin: cloning and expression in a heterologous fungal host. Mol Microbiol. 2002;46(4):1095-1105.
Crossref - Muccilli S, Restuccia C. Bioprotective role of yeasts. Microorganisms. 2015;3(4):588-611.
Crossref - Harman GE, Howell CR, Viterbo A, Chet I, Lorito M. Trichoderma species-opportunistic, avirulent plant symbionts. Nat Rev Microbiol. 2004;2(1):43-56.
Crossref - Tseng S-C, Liu S-Y, Yang H-H, Lo C-T, Peng K-C. Proteomic study of biocontrol mechanisms of Trichoderma harzianum ETS 323 in response to Rhizoctonia solani. J Agric Food Chem. 2008;56(16):6914-6922.
Crossref - Zhang D, Spadaro D, Valente S, Garibaldi A, Gullino ML. Cloning, characterization and expression of an exo-1, 3-β-glucanase gene from the antagonistic yeast, Pichia guilliermondii strain M8 against grey mold on apples. Biological Control. 2011;59(2):284-293.
Crossref - Zhang D, Spadaro D, Valente S, Garibaldi A, Gullino ML. Cloning, characterization, expression and antifungal activity of an alkaline serine protease of Aureobasidium pullulans PL5 involved in the biological control of postharvest pathogens. Int J Food Microbiol. 2012;153(3):453-464.
Crossref - Salazar O, Asenjo JA. Enzymatic lysis of microbial cells. Biotechnol Lett. 2007;29(7):985-994.
Crossref - Alasmar R, Ul-Hassan Z, Zeidan R, et al. Isolation of a Novel Kluyveromyces marxianus Strain QKM-4 and Evidence of Its Volatilome Production and Binding Potentialities in the Biocontrol of Toxigenic Fungi and Their Mycotoxins. ACS Omega. 2020;5(28):17637-17645.
Crossref - Spadaro D, Droby S. Development of biocontrol products for postharvest diseases of fruit: the importance of elucidating the mechanisms of action of yeast antagonists. Trends in Food Science & Technology. 2016;47:39-49.
Crossref - Klein MN, Kupper KC. Biofilm production by Aureobasidium pullulans improves biocontrol against sour rot in citrus. Food Microbiol. 2018;69:1-10.
Crossref - Chi M, Li G, Liu Y, et al. Increase in antioxidant enzyme activity, stress tolerance and biocontrol efficacy of Pichia kudriavzevii with the transition from a yeast-like to biofilm morphology. Biological Control. 2015;90:113-119.
Crossref
© The Author(s) 2021. Open Access. This article is distributed under the terms of the Creative Commons Attribution 4.0 International License which permits unrestricted use, sharing, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.