ISSN: 0973-7510
E-ISSN: 2581-690X
Multidrug resistant Pseudomonas aeruginosa is an alarming and emerging public health problem globally across the developing countries. Pseudomonas aeruginosa is still a major cause for nosocomial infection and approx 10-20% of these patients are admitted to the ICU’s. Bacterial isolates those are biofilm producers are more drug resistant than biofilm Non-producers. The aim of the present study was to evaluate the production of biofilm and β-lactamases (ESBL, MBL, AmpC) in multi drug resistant Pseudomonas aeruginosa isolated from ICU patients. The present cross-sectional prospective study was carried out in the Department of Microbiology, Santosh Medical College & Hospital, Ghaziabad, Uttar-Pradesh, India. A total of 115 isolates of P. aeruginosa were isolated from 502 clinical samples. After confirmation of MDR status of P. aeruginosa further processing for biofilm and beta lactamases was performed accordingly. Biofilm production was done by test tube method and tissue culture plate method along with phenotypic profiling of ESBL, MBL and AmpC was performed by disc potentiation test; IMP-EDTA combined disc test and Cefoxitin Cloxacillin Double Disc synergy test (CC-DDST) respectively. Out of 502 total human clinical samples 115 isolates were Pseudomonas aeruginosa giving the prevalence rate of 23%. Among 115 isolates of P. aeruginosa 60 (52%) were MDR phenotypes, Out of 60 MDR isolates 23 (38.3%) were ESBL producers, 22 (36.6%) were MBL producers, and 3(5%) were AmpC producers. Out of total 115 isolates 68(59%) isolates were biofilm producers and 47 (40.8%) were biofilm non-producers. Strict antibiotic policies with early detection of beta lactamases and detection of biofilm production should be performed regularly for all clinical isolates of Pseudomonas aeruginosa so as to guide antibiotic selection along with better management of severe infection in ICU patients.
Extended Spectrum beta lactamases (ESBL), Metallo betalactamases (MBL), Multidrug resistant, Biofilm
Multidrug resistant Pseudomonas aeruginosa is an alarming and emerging public health problem across the developing countries.1 It is mainly responsible for mortality in humans.2 Pseudomonas aeruginosa is still a major cause for nosocomial infection and approx 10-20% of these patients are admitted to the ICU’s.3 It is one of the most frequent gram negative non fermenting pathogen that is seen in ICU patients. It is known to be associated with Urinary tract infection, Surgical site infection, Pneumonia and bacteremia. The severity of infection increases when the patient’s immune system impedes.4
Biofilm is described as a structural community of bacterial cells bounded in self founded polymetric matrix adherent to biotic or abiotic surface.5 Biofilm formation is the key factor which is responsible for the chronic infection by Pseudomonas aeruginosa and other microorganisms. Biofilm is a complex composition of bacterial cells lodged in an extracellular matrix which is made up of proteins, extracellular DNA & exopolysaccharides. It is a safeguard for bacterial cells, which is very difficult to manage by antibacterial compounds.6 Bacterial isolates those are biofilm producers are more drug resistant than biofilm nonproducers. Some previous studies have shown that the Pseudomonas aeruginosa isolates that are biofilm producers are resistant to Ceftazidime, Ciprofloxacin & Tobramycin antibiotics at concentrations more than those necessary to kill planktonic bacteria.7,8
Antibacterial drug has less access due to impaired diffusion through biofilm matrix. Drug resistant strains of Pseudomonas aeruginosa show a high level of intrinsic resistance to antimicrobial drugs with the help of efflux pump, biofilm formation amino glycoside modifying enzymes and sometimes by mutation of chromosomal genes (ESBL & AmpC hyper expression).9 The potentiality of P. aeruginosa to produce variety of drug resistance mechanism has led to evolution of drug resistant phenotypes. This poses as a challenge to clinician for the treatment of such kind of severe infection. This type of situation draws attention for the detection of phenotypes those are producing different kind of mechanism for the drug resistance to avoid treatment failure and hospital acquired infection.10 The aim of the present study was to evaluate the production of biofilm and β-lactamases (ESBL, MBL, AmpC) in multi drug resistant Pseudomonas aeruginosa isolated from ICU patients.
The present cross-sectional prospective study was carried in the department of Microbiology, Santosh Medical College & Hospital, Ghaziabad, Uttar-Pradesh, India. The study was carried out for a period of two years i.e. from January 2019 to February 2021. Permission from Institutional Ethics Committee (IEC) was taken before carrying out the study (Reference No: SU/2021/092).3 Written informed consent was taken from all participants of the study.
Sample Collection and Processing
A total of 502 patient’s samples those were admitted in the ICU were enrolled in the study. Different clinical samples like ET aspirate, Blood, Pus, and Urine were collected with aseptic precaution in sterile universal container and were directly sent to the Microbiology laboratory as early as possible, samples were kept in refrigerator at 2-8°C temperature in case of inevitable situation. All Pseudomonas aeruginosa isolates were identified by conventional methods as per standard microbiology laboratory protocol and finally identified by observing the culture characteristic on routine laboratory culture media namely blood agar & MacConkey agar plates. Bacterial colonies showed Non-lactose fermenting pale colour colonies on MacConkey agar plates and were oxidase test positive and on nutrient agar pigmented and non-pigmented colonies with oxidase positive were found. Species level identification was performed with the help of manual biochemical test methods and finally pure isolates of Pseudomonas aeruginosa were used for further investigation. Standard operating procedure for the isolation and identification of bacteria were followed.11
Antimicrobial Susceptibility Testing (AST)
AST was performed for all clinical isolates by standard Kirby-Bauer disc diffusion method on Mueller Hinton agar (Hi-media labs, Mumbai, India). Pseudomonas aeruginosa control strain ATCC (American Type culture collection) 27853 were used during the study. Zone of inhibition was interpreted according to Clinical and Laboratory Standard Institute (CLSI) guidelines.12 MDR type is defined as P. aeruginosa. that are resistant to more than one antimicrobial agent in 3 or more antimicrobial categories.3
Phenotypic characterization of biofilm production
All bacterial isolates were tested by the following two methods for detection of biofilm formation (Tissue culture plate method and Tube adherence method):
Test tube method (Tube adherence method)
Tube method was performed according to Christensen et al. In this, loopful of bacterial culture (ATCC-27853) from overnight culture plate was inoculated into a test tube containing brain heart infusion broth (BHI) and incubated at 37°C for 24 hrs after incubation broth was discarded and cleaned/decanted with PBS (Phosphate buffer saline) pH (7.3) and dried at room temperature after drying tube were stained (treated) with 0.1% crystal violet for half an hour and then washed with water. Tubes were considered as positive if the side wall and bottom of the tube lined with visible film and reported as absent, Moderate or strong.13
Tissue culture plate method
TCP method was performed as described by Christensen et al.14 Bacterial colony from fresh culture plate of blood agar is inoculated into the glass test tube containing 2 ml of trypticase soya broth (TSB) with 1% glucose and incubated at 37°C for 24 hrs in stationary condition after incubation culture is diluted (1:100) with fresh TSB medium and 200μl from each tube is inoculated in to each wells of flat bottom polystyrene tissue culture plate without sealing the plate for proper aeration and in the similar way control organism were also inoculated in culture plate. TSB without bacterial cells is used as a negative control to check the sterility and non specific binding. After overnight incubation at 37°C content of each well was removed gently by tapping the plates on filter paper. The wells were washed with 200μl phosphate buffer saline (PBS) with pH 7.2 to remove the floating bacterial cells. After drying process fixation of wells was done by adding 2% sodium acetate for 5 minutes and stained with 1% crystal violet for 15 Minutes and then rinsed with deionized water. Plates were kept for drying at room temperature for 1 hour after which the optical densities was read with ELISA reader at 570 wave length optical density. Test was performed in triplicates and average of three OD values was taken. The OD values of each well were interpreted according to Table 1.
Table (1):
Specific growth rate of microalgae in normal medium and nitrogen enriched medium.
Mean OD value |
Adherence |
Biofilm formation |
|---|---|---|
<0.120 |
Weak |
Weak |
0.120-0.240 |
Moderate |
Moderate |
≥0.240 |
Strong |
High |
TCP: Tissue culture plate; OD: Optical densities.
Phenotypic detection of ESBL
Bacterial inoculum was prepared and lawn culture was made on MHA plates, after drying for 15 minutes disc of Ceftazidime and Ceftazidime+Calvulinic acid (disk potentiation test) were incorporated on MHA plates. After overnight incubation at 37°C plates were interpreted as ESBL positive if the zone size was ≥5mm for Ceftazidime+Calvulinic in comparison to zone size of Ceftazidime alone.15
Phenotypic detection of MBL
The IMP-EDTA combined disc test: Bacterial inoculum was prepared and lawn culture was made on MHA plates, after 15 minutes of drying the 2 Imipenem disc one with 10 µL of EDTA (750 µg) and the other disc without EDTA were placed on MHA culture plate 30mm apart and incubated overnight at 37°C, a ≥7mm increase in the zone size in IMP+EDTA disc was considered as MBL positive strain.16
Phenotypic detection of AmpC β-lactamase
Cefoxitin Cloxacillin Double Disc Synergy Test (CC-DDST)
The principle of this method is based on inhibitory effect of Cloxacillin on AmpC production. Bacterial inoculum was prepared and lawn culture was made on MHA plates, after 15 minutes of drying two antibiotic discs one of Cefoxitin (30ug) and other disc of Cefoxitin (30ug)/Cloxacillin (230ug) were placed on MHA culture plates 24mm apart with centers and incubated overnight at 37°C. A difference of ≥4mm in the inhibition zone of cefoxitin /cloxacillin and cefoxitin disc was considered as AmpC producers.17
Inhibitor-Based Method
Microbial inoculum of isolates were prepared in normal saline and turbidity was maintained with 0.5 McFarland standard and finally inoculated evenly on MHA plates then 2 Cefoxitin disc (30μg) with and without boronic acid (400 μg) were placed on dry MHA plate 30mm apart. After overnight incubation at 37°C aerobically a zone size of 5mm or more around the disc of Cefoxitin+boronic acid compared to the Cefoxitin disc alone was considered as AmpC positive isolates.18
All clinical samples received in microbiology laboratory were tested for the isolation, identification and Antibiotic sensitivity. A total of 115 isolates of P. aeruginosa were isolated from 502 clinical samples in the course of 2 year and all the clinical samples were non-duplicate. A total of 115 isolates of Pseudomonas aeruginosa was isolated from 502 clinical samples giving the prevalence rate of 23%. Out of 115 P. aeruginosa isolates 60 (52%) were MDR phenotypes, maximum number of MDR isolates were procure from endotracheal aspirates followed by urine, pus and blood among these samples ET aspirates & urine samples were considered for the majority of the positive isolates and it has been shown in Table 2. In this study qualitative and quantitative method was conducted for the detection of biofilm production. Out of 60 MDR isolates, 23( 38.3%) were ESBL producers, 22 (36.6%) were MBL producers, and 3 (5%) were AmpC producers.
Table (2):
Sample wise distribution of MDR Pseudomonas aeruginosa isolates.
Sample type |
MDR Pseudomonas aeruginosa (%) |
|---|---|
ET aspirate |
26(43) |
Pus |
12(20) |
Urine |
18(30) |
Blood |
2(3) |
BAL fluid |
2(3) |
Total |
60 |
Biofilm Production
In our study qualitative and quantitative method was conducted for the detection of biofilm production. Among 115 isolates of P. aeruginosa 109 (94.7%) isolates were producing biofilm and rest of 06 (5.2%) isolates were non biofilm producers. Out of total 60 MDR isolates 59 (98.3%) isolates were biofilm producers and 01 (1.6%) were biofilm non-producer. Among 60 MDR clinical isolates 59 (98.3%) were biofilm producers by tissue culture plate method and 53(88.3%) were biofilm producers by test tube method. Table 3 shows the comparison of Tube method and Tissue culture plate method results for biofilm production. Table 6 shows the comparison of results of biofilm detection by different methods documented in previous studies.
Table (3):
Comparison of Tube method and Tissue culture plate method results.
| Number of Isolates | Biofilm Formation | TCP n (%) | Tube method n (%) |
|---|---|---|---|
| N=60 | Strong | 44(73.3%) | 37(61.6%) |
| Moderate | 15(25%) | 16(13.9%) | |
| Weak/None | 01(1.6%) | 7(11.6%) |
Demographic distribution
Out of 60 MDR phenotypes, 41 were isolated from male patients and 19 were isolated from female patients and maximum numbers of cases were from the population between 31-50 years age group.
Drug resistance pattern of MDR P. aeruginosa
In our results, the highest resistance for MDRPA was found to be for Gentamicin followed by Ceftazidime, Cefepime, Ciprofloxacin, Amikacin, Piperacillin, Ticarcillin/Calvulinic acid Pipiracillin –Tazobactam and least resistance was found to be for Meropenem and Imipenem. Table 4 & 5 shows the resistance pattern of MDR Pseudomonas aeruginosa against various anti pseudomonal drugs and antibiotic resistance pattern of Pseudomonas aeruginosa in relation to biofilm production.
Table (4):
The resistance pattern of MDR Pseudomonas aeruginosa against various anti pseudomonal drugs.
Drug (Potency in microgram) |
MDR P. aeruginosa N (%) |
|---|---|
Colistin (10mcg) |
Nil |
Amikacin (30mcg) |
46(76%) |
Piperacillin -Tazobactam (100mcg/10mcg) |
23(38%) |
Piperacillin (100mcg) |
36(60%) |
Gentamicin (10mcg) |
51(85%) |
Meropenem (10mcg) |
10(16%) |
Imipenem (10mcg) |
11(18%) |
Ciprofloxacin (5mcg) |
48(80%) |
Ticarcillin/Calvulinic acid (75mcg/10mcg) |
29(48%) |
Azetronam (30mcg) |
46(76%) |
Cefepime (30mcg) |
48(80%) |
Ceftazidime (30mcg) |
52(86%) |
Polymyxin B (300 Units) |
Nil |
Table (5):
Antibiotic Resistance pattern of Pseudomonas aeruginosa in relation to biofilm production.
Drug (Potency in microgram) |
Biofilm Producers N (%) |
Non-Biofilm producer N (%) |
|---|---|---|
Colistin (10mcg) |
Nil |
Nil |
Amikacin (30mcg) |
30(65.2%) |
16(34.7%) |
Piperacillin -Tazobactam (100mcg/10mcg) |
19(82.6%) |
4 (17.3%) |
Piperacillin (100mcg) |
29(80.5%) |
7(19.4%) |
Gentamicin (10mcg) |
32(62.7%) |
19(37.2%) |
Meropenem (10mcg) |
8(80%) |
2(20%) |
Imipenem (10mcg) |
8(72.7%) |
3(27.2%) |
Ciprofloxacin (5mcg) |
30(62.5%) |
18(37.5%) |
Ticarcillin/Calvulinic acid (75mcg/10mcg) |
22(75.8%) |
7(24.1%) |
Azetronam (30mcg) |
28(60.8%) |
18(39.1%) |
Cefepime (30mcg) |
18(37.5%) |
30(62.5%) |
Ceftazidime (30mcg) |
23(44.2%) |
29(55.7%) |
Polymyxin B (300 Units) |
Nil |
Nil |
Table (6):
Comparison of results of biofilm detection by different methods documented in previous studies.
| No | Study | TCP method | TM method | |||||
|---|---|---|---|---|---|---|---|---|
| Strong | Moderate | Weak | Strong | Moderate | Weak | |||
| 1. | Mathur et al.19 | 14.4% | 39.4% | 46.7% | 11.8% | 29.6% | 58.5% | |
| 2. | Nagaveni et al.20 | 41.6% | 33.4% | 25% | 50% | 25% | 25% | |
| 3. | Vishnuprasad et al.21 | 52.4% | 14.2% | 33.3% | 52.4% | 33.3% | 14.2% | |
| 4. | Hassan et al.22 | 22.7% | 41% | 36.3% | 19% | 30% | 51% | |
| 5. | Panda et al.23 | 11.0% | 34.7% | 54.3% | 10.7% | 29% | 60.3% | |
| 6. | Ram et al.24 | 14.04% | 50.88% | 35.09% | 12.28% | 50.88% | 36.84% | |
| 7. | Triveda et al.25 | 21.8% | 17.1% | 16.4% | 10.9% | 10.9% | 7.8% | |
| 8. | Present Study | 68.6% | 26% | 5.2% | 65.3% | 22% | 12.5% | |
Correlation between biofilm and multi drug resistance profile
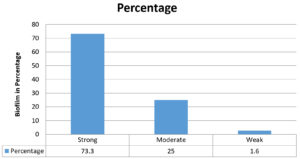
In our study we have performed qualitative and quantitative methods for biofilm detection. Among them, tissue culture method (Quantitative) is more sensitive and reliable as shown in Table 3/Fig. 1. According to tissue culture plate method out of 60 MDR P. aeruginosa isolates 44 (73.3%) isolates were strong biofilm producers and 15 (25%) were moderate biofilm producers and 01 (1.6%) were weak biofilm producers. As per test tube method out of 60 MDR P. aeruginosa isolates 37 (61.6%) isolates were strong biofilm producers and 16 (13.9%) were moderate biofilm producers and 07 (11.6%) were weak biofilm producers.
MDR P. aeruginosa is an emerging pathogen and has become a serious threat in recent years and the treatment of these resistant phenotypes is very challenging task for the clinicians.26 The prevalence rate of Pseudomonas aeruginosa in our study was 23% which was higher to 14.7% rate reported by Gill J.S. et al.3 However, Gupta R. et al.9 obtained the prevalence rate of 28%, while lower prevalence rate of 2.76% was reported by Senthamarai S. et al.27 in Tamilnadu. Prevalence rate of MDR Pseudomonas aeruginosa in our study was 52% however, Gill J.S. obtained the prevalence rate of 50% in their study. Saderi H. et al.28 reported a prevalence rate of 54.5% for MDR Pseudomonas aeruginosa in Iran while Mirzaei B. et al.29 in Tehran found the prevalence rate of 16.5% for MDR Pseudomonas aeruginosa. In the current study MDR Pseudomonas aeruginosa phenotypes were most commonly procured from lower respiratory tract followed by urine, pus and blood samples. Similar results were also reported by Gupta R. et al.9 However, Gill J.S. et al.3 reported urine and wound samples considered for the majority of the positive isolates. Our results were also agreed with Prakash V. et al.30 Among 115 isolates of P. aeruginosa 109 (94.7%) isolates were producing biofilm and rest of 06 (5.2%) isolates were non biofilm producers In the present study among 115 isolates of P. aeruginosa, 60 MDR isolates were reported among them 59 (98.3%) were biofilm producers and 01 (1.6%) were Non biofilm producers. These results were comparable to that of Gupta R. et al. However higher percentage of (85.72%) biofilm producers were also reported by Bankole et al.31 While lower results (29.1%) & (26.83%) were reported by Saha S. et al.32 and Kulkarni D.M et al.33 respectively. In our study among 59 MDR P. aeruginosa isolates maximum biofilm producing isolates were reported from blood samples, ET aspirates, BAL fluid and Urine samples, the similar results were also reported by Kulkarni D.M. et al. Comparable results were reported by Shrestha et al.34 and Gurung J. et al.35 In our study we found that male patients (68%) were predominant than female patients (32%) in case of MDRPA, these results were in agreement with Mirzaei et al.29 In our results, the highest resistance for MDRPA was found to be for Gentamicin followed by Ceftazidime, Cefepime, Ciprofloxacin, Amikacin, Piperacillin, Ticarcillin/Calvulinic acid Piperacillin –Tazobactam and least resistance was found to be for Meropenem and Imipenem, similar results were also obtained from study conducted by Biswal I et al.35 in burn patients. For MDRPA isolates, the drug of choice is carbapenems but increasing resistance towards Carbapenems is now a serious threat. In our study the resistance pattern for Imipenem & Meropenem was lowest as 18% and 16% respectively. The drug resistance patterns showed in the present study revealed that >50% isolates were resistant to fluoroquinolones, gentamicin, cephalosporin’s and aminoglycosides. The treatment and management options for such type of bacterial strains are limited which may result in treatment failures and thereby causing significant morbidity and mortality. The good efficacy of the carbapenems as it is an effective antibiotic in the management of nosocomial infections and it is found to be the precious weapon against MDRPA infections. In the current study, MDRPA isolates showed the lowest resistance to carbapenems, whereas Piperacillin alone showed a resistance rate of 60% whereas beta-lactam/beta-lactamase inhibitor Piperacillin/Tazobactam showed a lower resistance of 38% only, indicating that beta-lactamase inhibitor markedly increases the spectrum of activity of betalactams, which makes the combination drug the preferred choice against P. aeruginosa infections. In our study we reported the resistance pattern of biofilm producing clinical isolates of P. aeruginosa with maximum resistance against Carbapenems, Piperacillin-Tazobactam, Ticarcillin-Calvulinic acid, Monobactam group and Amikacin. Our results were in agreement with Kulkarni and Saha S. et al.32,33 respectively. In our study we found that the overall prevalence of ESBL, MBL and AmpC was 23 (38.3%), 22 (36.6%) and 3 (5%) respectively our results were agreed with Shrivastava G. et al.36 However, Sarkar S. et al.37 Production of multiple β-lactamases by
P. aeruginosa is therapeutic challenge and there is a need for urgent jurisdiction to control the spread of such type of resistant strains. Management and treatment of infections caused by Pseudomonas spp. is less complicated than drug resistant ones. The problem of bacterial drug resistance to commonly used antibiotics is very frequent globally as drug resistance is a greater problem in developing countries especially due to easy availability of antibiotics over the counter. To the best of our knowledge, this is the first study that includes information regarding drug resistance pattern of multi drug resistant biofilm producing clinical isolates of Pseudomonas aeruginosa among ICU patients. Also, the beta lactamase profiling has been included in this study which is a very crucial factor to be detected early for the better treatment of such infections. Tissue culture plate method is more sensitive and specific test as compared to Tube method as shown in Table 6.
There is a need for urgent jurisdiction to control the spread of such type of resistant strains and strict antibiotic policies and regular surveillance programme of antimicrobial resistance should be tailored to fend off the emergence of drug resistant Pseudomonas aeruginosa. Colistin & Polymyxin B still shows high sensitivity against multi drug resistant Pseudomonas aeruginosa phenotypes. Early detection of beta lactamases and detection of biofilm production should be performed regularly for all clinical isolates of Pseudomonas aeruginosa to guide antibiotic selection and for the better management of severe infection in ICU patients.
ACKNOWLEDGMENTS
The authors would like to thank Department of Microbiology, Santosh Medical College & Hospital, Ghaziabad, India for their support in carrying out the study.
CONFLICT OF INTEREST
The authors declare that there is no conflict of interest.
AUTHORS’ CONTRIBUTION
All authors listed have made a substantial, direct and intellectual contribution to the work, and approved it for publication.
FUNDING
None.
ETHICS STATEMENT
This study was approved by the Institutional Ethics Committee, Santosh Deemed to be University, India with reference No: SU/2021/092[3].
AVAILABILITY OF DATA
All datasets were analysed during this study are included in the manuscript.
- Abdulhaq N, Nawaz Z, Zahoor MA, Siddique AB. Association of biofilm formation with multi drug resistance in clinical isolates of Pseudomonas aeruginosa. EXCLI J. 2020;19:201-208.
Crossref - Hirsch EB, Tam VH. Impact of multidrug-resistant Pseudomonas aeruginosa infection on patient outcomes. Expert Rev Pharmacoecon Outcomes Res. 2010;10(4):441-451.
Crossref - Gill JS, Arora S, Khanna SP, Kumar KVSH. Prevalence of Multidrug-resistant, Extensively Drug-resistant, and Pan drug-resistant Pseudomonas aeruginosa from a Tertiary Level Intensive Care Unit. J Glob Infect Dis. 2016;8(4):155-159.
Crossref - Pagani L, Mantengoli E, Migliavacca R, et al. Multifocal detection of multidrug-resistant Pseudomonas aeruginosa producing the PER-1 extended-spectrum β-lactamase in northern Italy. J Clin Microbiol. 2004;42(6):2523-2529.
Crossref - Chaudhary BL, Bisht D, Faujdar SS. Biofilm Formation and its Association with Antibiotic Susceptibility Pattern inMethicillin-resistant Staphylococcus aureus Isolates. J Pure Appl Microbiol. 2021;15(4):2041-2049.
Crossref - Kamali E, Jamali A, Ardebili A, Ezadi F, Mohebbi A. Evaluation of antimicrobial resistance, biofilm forming potential, and the presence of biofilm-related genes among clinical isolates of Pseudomonas aeruginosa. BMC Res Notes. 2020;13(1):27.
Crossref - Anwar H, Costerton JW. Enhanced activity of combination oftobramycin and piperacillin for eradication of sessile biofilmcells of Pseudomonas aeruginosa. Antimicrob Agents Chemother. 1990;34(9):1666-1671.
Crossref - Moriarty TF, Elborn JS, Tunney MM. Effect of pH on the antimicrobial susceptibility of planktonic and biofilm-grown clinical Pseudomonas aeruginosa isolates. Br J Biomed Sci. 2007;64(3):101-104.
Crossref - Faujdar SS, Bisht D, Sharma A. Antibacterial potential of neem (Azadirachta indica) against uropathogens producing beta-lactamase enzymes: A clue to future antibacterial agent? Biomed Biotechnol Res J. 2020;4(3):232-238.
Crossref - Obritsch MD, Fish DN, MacLaren R, Jung R. National surveillance of antimicrobial resistance in Pseudomonas aeruginosa isolates obtained from intensive care unit patients from 1993 to 2002. Antimicrob Agents Chemother. 2004;48(12):4606-4610.
Crossref - Collee, J G, T J. Mackie, and J E. McCartney. Mackie & Mccartney Practical Medical Microbiology. New York: Churchill Livingstone, 1996. Print.14th edition.
- Clinical and Laboratory Standards Institute. Performance standards for antimicrobial susceptibility testing, 30th ed. CLSI supplement M100. Clinical and Laboratory Standards Institute, Wayne, PA. 2020.
- Christensen GD, Simpson WA, Bisno AL, Beachey EH. Adherence of slime-producing strains of Staphylococcus epidermidis to smooth surfaces. Infect Immun. 1982;37(1):318-326.
Crossref - Christensen GD, Simpson WA, Younger JA, et al. Adherence of cogulase negative Staphylococi to plastic tissue cultures:a quantitative model for the adherence of staphylococci to medical devices. J Clin Microbiol. 1985;22(6):996-1006.
Crossref - Rawat D, Nair D. Extended-spectrum β-lactamases in Gram Negative Bacteria. J Glob Infect Dis. 2010;2(3):263-74.
Crossref - Faujdar SS, Bisht D, Sharma A. Antibacterial activity of Syzygium aromaticum (clove) against uropathogens producing ESBL, MBL, and AmpC beta-lactamase: Are we close to getting a new antibacterial agent? J Family Med Prim Care. 2020;9(1):180-186.
Crossref - Polsfuss S, Bloemberg GV, Giger J, Meyer V, Bottger EC, Hombach M. Practical approach for reliable detection of AmpC beta-lactamase-producing Enterobacteriaceae. J Clin Microbiol. 2011;49(8):2798-2803.
Crossref - Thomson KS. Controversies about extended-spectrum and AmpC beta-lactamases. Emerg Infect Dis. 2001;7(2):333-336.
Crossref - Mathur T, Singhal S, Khan S, Upadhyay DJ, Fatma T, Rattan A. Detection of biofilm formation among the clinical isolates of Staphylococci: An evaluation of three different screening methods. Indian J Med Microbiol. 2006;24(1):25-29.
Crossref - Nagaveni S, Rajeshwari H, Oli AK, Patil SA, Chandrakanth RK. Evaluation of biofilm forming ability of the multidrug resistant Pseudomonas aeruginosa. Bioscan. 2010;5:563-566.
- Prasad SV, Ballal M, Shivananda PG. Slime production a virulence marker in Pseudomonas aeruginosa strains isolated from clinical and environmental specimens: A comparative study of two methods. Indian J Pathol Microbiol. 2009;52(2):191-193.
Crossref - Hassan A, Usman J, Kaleem F, Omair M, Khalid A, Iqbal M. Evaluation of different detection methods of biofilm formation in the clinical isolates. Braz J Infect Dis. 2011;15(4):305.
Crossref - Panda PS, Chaudhary U, Dube SK. Comparison of four different methods for detection of biofilm formation by uropathogens. Indian J Pathol Microbiol. 2016;59(2):177-179.
Crossref - Ram Mohan Reddy, K. Tube Adherence Test as a Screening Tool for Detection of Biofilm Formation among Staphylococcus aureus. Int J Curr Microbiol App Sci. 2017;6(8):1325-1329.
Crossref - Triveda L, Gomathi S. Detection of biofilm formation among the clinical isolates of Enterococci: An evaluation of three different screening methods. Int J Curr Microbiol App Sci. 2016;5(3):643-650.
Crossref - Pfeifer Y, Cullik A, Witte W. Resistance to cephalosporins and carbapenems in Gram-negative bacterial pathogens. Int J Med Microbiol. 2010;300(6):371-379.
Crossref - Senthamarai S, Reddy ASK, Sivasankari S, et al. Resistance Pattern of Pseudomonas aeruginosa in a Tertiary Care Hospital of Kanchipuram, Tamilnadu, India. J Clin Diagn Res. 2014;8(5):30-32.
Crossref - Saderi H, Owlia P. Detection of Multidrug Resistant (MDR) and Extremely Drug Resistant (XDR) P. Aeruginosa Isolated from Patients in Tehran, Iran. Iran J Pathol. 2015;10(4):265-271. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC4539747/
- Mirzaei B, Bazgir ZN, Goli HR, Iranpour F, Mohammadi F, Babaei R. Prevalence of multi-drug resistant (MDR) and extensively drug-resistant (XDR) phenotypes of Pseudomonas aeruginosa and Acinetobacter baumannii isolated in clinical samples from Northeast of Iran. BMC Res Notes. 2020;13(1):380.
Crossref - Prakash V, Mishra PP, Premi HK, Walia A, Dhawan S, Kumar A. Increasing incidence of multidrug resistant Pseudomonas aeruginosa in inpatients of a tertiary care hospital. Int J Res Med Sci. 2014;2(4):1302-1306.
Crossref - Bankole OO, Vasanthi R, et al. Molecular methods for detecting biofilm producing multidrug resistant clinical isolates. Ann Trop Med & Public Health. 2020;23(S13A):SP231319.
Crossref - Saha S, Devi KM, Damrolien S, Devi KS, Krossnunpuii, Sharma KT. Biofilm production and its correlation with antibiotic resistance pattern among clinical isolates of Pseudomonas aeruginosa in a tertiary care hospital in north-east India. Int J Adv Med. 2018;5(4):964-968.
Crossref - Kulkarni DM, Nilekar SL, Vidhya T. Association of biofilm production in ESBL and MBL producing clinical isolates of Pseudomonas aeruginosa. Trop J Pathol Microbiol. 2020;6(2):174-180.
Crossref - Shrestha R, Nayak N, Bhatta DR, Hamal D, Subramanya SH, Gokhale S. Drug Resistance and Biofilm Production among Pseudomonas aeruginosa Clinical Isolates in a Tertiary Care Hospital of Nepal. Nepal Medical College Journal. 2019;21(2):110-116.
Crossref - Biswal I, Arora BS, Kasana D, Neetushree. Incidence of multidrug resistant pseudomonas aeruginosa isolated from burn patients and environment of teaching institution. J Clin Diagn Res. 2014;8(5):26-29.
Crossref - Shrivastava G, Bhatambare GS, Patel KB. Evaluation of prevalence and antibiogram of multi drug resistant, extensively drug resistant and pan drug resistant Pseudomonas aeruginosa in patients visiting a tertiary care hospital in central India. CHRISMED J Health Res. 2014;1(3):145-149.
Crossref - Sarkar S, Dutta S, Namhata A, Banerjee C, Sengupta M, Sengupta M. Beta-lactamase Profile and Biofilm Production of Pseudomonas aeruginosa isolated from a Tertiary Care Hospital in Kolkata, India. J Clin Diagn Res. 2020;14(10):22-27.
Crossref
© The Author(s) 2022. Open Access. This article is distributed under the terms of the Creative Commons Attribution 4.0 International License which permits unrestricted use, sharing, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.