ISSN: 0973-7510
E-ISSN: 2581-690X
This study aims to isolate and characterize efficient biohydrogen generating facultative anaerobic bacteria from various samples, viz., biogas plant (BGP), municipal sewage (MS), and dairy industry treatment plant (DTP). The physicochemical properties of various untreated anaerobic sludge samples reflect the anoxic state and appropriateness of the substrate for separating biohydrogen generating bacteria. The biohydrogen producing bacterial strains were separated from methanogens using the heat-treatment method. The facultative anaerobic bacterial load of heat-treated test samples was determined viz., 27.2±0.57×106 (BGP), 21.8±0.43×106 (MS), and 18.6±0.92×106 (DTP) CFU mL-1 (Colony forming unit), which decreased from the total anaerobic bacterial load of untreated anaerobic sludge viz., 32.1±0.28×106 (BGP), 42.2±0.16×106 (MS), and 34.7±0.12×106 (DTP) CFU mL-1. The 28 predominant bacterial isolates strains were isolated from the heat-treated test samples. All 28 bacterial strains were identified using microscopic and biochemical techniques. Biohydrogen producing potential bacterial strains were screened using the Hungate technique with glucose as a carbon source. Among them, 12 strains were capable of producing biohydrogen, among these 5 strains being excellent biohydrogen producers. Based on the16s rRNA molecular sequencing, the 5 selected biohydrogen generating organisms were authenticated as viz., Salmonella bongori (MZ636759), Escherichia coli (MZ636716), Staphylococcus hominis (MZ636713), Yersinia enterocolitica (OM009292), and Shewanella oneidensis (MZ636800). The gas composition study by GC-TCD in a fermentative medium shows that Shewanella oneidensis (MZ636800) could produce the best biohydrogen (111.4±8.3 mLH2/L), followed by Salmonella bongori (MZ636759) with 98.1±2.9 mL H2/L and Escherichia coli (MZ636716) with 86.7±6.2 mLH2/L.
Biohydrogen, Facultative Anaerobic Bacteria, Anaerobic Sludge, Biogas Plant, Municipal Sewage, Dairy Industry
Global energy consumption has increased in recent decades due to population expansion and industrialization.1 Furthermore, the scarcity of fossil fuels, the high volume of price volatility, and the substantial environmental effect have focused global attention on exploring alternative fuels.2 Renewable energy can be a potential solution to the many issues connected with fossil fuels. Biofuels are promising and sustainable renewable energy sources since they are green, pollution-free, simple to generate, highly effective, and environmentally friendly.3 The liquid and gaseous biofuels include bio-ethanol, bio-butanol, bio-diesel, bio-oil, bio-gas, bio-methane, bio-ethane, bio-butane, and bio-hydrogen.4
Biohydrogen production technology can meet a portion of the world’s energy requirements.5-6 Under dark fermentation and photo fermentation conditions, microbes can be employed to make biohydrogen.7 Several researchers have used Industrial effluent from the sugar industry, beverage industry, chemical industry, palm oil effluent, and distillery sector wastewater as a source for biohydrogen generation8 as a byproduct during an acidogenic stage of anaerobic digestion. A dark fermentation technique has been developed to improve biohydrogen generation; however, it is challenging to employ commercially.9 Substrate composition, nutrient availability, reactor modality, bacterial consortia, and yield all impact the efficiency and utility of biohydrogen generation through fermentation.10 The substrate is essential in the metabolism of bacteria throughout the biohydrogen production process, and the carbon to nitrogen ratio may be tuned to increase yield.11
A wide array of bacteria, including anaerobes such as C.butyricum,12 E.asburiae,13 facultative anaerobes such as E.coli,14 and aerobes B.coagulans have a prominent role in a fermentative biohydrogen generation.15 For significant biohydrogen production by dark fermentation, microbial populations from diversified anaerobic sludge and similar substrates (soil, sediment, and compost) have been studied well.16 Biohydrogen synthesis might allow the sector to be sustainable concerning energy production. When biohydrogen is burnt, the maximum power could be derived, and this thermal power may be conveniently transferred to electric power energy. Hydrogen gas emits pure water on combustion, which is harmless and does not lead to global warming or pollution.17 Because of the above facts, the present investigation screened biohydrogen generating microorganisms from different anaerobic sludges through enriched techniques. The most biohydrogen evolving facultative anaerobic bacteria were chosen and employed for further research. Conventional microbial characterization and molecular sequence (16S rRNA) analysis were used to authenticate the selected bacterial cultures having better biohydrogen production, and GC-TCD was used to assess the bacterial biohydrogen generation ability. To overcome the energy crisis for power generation, biohydrogen conversion into the electrical current is highly recommended which can be achieved by microbial electrolysis cells. The present study focused to isolate novel potential biohydrogen producing bacteria from different anaerobic sludge samples. The present study determines that the microbial strains E.coli, S.bongori, S.hominis, S.oneidensis and Y.enterocolitica able to carry fumarate lyase pathway which leads to hydrogen production. The study also reveals that rather than using S.bongori, S.oneidensis and Y.enterocolitica as mixed culture in biohydrogen production, pure culture leads an effective production.
Anaerobic Sludge Sample Collection
For the isolation of facultative anaerobic biohydrogen producing bacterial strains, different untreated anaerobic sludge from a biogas plant (BGP), municipal sewage (MS), and a dairy industry treatment plant (DTP) were used. The untreated anaerobic sludge samples from the biogas plant at The Gandhigram Rural Institute (Deemed to be University), Gandhigram; municipal sewage sludge from Pallapatti, and dairy industry sludge from SPS Dairy and Food Industry, Nagaiyagoundanpatti, Dindigul District, Tamil Nadu, India, were brought to the lab in sterile, airtight serum bottles under aseptic condition and preserved at 4°C for future research.
Physiochemical Characteristics of Anaerobic Sludge
Physiochemical characteristics of untreated anaerobic sludge in BGP, MS, and DTP such as pH, COD (Chemical Oxygen Demand), TSS (Total Suspended Solid), TS (Total Solids), VSS (Volatile Suspended Solids), Alkalinity, VDS (Volatile dissolved solids), Phosphates, Sodium, Calcium, and Nitrogen were analyzed by standard methods as described in APHA18 manual and performed in triplicate. The statistical analysis of the data was performed using Microsoft Excel 2016.
Enumeration of the Total Microbial Population from Untreated Anaerobic Sludge Samples
The total bacterial population of untreated anaerobic sludge from BGP, MS, and DTP was counted using the standard plate count method.19 A sludge sample of 1g was aseptically suspended in 100mL sterile normal saline (0.85% NaCl), agitated thoroughly, and labeled as 10-2 dilution. Under aseptic conditions, 1mL from 10-2 dilution stock was serially diluted to 10-6 dilution using sterile distilled water (9ml in each tube). The total microbial population of anaerobic sludge was investigated by counting the population of bacteria, fungi, and actinomycetes using the pour plate technique. 1mL sample from respective dilutions viz., 10-5 & 10-6 (for bacteria); 10-3 & 10-4 (for fungi), and 10-2 & 10-3 (for actinomycetes) was transferred into sterile petri plates and poured specific growth agar media viz., bacteria – nutrient agar, fungi – Martin’s rose bengal agar, and Actinomycetes – Kenknights’ agar and mixed well before the media gets solidification. The inoculated petri dishes were incubated for 24 hours at 37°C to observe bacterial growth, for 5 days at 28°C for fungi growth, and for 7 days at 28°C for actinomycetes growth.
Isolation of Facultative Anaerobic Bacteria from Heated Treated Anaerobic Sludge
The predominant facultative anaerobic bacterial strains from heat-treated anaerobic sludge of BGP, MS, and DTP were isolated by spread plate technique.16-20 A sludge sample of 10g was suspended in 90mL of distilled water and shaken well. The dissolved samples were further heated by use of the hot-air oven at the temperature of 100°C for 45 min to destroy cell methanogens and cooled to room temperature before plating.21 For selective biohydrogen producing facultative anaerobic bacterial isolation, 10ml of each heat-treated sludge sample was enriched at room temperature by adding 90mL nutrient broth in anaerobic conditions for 3 days.21 From each heat-treated sludge enriched in nutrient broth, a 0.1mL sample was aseptically inoculated on nutrient agar and spread over the plate with an L-rod. The inoculated Petri dishes were incubated in an anaerobic jar purged with carbon dioxide under room temperature conditions for 24 hours. The predominant bacterial colonies were isolated from heat-treated sludge of BGP, MS, & DTP sludge and preserved as a pure culture for future investigation.
Facultative Anaerobic Bacterial Identification
Selected 28 predominant bacterial cultures were identified through colony morphology, gram staining, and biochemical characteristics. The biochemical tests include IMVIC and glucose fermentation were carried out by following Bergy’s Manual of Determinative bacteriology Eggerth.22
Screening of Biohydrogen Producing Facultative Anaerobic Bacteria
All the 28 selected facultative anaerobic bacterial isolates from heat-treated anaerobic sludge of BGP (9 strains), MS (9 strains), and DTP (10 strains) were qualitatively screened for biohydrogen production by the Hungate method.23 For the screening test, the basal fermentation medium containing (3 g/L-1) NH4HCO3; (0.125 g/L-1) KH2PO4; (0.016 g/L-1) MnSO4. 6H2O; (0.100 g/L-1) MgCl2.6H2O trace elements: (0.025 g/L-1) FeSO4.7H2O; (0.002 g/L-1) CoCl2.5H2O; (0.005 g/L-1) CuSO4.5H2O;(6.72 g/L-1) NaHCO3, and (5 g/L-1) glucose as a carbon source were used. The sterile serum bottle of 100mL was filled aseptically with 50 mL of basal medium to which 1mL of 24 hours fresh bacterial cultures grown in nutrient broth was inoculated. After inoculation, the bottle was capped with an elastomer rubber bung and tightened with an aluminum cap using the crimper. The Nitrogen and CO2 gas were sparged for 5 minutes via a fitted needle on the closed serum bottle’s top. The inoculated container was incubated at 37°C for 48 hours and estimated for biohydrogen production by the Hungate method using a sterile glass syringe.
Gas Compositional Analysis by GC-TCD
The 5 selective potential biohydrogen-producing bacterial strains from the screening test were further analyzed for gas production by Gas Chromatography with Thermal Conductivity Detector (GC-TCD).17 The gas composition (H2, CO2, and O2) produced by potential biohydrogen producing facultative anaerobic bacteria in the basal fermentation medium’s headspace was evaluated using GC-TCD (SHIMADZU GC-2014, Japan). Chromatography: Gas Chromatography (GC); Detector: Thermal conductivity Detector (TCD); Column: Stationary Phase – 80/100 mesh of Porapak Q; Temperature: 80°C, 100°C, and 150°C; Carrier gas: Mobile Phase – Nitrogen (N); Flow rate: 20 mLmin-1; Sample Volume: 1µL were used for gas analysis.
Cumulative biohydrogen production (ml/L-1) was calculated through a modified Gompertz equation to interpret the characteristics of hydrogen produced from the batch test,17 which can be represented as follows:
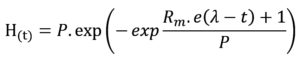 Cumulative H2 production in H(t); H2 production potential in P; the maximum H2 production rate in Rm; constant value 2.71828 in e; lag-phase in (l); time (t). The corresponding values of P, Rm, and l for each batch were carried out by sigmaplot software.
Cumulative H2 production in H(t); H2 production potential in P; the maximum H2 production rate in Rm; constant value 2.71828 in e; lag-phase in (l); time (t). The corresponding values of P, Rm, and l for each batch were carried out by sigmaplot software.
SEM Imaging Analysis for Selected Biohydrogen Producing Bacterial Strains
The morphological structures of five selected biohydrogen producing bacterial strains were visualized by Scanning Electron Microscope (SEM).24-25 A liquid sample of a biohydrogen generating bacterial culture was mounted on a glass surface and fixed for 4 hours with 4 % glutaraldehyde. The fixed culture was then dried in ethyl alcohol with different concentrations viz., 30, 50, 70, 80, and 100% (v/v) for 5 minutes each at room temperature. The dehydrated bacterial culture was passed under the scanning electron microscope (SEM Vega 3 Tescan model).
Molecular Characterization of Potential Biohydrogen Producing Facultative Anaerobic Bacteria
The 5 potential biohydrogen producing facultative anaerobic bacterial strains, after confirmation by GC-TCD, were subjected to 16S rRNA sequencing for genetic level identification at CTBC (Central for Tropical Biodiversity conservation), Malappuram, Kerala, India. The resulting molecular sequences were analyzed by comparing the reported public data in NCBI and Blast algorithm to verify their individuality and integrity. The gene sequences were deposited into the genebank, and accession numbers were assigned. Sequences have been aligned and further developed to understand the variation among the microbial group and species identification by framing a phylogenetic tree through the MEGA program [version 5.0] based on the neighbor-joining method.
Physiochemical Characteristics of Untreated Anaerobic Sludge
The physiochemical parameters of untreated anaerobic sludge from BGP, MS, and DTP were analyzed and recorded (Table 1). The pH significantly impacts the degradation of organic matter by altering the efficiency of hydrolases.26 Research has found that methanogenesis proceeds effectively in an anaerobic reactor with a pH of 6.5 to 8.2, whereas hydrolysis and acidogenesis occur at around 5.5 and 6.5, respectively.27-28 In this study, the pH of untreated anaerobic sludge samples was found to be about 9.16±0.3 (BGP), 7.23±0.08 (MS), and 8.3±0.08 (DTP) (Table 1). Zhao et al.,29 have already reported the pH of 8.5 in the anaerobic sludge of the biogas plant.
Table (1):
Physiochemical characteristics of anaerobic sludge of BGP, MS, and DTP.
| Parameters | Anaerobic sludge | ||
|---|---|---|---|
| Biogas plant | Municipal sewage | Dairy industry | |
| pH | 9.16±0.3 | 7.23±0.08 | 8.3±0.08 |
| COD | 101.2±0.9 | 55.1±0.6 | 121.4±0.5 |
| Alkalinity | 62.6±0.3 | 76.2±0.8 | 30.2±0.2 |
| TS | 38.6±0.7 | 21.3±0.3 | 50.3±0.2 |
| TDS | 82.1±0.9 | 162.1±0.6 | 131.2±0.7 |
| TSS | 34.6±0.4 | 13.7±0.9 | 19.4±0.4 |
| VS | 14.6±0.7 | 8.3±0.3 | 9.2±0.6 |
| VSS | 19.3±0.4 | 53.2±0.03 | 23.7±0.2 |
| Phosphates | 0.28±0.04 | 0.97±0.05 | 0.45±0.1 |
| Calcium | 0.364±0.3 | 0.86±0.2 | 0.96±0.01 |
| Nitrogen | 3.9±0.1 | 2.4±0.06 | 4.5±0.2 |
| Sodium | 1.4±0.9 | 4.6±0.7 | 2.2±0.3 |
Note: Values are Mean of three replicates ± Standard Error / All the parameters are in mg/L-1 except pH.
TSS (Total Suspended Solid) determines the efficiency of anaerobic sludge treatment processes. In the present study, the biogas plant shows a high concentration of TSS 34.6±0.4 (Table 1), and these results are very close to the study of Wetts et al.,30 where the TSS estimation of the biogas plant was found as 36.0 mg/L-1. The present study recorded TSS and TS of municipal sewage sludge as TSS 13.7±0.9 mg/L-1 and TS 21.3±0.3 mg/L-1, respectively. In an earlier study, Farhat et al.,31 evaluated the municipal sewage and reported 5.2±0.2 mg/L-1 TSS and TS 5.8±0.21 mg/L-1. In the dairy sector, anaerobic sludge has high levels of COD 121.4±0.5 mg/L-1 (Table 1), which is selected to calculate the oxygen equivalent of the organic carbon content of a sensitive substance to oxidation by a powerful chemical oxidizer. Anaerobic treatment, on the other hand, does not utilize oxygen. The anaerobic sludge of municipal sewage shows high concentrations of COD 55.1±0.6 mg/L-1 (Table 1) and comparable outcomes have been recorded by Borowski et al.,32 and they revealed high concentration of COD 41.06±13.30 mg/L-1 in municipal sewage sludge.
The Total Microbial Population of Untreated Anaerobic Sludge
The total microbial count of bacteria, fungi, and actinomycetes from three untreated anaerobic sludge samples was enumerated using the spread plate technique, and the results are recorded in Table 2.
Table (2):
Total microbial population of untreated anaerobic sludge of BGP, MS, and DTP.
| Anaerobic sludge samples |
Microbial population | ||
|---|---|---|---|
| Bacteria x106 (CFU mL-1) | Fungi x103 (CFU mL-1) | Actinomycetes x102 (CFU mL-1) | |
| Bio-gas plant(BGP) | 35.2±0.57 | 16.2± 0.62 | 1.4±0.34 |
| Municipal sewage(MS) | 55.8±0.43 | 9.8±0.23 | 2.5±0.54 |
| Dairyindustry treatment plant(DTP) | 68.6±0.92 | 8.4±0.61 | 1.6±0.42 |
Note: Values are Mean of three replicates ± Standard Error.
This study shows that the bacterial load was higher in dairy sludge, 68.6±0.92 CFU mL-1. In comparison, the fungal population was increased in biogas plant sludge at 16.2±0.62 CFU mL-1, and the actinomycetes population was higher in municipal sewage sludge at 2.5±0.54 CFU mL-1 (Table 2). Mukherjee et al.,33 reported the cell density load. The bacteria profile of raw dairy effluent revealed a significant concentration of anaerobic and aerobic bacteria of 5±0.02 Log CFU mL-1 and 4±0.12 Log CFU mL-1, respectively.
Isolation of Facultative Anaerobic Bacteria from Heat-treated Anaerobic Sludge
The total bacterial population was enumerated from heat-treated anaerobic sludge samples of BGP, MS & DTP, which shows a high difference in colony count from the whole bacterial load of the untreated sludge samples (Table 3).
Table (3):
Total facultative anaerobic bacterial population of untreated and heat-treated anaerobic sludge of BGP, MS, and DTP.
| Facultative anaerobic bacterial population (106 × CFUmL-1) | ||
|---|---|---|
| Anaerobic sludge Sample | Untreated anaerobic sludge samples | Heat-treated anaerobic sludge samples |
| Bio-gas plant (BGP) | 32.1±0.28 | 27.2±0.57 |
| Municipal sewage (MS) | 42.2±0.16 | 21.8±0.43 |
| Dairy industry treatment plant (DTP) | 34.7±0.12 | 18.6±0.92 |
Note: Values are Mean of three replicates ± Standard Error.
The total population of facultative anaerobic bacteria in heat-treated anaerobic sludge samples of (BGP), (MS), and (DTP) was recorded with a cell count of 27.2±0.57 (BGP), 21.8±0.43 (MS), and 18.6±0.92 (DTP) CFU mL-1 respectively (Table 3). The bacterial count in pretreated sludge indicates the chances of methanogens and hydrogen-consuming bacteria.16 Similar to these results, Alibardi et al.,34 evaluated the microbial community of anaerobic sludge before and after heat-shock pretreatment at 100°C for 30 minutes and reported 1.3×107(CFU/g-1) and 1.0×105(CFU/g-1) respectively. Anaerobic sludge pretreatment is one of the potential techniques for selective isolation of hydrogen producers by inhibiting the hydrogen consumer.16-36 This study isolated 28 predominant facultative anaerobic bacteria from pretreated anaerobic sludge samples, including 9 strains from BGP, 9 strains from MS, and 10 strains from DTP.
Identification of Facultative Anaerobic Bacteria
Morphological characterization of 28 bacterial isolates by gram staining technique shows that the 15 strains were gram-negative, and the other 13 strains were gram-positive (Table 4). Among 10 bacterial isolates from biogas slurry, 8 strains were rod, and 2 strains were cocci in shape. In municipal sewage sludge, all 9 strains in rod shape and dairy industries reported 7 strains in rod and 3 strains in cocci. A biochemical study of microorganisms is essential to identify their genus and species of bacterial isolates.37 In this study, 28 selected bacterial isolates were identified through various biochemical characterizations, and the outcomes were recorded in (Table 4).
Table (4):
Morphological and Biochemical Characteristics of facultative anaerobic bacteria of anaerobic sludge samples.
| Isolates | Sources | Morphological characteristics | Biochemical characteristics | Name of the organisms identified | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Colony | Shape | Gram’s stain | Motility | IMVIC | Catalase | Sugar fermentation | |||||||
| Glucose | Sucrose | Lactose | Mannitol | ||||||||||
| BGP1 | Biogas Plant | Circular, entire, and umbonate | Rod | – | Motile | -/-/+/+ | + | + | + | – | + | Serratia marcescens | |
| BGP2 | Irregular, undulate, and opaque | Rod | + | Motile | -/+/+/+ | + | + | + | + | + | Bacillus licheniformis | ||
| BGP3 | Entire, convex, and circular | Cocci | + | Non-motile | -/-/+/- | – | + | + | + | + | Enterococcus Faecalis | ||
| BGP4 | Entire, convex, and circular | Rod | – | Motile | -/+/-/+ | + | + | – | – | + | Salmonella bongori | ||
| BGP5 | Circular, convex, and entire | Rod | – | Motile | -/-/-/+ | + | – | – | – | + | Pseudomonas spp | ||
| BGP6 | Entire, convex, and entire | Cocci | + | Non-motile | -/-/+/- | – | + | – | + | + | Enterococcus faecium | ||
| BGP7 | Circular, convex, and entire | Cocci | + | Non-Motile | -/+/+/- | + | – | – | – | – | Micrococcus spp | ||
| BGP8 | Circular, raised, and entire | Rod | + | Non-motile | -/-/-/- | – | + | + | + | – | Lactobacillus spp | ||
| BGP9 | Circular, entire, and opaque | Rod | – | Motile | -/+/-/+ | + | + | – | – | – | proteus mirabilis | ||
| MS1 | Municipal Sewage | Circular, raised, and entire | Rod | + | Non-motile | -/-/-/- | – | + | + | + | – | Lactobacillus spp | |
| MS2 | Circular, convex, and entire | Rod | – | Non-motile | -/+/-/- | + | + | – | – | + | yersinia spp | ||
| MS3 | Circular, entire and raised | Rod | – | Motile | +/+/-/- | + | + | + | + | + | E. coli | ||
| MS4 | Circular, convex, and translucent | Rod | – | Non-motile | +/+/-/- | + | – | – | – | + | Shigella spp | ||
| MS5 | Entire, convex, and opaque | Rod | – | Motile | -/-/+/+ | + | + | – | + | + | Enterobacter aerogene | ||
| MS6 | Circular, raised, and entire | Rod | + | Non-motile | -/-/-/- | – | + | + | + | – | Lactobacillus spp | ||
| MS7 | Circular, entire and raised | Rod | – | Motile | +/+/-/- | + | + | + | + | + | E.coli | ||
| MS8 | Entire, convex, and circular | Rod | – | Motile | -/+/-/- | + | + | – | – | + | Salmonella spp | ||
| MS9 | Circular, entire, and convex | Rod | – | Motile | +/+/-/+ | + | + | – | – | – | Proteus spp | ||
| DTP1 | Dairy Industry Treatment Plant | Circular, convex, and entire | Rod | – | Motile | -/-/-/+ | + | – | – | – | – | Pseudomonas spp | |
| DTP2 | Circular, entire and opaque | cocci | + | Non-motile | -/-/-/- | + | + | + | + | – | Staphylococcus spp | ||
| DTP3 | Circular, raised, and entire | Rod | + | Non-motile | -/-/-/- | – | + | + | + | – | Lactobacillus spp | ||
| DTP4 | Circular, entire, and convex | Rod | – | Motile | +/+/-/+ | + | – | – | – | – | Proteus spp | ||
| DTP5 | Opaque, convex, and circular | Rod | – | Motile | +/+/+/+ | + | + | + | – | + | Aeromonas hydrophila | ||
| DTP6 | Circular, entire and convex | cocci | + | Non-motile | -/+/+/+ | + | + | + | + | + | staphylococcus auerou | ||
| DTP7 | Circular, convex, and smooth | Rod | – | Motile | -/-/-/+ | + | + | + | – | + | Shewanella spp | ||
| DTP8 | Circular, entire and convex | Rod | + | Motile | -/-/+/+ | + | + | + | – | – | Bacillus cereus | ||
| DTP9 | Circular, convex, and entire | Cocci | + | Non-motile | -/+/+/- | + | – | – | – | – | Micrococcus spp | ||
| DTP10 | Circular, raised, and entire | Rod | + | Non-motile | -/-/-/- | – | + | + | + | – | Lactobacillus acidophilus | ||
+ Negative; – Positive; IMVIC- Indole/Methyl red/Voges proskauer/Citrate
Screening of Facultative Anaerobic Bacteria for Biohydrogen Production
All the 28 facultative anaerobic bacterial isolates from different anaerobic sludge samples were screened for biohydrogen production under fermentative test by the Hungate method. Among the isolates, 12 strains include E.coli, Salmonella spp, Enterococcus spp, Staphylococcus spp, Proteus spp, yersinia spp, Klebsiella, B.cereus, Micrococcus, S.oneidensis, E.aerogene, and Pseudomonas spp were able to produce better biohydrogen. Earlier studies also reported various biohydrogen producing microorganisms from different samples, viz., Proteus in oil refineries effluent,38 Pseudomonas in lactate wastewater,39 E.Faecium in anaerobic digested sludge,40 E.aerogenes & E.cloacae in domestic wastewater,41 B firmus in municipal sludge Sinha and Pandey,42 Clostridium spp., B.megaterium, Staphylococcus spp, B.subtilis & Lactobacillus spp in granular sludge,43 and Klebsiella in anaerobic sewage sludge.44 Morra et al.,45 investigated different effective biohydrogen producing bacterial strains viz., C.beijerinckii, L.plantarum, E.devriesei, and S.hominis from digestate plant of biohydrogen pilot-scale plant. Based on the biohydrogen producing ability, 5 bacterial strains were selected based on potential biohydrogen producers and were further characterized through GC-TCD.
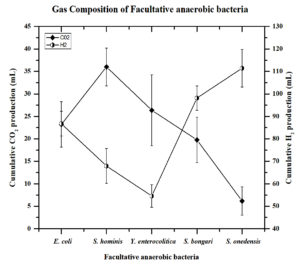
Figure 1. H2 and CO2 composition of facultative anaerobic bacteria in the fermentative medium by GC-TCD.
Gas Compositional Analysis by GC-TCD
The 5 efficient biohydrogen generating bacteria were studied for biohydrogen generation in a fermentative medium with glucose as a carbon source at 37°C and estimated by GC-TCD. Cumulative hydrogen production and CO2 production were evaluated, and the results are recorded in (Figure 1). The volume of gas and gas analysis with gas chromatography reveals that out of the 5 isolates, S.oneidensis shows the maximum cumulative H2 production of 111.4±8.3 mLH2/L, while the cumulative H2 production of the other four strains, viz., S.bongori, E.coli, S.hominis, and Y.enterocolitica was estimated as 98.1±2.9 mLH2/L, 86.7±6.2 mLH2/L, 67.9±7.7 mLH2/L, and 54.5±5.3 mLH2/L respectively. The Carbon dioxide (CO2) yield in the fermentative medium was higher for S.hominis 36.8±4.7 mLCO2/L than for other strains such as Y.enterocolitica (26.3±6.8 mLCO2/L), S.oneidensis (6.1±3.13 mLCO2/L), S.bongori (19.7±5.3 mLCO2/L) and E. coli 23.2±5.2 mLCO2/L (Table 1).
Table (5):
Productive yield of H2 and CO2 of facultative anaerobic bacteria by GC-TCD.
Facultative anaerobic bacteria |
Maximum Hydrogen production rate (mLH2/L) |
Maximum CO2 production rate (mL CO2/L) |
Hydrogen productivity (mol-H2/mol-glucose consumed) |
CO2productivity (mol-CO2 /mol-glucose consumed) |
|---|---|---|---|---|
Escherichia coli |
0.53± 4.3 |
0.13± 2.3 |
0.62±5.9 |
0.08±5.9 |
Staphylococcus hominis |
0.48± 2.6 |
0.28± 4.3 |
0.42±3.1 |
0.12±3.1 |
Yersinia enterocolitica |
0.34± 5.2 |
0.17± 1.3 |
0.78±2.8 |
0.09±2.8 |
SalmonellBongari |
0.62± 2.2 |
0.11± 3.8 |
0.72±1.2 |
0.02±1.2 |
Shewahellaonedensis |
0.76± 4.2 |
0.33± 5.8 |
0.88±2.8 |
0.03±2.8 |
Note: Values are Mean of three replicates ± Standard Error.
The gas composition of five potential biohydrogen producing strains, including S.bongori from the biogas plant, S.oneidensis, and E.coli from the dairy industry, S.hominis, and Y.enterocolitica from municipal sewage, were estimated by GC-TCD (Table 5). The maximum H2 (0.76±4.1 mLH2/L) and CO2 production rate (0.13±2.4 mLH2/L) along with better H2 yield (0.88±2.8 mol-H2/mol-glucoseconsumed) were achieved in the S.oneidensis. Minimal hydrogen production rate (0.62 ±2.1 mLH2/L) and H2 yield were (0.72±1.2 mol-H2/mol-glucoseconsumed) attained in S.bongori. The lowest CO2 production rate (0.17±1.2 mLCO2/L) and CO2 yield (0.09±2.5 mol-CO2/mol-glucoseconsumed) were measured in Y.enterocolitica. During H2 yield in a fermentative medium, CO2 production rate (0.13±2.5 mLCO2/L) and CO2 yield (0.08±5.9 mol-CO2/mol-glucoseconsumed) were less for the strain E.coli. Biohydrogen production from different carbon sources by E.coli showed the maximum H2 yield of 0.55±0.05 mol H2/mol substrate.14 The mixed biohydrogen producing culture, namely, Enterobacteria, CFB group bacteria, S.bongori, B.goodwinii, E.amylovora, Sulfurospirillum sp, T.thiocaminus, and H.thermophila yield maximum hydrogen production of 25.3 and 11.1 ml/day at pH 5.0.46
SEM Observation of Hydrogen-producing Bacteria
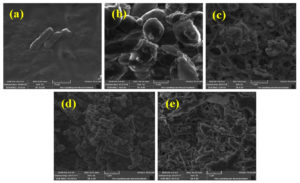
SEM micrographs of the five efficient biohydrogen producing strains are depicted in Figure 2. The bacterial strains such as E.coli, S.oneidensis, S.bongori, and Y.enterocolitica were rod-shaped, and S.hominis was spherical in shape (Figure 2).
Figure 2. SEM micrographs of bacteria (a) E.coli (b) S.hominis (c) S.oneidensis (d) S.bongori and (e) Y.enterocolitica.
Molecular Sequencing
The 5 bacterial strains with high potential to produce biohydrogen were determined by analyzing molecular sequences of 16S rRNA (Figure 3). The reported 16S rRNA gene sequences were arranged via the program BLAST in the study.
Figure 3. 16S rRNA sequence of five potential biohydrogen producing facultative anaerobic bacterial isolates.
Gene sequence analysis of the 16S rRNA gene revealed a resemblance of 98 % for S.oneidensis, 96% for S.bongori, 96 % for E. coli, 97% for Y.enterocolitica, and 94% for S.hominis. The MEGA 5.0 program was used to construct a phylogenetic tree (Figure 3). The accession number for 16S rRNA sequence submitted to genebank database viz., S.oneidensis (MZ636800), S.bongori (MZ636759), E.coli (MZ636716), Y.enterocolitica (OM009292) and S.hominis (MZ6368713).
Biohydrogen is a widely known fuel for future energy demand and is economically feasible. Biohydrogen production faces limitations like slow substrate conversion, the effect of carbon-rich acid intermediates, change in the redox system, and buffering capacity that lowers the hydrogen production. In the present work, to achieve excellent substrate conversion efficiency in biohydrogen, facultative anaerobic bacteria possessing potential catalytic activity from different anaerobic sludge were isolated. The 28 facultative anaerobic bacterial strains were isolated from heat-treated anaerobic sludge samples from BGP, MS & DTP. Among the selected bacterial strains, 5 strains such as S.bongori, E.coli, S.hominis, Y.enterocolitica, and S.oneidensis were found to be potential H2 producers. The study also reveals that S.oneidensis (MZ636800) is a high potential bio-H2 producer that could produce 111.4±8.3 mLH2/L biohydrogen gas by dark fermentation. Further research might lead to economic utilization of the selected biohydrogen producing bacterial isolate, S.oneidensis.
ACKNOWLEDGMENTS
The author would like to thank Head of Biology Department, GRI-DTBU for granting the required experimental resources and assistance. Authors also thankful to the Central Instrument Facility for providing the SEM facility (GRI-DTBU) and the GRI General Library for supplying the various softwares for improving the English language and for similarity check screening. We thank the Laboratory of Bioenergy and Bioremediation, Microbiology Department, Alagappa University Karaikudi, Tamil Nadu, India for facilitating with GC-TCD instrument for analysis.
CONFLICT OF INTEREST
The authors declare that there is no conflict of interest.
AUTHORS’ CONTRIBUTION
All authors listed have made a substantial, direct and intellectual contribution to the work, and approved it for publication.
FUNDING
None.
DATA AVAILABILITY
All data sets assessed during this research are reported in the article, and the 16s rRNA sequences of Accession No. MZ636759, MZ636716, MZ636800, MZ6368713, and OM009292 are accessible in National Center for Biotechnology Information (NCBI) Nucleotide databases.
ETHICS STATEMENT
Not applicable.
- Akia M, Yazdani F, Motaee E, Han D, Arandiyan H. A review on conversion of biomass to biofuel by nanocatalysts. Biofuel Res J. 2014;1(1):16-25.
Crossref - Ibarra-Gonzalez P, Rong BG. A review of the current state of biofuels production from lignocellulosic biomass using thermochemical conversion routes. Chin J Chem Eng. 2019;27(7):1523-1535.
Crossref - Sekoai PT, Ouma CNM, du Preez SP, et al. Application of nanoparticles in biofuels: An overview. Fuel. 2019;237:380-397.
Crossref - Ladole MR, Mevada JS, Pandit AB. Ultrasonic hyperactivation of cellulase immobilized on magnetic nanoparticles. Bioresource Technol. 2017;239:117-126.
Crossref - Heo E, Kim J, Cho S. Selecting hydrogen production methods using fuzzy analytic hierarchy process with opportunities, costs, and risks. Int J Hydrog Energy. 2012;37(23):17655-17662.
Crossref - Ren X, Dong L, Xu D, Hu B. Challenges towards hydrogen economy in China. Int J Hydrog Energy. 2020;45(59):34326-34345.
Crossref - Argun H, Dao S. Hydrogen gas production from waste peach pulp by dark fermentation and electrohydrolysis. Int J Hydrog Energy. 2016;41(27):11568-11576.
Crossref - Boodhun BSF, Mudhoo A, Kumar G, Kim SH, Lin CY. Research perspectives on constraints, prospects and opportunities in biohydrogen production. Int J Hydrog Energy. 2017;42(45):27471-27481.
Crossref - Kumar G, Sivagurunathan P, Pugazhendhi A, et al. A comprehensive overview on light independent fermentative hydrogen production from wastewater feedstock and possible integrative options. Energy Conversion and Management. 2017;141:390-402.
Crossref - Mota VT, Ferraz Junior ADN, Trably E, Zaiat M. Biohydrogen production at pH below 3.0: Is it possible? Water Research. 2018;128:350-361.
Crossref - Hernandez-Mendoza CE, Moreno-Andrade I, Buitron G. Comparison of hydrogen-producing bacterial communities adapted in continuous and discontinuous reactors. Int J Hydrog Energy. 2014;39(26):14234-14239.
Crossref - Jiang D, Fang Z, Chin SX, Tian XF, Su TC. Biohydrogen Production from Hydrolysates of Selected Tropical Biomass Wastes with Clostridium Butyricum. Sci Rep. 2016;6:27205.
Crossref - Alvarez-Guzman CL, Cisneros-de la Cueva S, Balderas-Hernandez VE, Smolinski A, De Leon-Rodriguez A. Biohydrogen production from cheese whey powder by Enterobacter asburiae: Effect of operating conditions on hydrogen yield and chemometric study of the fermentative metabolites. Energy Reports. 2020;6:1170-1180.
Crossref - Taifor AF, Zakaria MR, Mohd Yusoff MZ, Toshinari M, Hassan MA, Shirai Y. Elucidating substrate utilization in biohydrogen production from palm oil mill effluent by Escherichia coli. Int J Hydrog Energy. 2017;42(9):5812-5819.
Crossref - Lertsriwong S, Glinwong C. Newly-isolated hydrogen-producing bacteria and biohydrogen production by Bacillus coagulans MO11 and Clostridium beijerinckii CN on molasses and agricultural wastewater. Int J Hydrog Energy. 2020;45(51):26812-26821.
Crossref - Murugan RS, Dinesh GH, Swetha TRA, et al. Acinetobacter junii AH4-A potential strain for bio-hydrogen production from dairy industry anaerobic sludge. J Pure Appl Microbiol. 2018;12(4):1761-1769.
Crossref - Dinesh GH, Sundaram K, Mohanrasu K, et al. Optimization (substrate and pH) and anaerobic fermentative hydrogen production by various industrial wastes isolates utilizing biscuit industry waste as substrate. J Pure Appl Microbiol. 2018;12(3):1587-1595.
Crossref - Federation WE. APHA, AWWA, WEF. Standard Methods for examination of water and wastewater. Anales de Hidrologia Medica. 2012;5(2):185-186-186.
- Campos CR, Mesquita VA, Silva CF, Schwan RF. Efficiency of physicochemical and biological treatments of vinasse and their influence on indigenous microbiota for disposal into the environment. Waste Management. 2014;34(11):2036-2046.
Crossref - Niu K, Zhang X, Tan WS, Zhu ML. Characteristics of fermentative hydrogen production with Klebsiella pneumoniae ECU-15 isolated from anaerobic sewage sludge. Int J Hydrog Energy. 2010;35(1):71-80.
Crossref - Mechery J, Biji B, Thomas DM, Sylas VP. Biohydrogen production by locally isolated facultative bacterial species using the biomass of Eichhornia crassipes: effect of acid and alkali treatment. Energy, Ecology, and Environment. 2017;2(5):350-359.
Crossref - Eggerth AH. The Gram-positive Non-spore-bearing Anaerobic Bacilli of Human Feces. J Bacteriol. 1935;30(3):277-299.
Crossref - Miller TL, Wolin MJ. A Serum Bottle Modification of the Hungate Technique for Cultivating Obligate Anaerobes. Appl Microbiol. 1974;27(5):985-987.
Crossref - Badiei M, Jahim JM, Anuar N, Sheikh Abdullah SR, Su LS, Kamaruzzaman MA. Microbial community analysis of mixed anaerobic microflora in suspended sludge of ASBR producing hydrogen from palm oil mill effluent. Int J Hydrog Energy. 2012;37(4):3169-3176.
Crossref - Kumari S, Das D. Improvement of biohydrogen production using acidogenic culture. Int J Hydrog Energy. 2017;42(7):4083-4094.
Crossref - Mathew AK, Bhui I, Banerjee SN, et al. Biogas production from locally available aquatic weeds of Santiniketan through anaerobic digestion. Clean Technol Environ Policy. 2015;17(6):1681-1688.
Crossref - Lee DH, Behera SK, Kim JW, Park HS. Methane production potential of leachate generated from Korean food waste recycling facilities: A lab-scale study. Waste Management. 2009;29(2):876-882.
Crossref - Goswami R, Chattopadhyay P, Shome A, et al. An overview of physico-chemical mechanisms of biogas production by microbial communities: A step towards sustainable waste management. 3 Biotech. 2016;6(1):72.
Crossref - Zhao HY, Li J, Liu JJ, Lu YC, Wang XF, Cui ZJ. Microbial Community Dynamics During Biogas Slurry and Cow Manure Compost. J Integr Agrice. 2013;12(6):1087-1097.
Crossref - Wett B, Schoen M, Phothilangka P, Wackerle F, Insam H. Model-based design of an agricultural biogas plant: Application of Anaerobic digestion model No. 1 for an improved four chamber scheme. Water Sci Technol. 2007;55(10):21-28.
Crossref - Farhat A, Asses N, Ennouri H, Hamdi M, Bouallagui H. Combined effects of thermal pretreatment and increasing organic loading by co-substrate addition for enhancing municipal sewage sludge anaerobic digestion and energy production. Process Saf Environ Prot. 2018;119:14-22.
Crossref - Borowski S, Domanski J, Weatherley L. Anaerobic co-digestion of swine and poultry manure with municipal sewage sludge. Waste Management. 2014;34(2):513-521.
Crossref - Mukherjee S, Kundu R, Narula R. Environmental Biotechnology For Soil and Wastewater Implications on Ecosystems. 2020.
Crossref - Alibardi L, Favaro L, Lavagnolo MC, Basaglia M, Casella S. Effects of heat treatment on microbial communities of granular sludge for biological hydrogen production. Water Sci Technol. 2012;66(7):1483-1490.
Crossref - Mu Y, Yu HQ, Wang G. Evaluation of three methods for enriching H2-producing cultures from anaerobic sludge. Enzyme and Microbial Technology. 2007;40(4):947-953.
Crossref - Assawamongkholsiri T, Reungsang A, Pattra S. Effect of acid, heat and combined acid-heat pretreatments of anaerobic sludge on hydrogen production by anaerobic mixed cultures. Int J Hydrog Energy. 2013;38(14):6146-6153.
Crossref - Tandon M, Thakur V, Tiwari KL, Jadhav SK. Enterobacter ludwigii strain IF2SW-B4 isolated for bio-hydrogen production from rice bran and de-oiled rice bran. Environ Technol Innov. 2018;10:345-354.
Crossref - Porwal S, Kumar T, Lal S, et al. hydrogen and polyhydroxybutyrate producing abilities of microbes from diverse habitats by the dark fermentative process. Bioresource Technol. 2008;99(13):5444-5451.
Crossref - Ziara RMM, Miller DN, Subbiah J, Dvorak BI. Lactate wastewater dark fermentation: The effect of temperature and initial pH on biohydrogen production and microbial community. Int J Hydrog Energy. 2019;44(2):661-673.
Crossref - Yin Y, Wang J. Characterization and Hydrogen production performance of a novel strain Enterococcus faecium INET2 isolated from gamma irradiated sludge. Int J Hydrog Energy. 2016;41(48):22793-22801.
Crossref - Abd-Alla MH, Gabra FA, Danial AW, Abdel-Wahab AM. Enhancement of biohydrogen production from sustainable orange peel wastes using Enterobacter species isolated from domestic wastewater. International Journal of Energy Research. 2019;43(1):391-404.
Crossref - Sinha P, Pandey A. Biohydrogen production from various feedstocks by Bacillus firmus NMBL-03. Int J Hydrog Energy. 2014;39(14):7518-7525.
Crossref - Pachiega R, Sakamoto IK, Varesche MB, Hatanaka RR, de Oliveira JE, Maintinguer SI. Obtaining and Characterization of Mesophilic Bacterial Consortia from Tropical Sludges Applied on Biohydrogen Production. Waste and Biomass Valorization. 2019;10(6):1493-1502.
Crossref - Xiao L, Deng Z, Fung KY, Ng KM. Biohydrogen generation from anaerobic digestion of food waste. Int J Hydrog Energy. 2013;38(32):13907-13913.
Crossref - Morra S, Arizzi M, Allegra P, et al. Expression of different types of [FeFe]-hydrogenase genes in bacteria isolated from a population of a bio-hydrogen pilot-scale plant. Int J Hydrog Energy. 2014;39(17):9018-9027.
Crossref - Dursun N, Gulsen H. Evaluation of industrial waste black cumin (Nigella Sativa) for biohydrogen production without pretreatment. Environ Develop Sustain. 2021;
Crossref
© The Author(s) 2022. Open Access. This article is distributed under the terms of the Creative Commons Attribution 4.0 International License which permits unrestricted use, sharing, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.