ISSN: 0973-7510
E-ISSN: 2581-690X
Growth promoting Rhizobium spp are frequently used as biofertilizers for agricultural cropping system. Furthermore, Isolation, screening and biochemical characterization of Rhizobium for a specific plant is necessary to examine ability of isolated bacteria to affect the growth and development of host plant in various ways. The current study was aimed to isolate plant specific rhizobacterial strains which are compatible with lentil (Lens culinaris Medik.L.) plant. 20 bacterial isolates have been isolated from root nodules of lentil from various agro ecological area and their biochemical characterization was performed by different plant growth promotion activities. The result showed that, among 20 isolates, four isolates have vigorous plant growth promoting activities. Four bacterial strains were able to solubilise phosphorous along with hormone production. Moreover, among four bacterial strains, two strongly produced HCN and siderophore in vitro. Subsequently, all selected bacterial isolates were inoculated in lentil seeds of variety HUL57 to study germination percentage and vigour index of the crop. Out of four isolates 26N isolate performed best growth promotion activities on lentil seedlings. Finally, on the basis of performance of bacteria on plant, four isolates were characterized using molecular approach of species identification such as 16S rRNA sequencing.
Lentil, Rhizobium spp, PGPR, 16SrRNA Sequencing, Seed Germination, Vigour Index
In rhizosphere several beneficial micro-organisms viz, bacteria fungi actinomycetes etc. are found which promote the plant growth naturally. In natural rhizospheric zone various growth promoting microbes having drastic growth promotion activities and potential biocontrol ability have been found.1 Moreover, excessive use of chemical fertilizers in agriculture soil enhanced soil salinity, heavy metal accumulation and water eutrophication.2 Judicious use of chemical fertilizer not only affects the physiochemical properties of soil but also adversely affects rhizospheric microbial population. For obtaining high crop yield use of sodium and potassium containing chemical fertilizers make a negative impact on soil pH, soil structure, deterioration and the increasing feature of acid.3 Direct plant growth promotion activity including mineral solubilization (P, K and Zn) phytohormone production (IAA, GA3 and cytokinins) while indirect mechanism including hydrogen cyanide (HCN), siderophore, antibiotic production along with induced systemic resistance activities.4 Rhizobacteria are reported to produce diverse range of bioactive chemicals like organic volatile compounds, antibiotics and several defensive enzymes which promote growth of plant and provide protection against many phytopathogens including Rhizoctonia spp, Aspergillus spp.5,6
Particularly in dicots, IAA specifically induces root formation, root elongation and enhances surface volume of root thereby helps in easily uptake of nutrients from rhizospheric region.7 Legumes and the rhizosphere provide most of the nutritional requirements of nodule bacteria and enhances the Rhizobium population several folds during plant growth. Rhizobium–legume associations are usually host specific, and a given rhizobial strain can infect only a limited number of hosts.8 Rhizobia are Gram-negative bacteria able to establish symbiotic relationships with legume species by eliciting root nodules. Pisum sativum and legumes such as red pea (Lathyrus cicera), faba beans (Vicia faba) and lentil (Lens culinaris), can be nodulated by several species of the genus Rhizobium, such as Rhizobium leguminosarum, R. pisi, R. fabae, R. laguerreae, R. bangladeshense, R. lentis, R. binae and R. anhuiense.9
Extracellular enzymatic activity of PGPR’s including cellulase, amylase and chitinase protect crop from various biotic stresses furthermore, catalase and oxidase enzymes protect crop from drought, salinity, heavy metal etc.10,11
Lentil (Lens culinaris Medik) is an important and popular legume mainly in Central and Southwest Asia, Southern Europe, North Africa and Ethiopian countries,12 due to its grain human consumption, phytochemical content, adaptation to arid and semi-arid climate and ability to fix atmospheric nitrogen through a symbiosis with rhizobial bacteria.13 Lentil seeds represent a low-cost source of protein and starch, with the advantage of being resistant to starch when compared to cereal, root and tuber starch.14
In the view of this experiment current research was carried out to investigate the effects of potent plant growth promoting bacterial isolates on lentil plant growth promotion and isolated bacteria exhibited higher germination % and vigour index without application of chemical fertilizers which proves them to be used as bioinoculant for accelerated growth and development of lentil as well as other crops.
Field site and sample collection
Root nodules along with lentil plant were collected from different agroclimatic zones of lentil growing field including districts of Chhattisgarh, Uttar pradesh and samples were kept in sterilized plastic bags at 4°C for further use. Samples were collected from 45 days old lentil plants and nodules were brought to the laboratory immediately and washed with tap water to remove adhering soil particles and to ensure root system free from soil. Root nodules were surface sterilized with 70% ethanol for 1 minute followed by sterilizing with 0.2% HgCl2 for 30 seconds. Nodules were washed subsequently in sterile water in order to remove excess of chemical residues, finally YEMA (Yeast extract mannitol agar) nutrient media was used for streaking of bacteria and isolation of single colony. Pure culture was obtained by repeated streaking of isolated single colonies on YEMA plates. Pure culture was stored in YEMA slants until further use at 4°C in refrigerator.
Biochemical characterization
Phosphate solubilization
Qualitative assay of phosphate solubilizing activity of bacteria was measured on Pikovskaya’s Agar Medium containing; yeast extract 0.5 g, dextrose 10 g, Ca3(PO4)2 5 g, (NH4)2SO4 0.5 g, KCl 0.2 g, MgSO4 0.1 g, MnSO4 0.1mg, FeSO4 0.1 mg, and agar 15 g in 1000ml of distilled water. Clear transparent zone (halo zones) around the bacterial colony made confirmation of phosphate solubilization activity within 24 to 72 h.15
Auxin production
Auxin synthesis by bacteria was estimated using the Salkowski reagent (12 g of FeCl3/ L in 7.9 M H2SO4). Yeast extract mannitol broth containing tryptophan inoculated with bacterial broth was incubated at 30°C for 48 h in BOD incubator. 1ml bacterial broth of each culture was centrifuged at 10,000 rpm for 10 min and the supernatant was collected and then 2 ml Salkowski reagent was added, followed by incubation for 1 h at dark room. Appearance of pink colour indicated IAA production. For the quantification of IAA, absorbance was taken at 530 nm by using UV/visible spectrophotometer.16
Standard curve of IAA
Standard curve was made by using IAA solution in 0-100 μg/ml concentration. After making volume to 1ml using distilled water followed by adding Salkowski’s reagent (2 ml), total volume was made to 4 ml and incubated for 25 minutes at room temperature. Standard curve was plotted with the different readings obtained by taking absorbance at 530 nm.
The production of IAA was calculated by equation (y = mx + c) in µg /ml.
Where
Y = O.D. of Rhizobium and PGPR culture.
m = O.D. of blank solution
x = amount of IAA produced by Rhizobium and PGPR strains
c = Zero (constant)
Hydrogen cyanide production
HCN production was evaluated for bacterial isolates on YEM (Yeast extract mannitol) broth containing 4.4g with glycine. Whatman filter paper No.1 was soaked with picric acid (0.05%) solution in 2% sodium carbonate and placed in the test tube sealed with parafilm and incubated at 30°C for 48-72 h. A colour change of the filter paper from deep yellow to reddish-brown colour was considered as an indication of HCN production.17
Siderophore production
Siderophore production was determined following protocol of Schwyn and Neilands.18 1 μl of bacterial culture raised overnight in Luria broth was spotted on Chrome Azurol S agar plates and incubated at 30±2°C for 48 h to 72h. The plates were observed for the development of an orange halo colour zone around the bacterial colony. Qualitative assay was further confirmed by CAS broth. The assay was carried out by mixing the culture supernatant (0.5 ml) with 0.5 ml CAS reagent and the absorbance was measured at 630 nm against a reference consisting of uninoculated liquid medium.19
Extracellular enzymatic activities
Isolated rhizobacteria was identified primarily by microscopic observation viz, gram reaction, colony morphology, pigmentation, mobility and cell shape20 followed by several biochemical and enzymatic test viz, catalase,21 amylase,22 cellulase,23 chitinase24 and oxidase25 test was done by the reference protocol.
DNA isolation and 16S rRNA Sequencing
For molecular characterization bacterial genomic DNA was isolated and subjected to PCR amplification. Finally, amplified DNA of bacteria sent to Anuvanshiki (OPC) PVT LTD New Delhi, for 16S r RNA sequencing by using universal forward 27F 5’-AGAGTTTGATCCTGGCTCAG-3’ and reverse primer 1492R 5’- GGTTACCTTGTTACGACTT-3’ under standard condition.
Pot experiment
All the four promising rhizospheric bacterial isolates were inoculated with Lentil seeds by seed biopriming for observing plant growth promoting traits. Seeds of lentil were surface sterilized by 0.2% HgCl2 for 2 min followed by rinsing in sterile double distilled water for 10 min. Seeds were further soaked in 10 ml of Yeast extract mannitol (YEM) broth inoculated with bacteria containing 109 CFU/ml and kept in rotatory shaker at 30°C for overnight. Further bioprimed seeds were sown in pots containing sterilized soil. Each pot had a soil composition of 40% sand, 28% silt & 32% clay. Round pots used were having dimensions of length 3.5 inch and width 3 inch. Subsequently, 10 plants per pot were maintained at 60% water holding capacity and placed in a temperature-controlled plant growth chamber with 12h light/12h dark at constant temperature of 25°C with 60% relative humidity of the air. At the end of experiment, plants were uprooted and seed germination and vigour index were calculated by towel method.26
Data Analysis
Data were recorded in triplicates for biological and experimental replicates. The statistical analysis was performed using SPSS 16.00. Means were compared using the least significant difference (LSD) test of analysis of variance (ANOVA) at p = 0.05. The results are reported as mean ± standard deviation.
Isolation and morphological characterization of bacterial isolates
Four selected growth promoting bacteria viz Rhizobium leguminosarum (26N), Rhizobium spp (LN3), Rhizobium spp (LCG6), Rhizobium pusense (LCG5) were isolated from root nodules of lentil (Table 1) and their morphological evaluation was confirmed by variation in colony appearance, margin, elevation, shape, size, colour, pigmentation and colony surface (Table 2).
Table (1):
Isolation of bacteria from rhizospheric soil from different locations.
No. |
Strain |
Location |
Rhizospheric soil associated with Crops |
|---|---|---|---|
1 |
26N |
District Dhamtari, Chhattisgarh ( 20.6118° N, 81.7787° E) |
Lentil (lens culinaris Medik.L.) |
2 |
LN3 |
District Balod, Chhattisgarh (20.7750° N, 81.2519° E) |
Lentil (lens culinaris Medik.L.) |
3 |
LCG6 |
District Rajnandgaon, Chhattisgarh (21.1346° N, 80.8987° E) |
Lentil (lens culinaris Medik.L.) |
4 |
LCG5 |
District Durg, Chhattisgarh (21.1623° N, 81.4279° E) |
Lentil (lens culinaris Medik.L.) |
Molecular Identification
Rhizobacterial strains were confirmed by 16S rRNA sequencing. Nucleotide sequences of 26N obtained through 16S rRNA sequencing were subjected for homology search using nucleotide BLAST homology search tool at NCBI. It showed maximum sequence similarity of 100% with Rhizobium leguminosarum bv viciae. Further sequences of this species were deposited in Genbank with accession number ON159934. Isolate LN3 was deposited with accession number OL873220 which showed maximum sequence similarity of 100% with Rhizobium spp., LCG6 was submitted with the accession number OL884354 which showed 100% similarity with Rhizobium spp and LCG5 showed 100 % sequence similarity with Rhizobium pusense and submitted with the accession number of OL873321 (Table 2 & 3).
Table (2):
Morphological characterization of selected strains.
S. No. |
Bacteria |
Strains |
Gram stain |
Motility |
Colony shape |
Colour |
Surface |
Margin |
Accession number |
|---|---|---|---|---|---|---|---|---|---|
1 |
Rhizobium leguminosarum |
26N |
-ve |
Motile |
Convex |
Pale pink |
Mucilaginous |
Entire |
ON159934 |
2 |
Rhizobium spp |
LN3 |
-ve |
Motile |
Concave |
Whitish to pale pink |
Smooth |
Entire |
OL873220 |
3 |
Rhizobium spp |
LCG6 |
– ve |
Motile |
Irregular |
Creamy white |
Mucilaginous |
Entire |
OL884354 |
4 |
Rhizobium pusense |
LCG5 |
– ve |
Motile |
Irregular |
white |
Mucilaginous |
Entire |
OL873321 |
Table (3):
Molecular characterization of growth promoting bacteria.
No. |
PGPR’s |
Strains |
Sequence Similarities |
Accession number |
|---|---|---|---|---|
1 |
Rhizobium leguminosarum |
26N |
100% |
ON159934 |
2 |
Rhizobium spp |
LN3 |
100% |
OL873220 |
3 |
Rhizobium spp |
LCG6 |
100% |
OL884354 |
4 |
Rhizobium pusense |
LCG5 |
100% |
OL873321 |
Table (4):
Biochemical evaluation of selected growth promoting bacteria.
No. |
PGPR |
Phosphate solubilization (cm) |
Indole acetic acid (µg/ml) |
Ammonia Production |
Hydrogen cyanide production |
Siderophore production |
|---|---|---|---|---|---|---|
1 |
LUP2 |
2.1±0.05 |
4.76±0.07 |
+ |
++ |
+ |
2 |
UP4N |
0.00±0 |
71.96±1.6 |
– |
– |
– |
3 |
LN2 |
2.54±0.03 |
5.06±0.07 |
– |
– |
– |
4 |
19N |
3.7±0.04 |
53.23±0.5 |
+ |
++ |
++ |
5 |
20NII |
1.61±0.01 |
59.14±2 |
– |
– |
– |
6 |
LUP4 |
0.00±0 |
59.45±3.2 |
++ |
– |
– |
7 |
18N |
0.00±0 |
51±2.1 |
– |
– |
++ |
8 |
1N |
0.00±0 |
4.45±0.07 |
– |
– |
– |
9 |
UP6N |
1.55±0.02 |
71.60±0.7 |
– |
– |
– |
10 |
LN4Y |
0.00±0 |
71.81±0.5 |
– |
– |
– |
11 |
LN3 |
3.12±0.05 |
96.63±4.5 |
+++ |
+++ |
+++ |
12 |
LN4P |
1.4±0.01 |
63.77±2.9 |
– |
– |
+ |
13 |
LCG6 |
3.12±0.05 |
92.41±5 |
++ |
++ |
+++ |
14 |
26N |
3.24±0.03 |
102.34±5 |
+++ |
+++ |
+++ |
15 |
LCG5 |
3.7±0.03 |
83.91±5 |
– |
+ |
+++ |
16 |
UP8N |
1.56±0.06 |
67.38±4.6 |
– |
– |
– |
17 |
27N |
2.1±0.02 |
64.29±2.5 |
++ |
+ |
++ |
18 |
UP2N |
0.00±0 |
3.83±2.5 |
+ |
+ |
+ |
19 |
LCG2 |
1.67±0.07 |
70.11±1.8 |
– |
– |
|
20 |
LNBHU |
0.9±0.02 |
54.66±0.9 |
– |
– |
– |
Biochemical characterization
Phosphorus solubilization
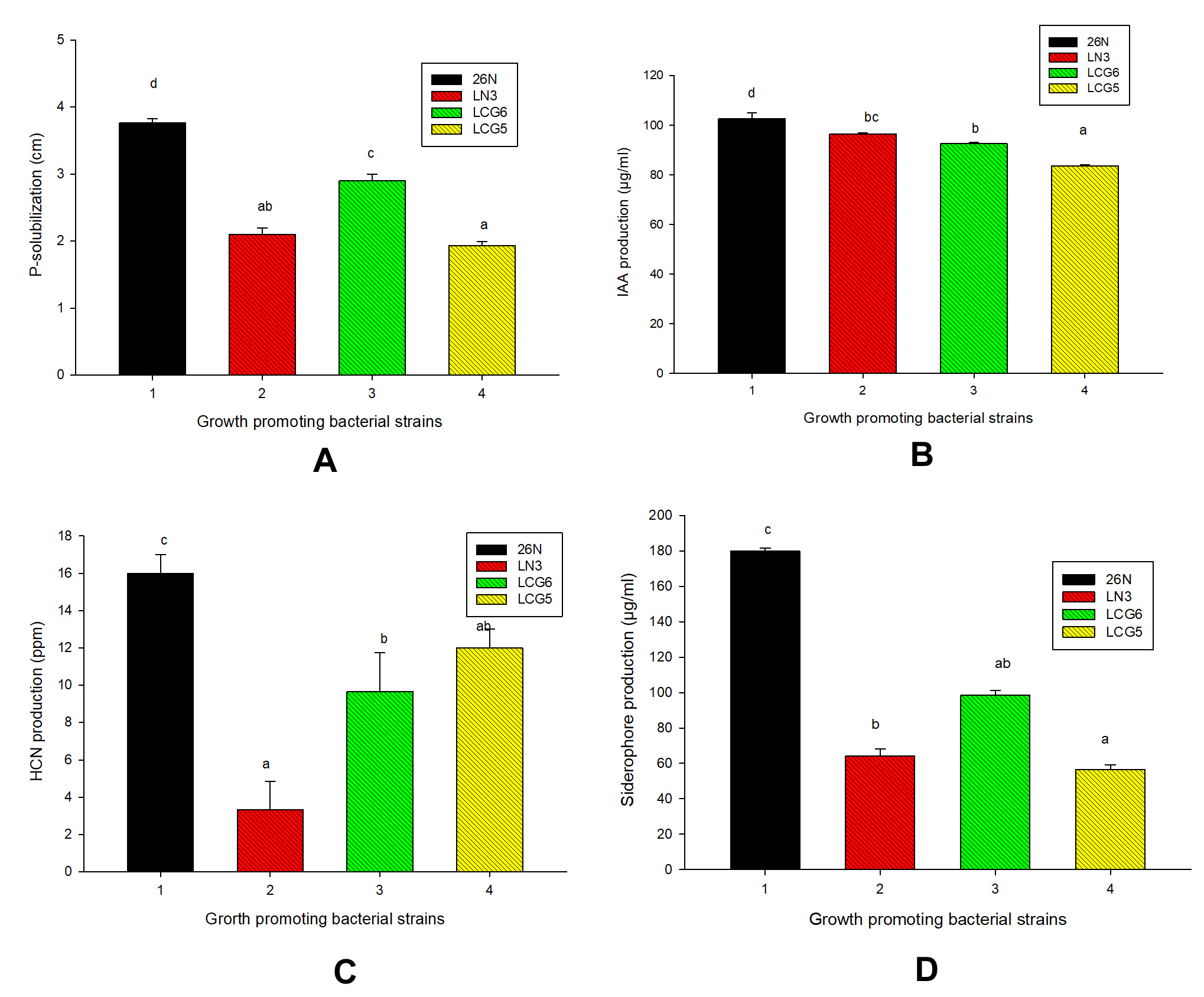
Among 20 bacterial isolates 75% isolates have ability to solubilize inorganic phosphorous in vitro (Table 4) and moreover, 4 bacterial strains (26N, LN3, LCG6 and LCG5) have greater potential to solubilize phosphorous. Among four bacterial isolates highest phosphorous was solubilized by 26N strain (3.7cm) followed by other strains under study (Table 4 & Figure 1A, 2A).
Figure 1. Biochemical evaluation of selected strains of Rhizobium spp: (A) P-solubilization, (B) IAA production, (C) HCN production, (D) Siderophore production
Indole acetic acid production
IAA is another important trait for selection of PGPR’s and here moderate to strong IAA production was recorded by selected isolates. Among four isolates highest production of IAA was shown by 26N (102 µg/ml) followed by LN3 (96 µg/ml), LCG6 (92 µg/ml) and LCG5 (83 µg/ml) (Figure 1B & 2B).
Hydrogen cyanide production
Along with IAA production HCN production indirectly influences plant growth. HCN is a volatile organic compound (VOC) that display antifungal activity against phytopathogens or acts as an inducer of plant resistance by blocking electron transport chain (ETC). In this study, all the four bacterial strains found to able to produce HCN and strong HCN production was shown by 26N on the basis change in colour of filter paper (Figure 1 C & 2C). Quantified form of highest HCN production observed by 26N (16ppm), followed by LN3 (3ppm), LCG6 (9ppm) and LCG5 (12ppm) (Table 4).
Siderophore production
In CAS media and broth 26N, LN3, LCG6 and LCG5 strains showed positive response for siderophore production (Table 4). Furthermore, among four rhizobacteria maximum siderophore production shown by 26N (18±0.4mm) followed by other strains under study (Figure 1D). For the quantitative estimation of siderophore production highest production was recorded for 26N (178 µg/ml), followed by LCG6 (101 µg/ml), LN3 (68 µg/ml), and LCG5 (54 µg/ml) (Figure 2D).
Figure 2. Biochemical evaluation of growth promoting Rhizobium spp. (A) P-solubilization, (B) IAA production, (C) HCN production, (D) Siderophore production
Extracellular enzyme production
Selected strains of growth promoting bacteria posses extracellular enzymatic activity which play an important role in protection of crop against various phytopathogenic fungi and environmental stresses. All selected strains of bacteria were able to produce catalase and oxidase enzyme (Table 5). 26N and LCG5 strains strongly produced CAT and oxidase enzymes while other two strains showed moderate production of CAT along with oxidase. In addition, a-amylase enzyme in the aleurone layer helps in hydrolyzing the endospermic starch into smaller sugar molecules, which provide energy for enhancing vigour index parameter. Amylase along with cellulase was strongly produced by 26N strain (Table 5). Chitinase enzyme serves various functions such as antagonistic activity against phytopathogenic fungi by degradation of fungal cell wall. Among four strains two strains were able to produce chitinase enzyme. Strongly chitinase was produced by 26N strain while moderate chitinase production was recorded for LCG5 (Table 5).
Table (5):
Extracellular enzyme production by selected bacteria.
PGPR’s |
Catalase |
Amylase |
Cellulase |
Chitinase |
Oxidase |
|---|---|---|---|---|---|
Rhizobium spp |
+++ |
+++ |
+++ |
+++ |
+++ |
Neorhizobium
huautlense |
+ |
+ |
++ |
– |
++ |
Sinorhizobium saheli |
++ |
++ |
++ |
– |
+ |
Sinorhizobium saheli |
+++ |
– |
– |
++ |
+++ |
Effect of Rhizobium strains on lentil growth promotion
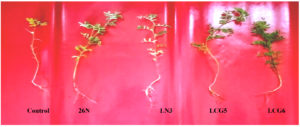
In this present study, it was found that biopriming of lentil seed with selected PGPR strains showed higher root length, shoot length, root weight, shoot weight and vigour index after 21 days of seed germination.
Figure 3. Effect of selected strains of growth promoting Rhizobium spp on Lentil. Root lenght (A), Shoot length (B), Fresh root weight (C), Fresh shoot weight (D), Germination percentage (E), Vigour index (F)
Lentil seeds treated with selected bacterial strains exhibited significant improvement in root and shoot length in comparison to control plant. Those plants which have not received bacterial treatments are considered as control plant. A control plant is used to compare those treated with bacteria (Figure 4). Highest root length and shoot length of lentil crop was observed for plant treated with 26 N strain followed by other strains (Figure 3A & B). When compared to control plant, maximum increment in root and shoot length were recorded for plants treated with strains 26N (63 & 90%) followed by LN3 (45 & 42%), LCG6 (21 & 31%) and LCG5 (27 & 13%).
Among four bacterial strains highest percentage increment of root and shoot weight was observed for plants treated with strain 26N (103 & 73%) followed by other bacterial strains under study (Figure 3C & D).
Extensive enhancement in the percentage seed germination was shown after treatment with various PGPR’s. Significant improvement in lentil seed germination percentage under pot study ranged from 15 to 58% after seed biopriming by selected growth promising bacteria and furthermore maximum vigour index in lentil was observed after PGPR inoculation. After seed biopriming highest vigour index was recorded for plants treated with 26N (1763%), followed by LN3 and lowest was observed for control (414%) plant. In comparison to control plant, 26N strain showed 332% more vigour index (Figure 3E & F).
Rhizobium is soil bacteria that colonize root nodule of lentil plant symbiotically and induces the plant growth through wide range of biological mechanism. The pragmatic investigations of this plant-microbiome association develop our understanding in profiling microbial communities and their relationship like mutualistic, commensalistic, and parasitic microbiota with host plant.27,28 Moreover, due to rich availability of nutrients in the rhizospheric soil, plant rhizosphere is known to be perfect ecological niche for diverse range of soil microorganisms.29,30
The isolated bacterial strains from root nodule were screened for both direct and indirect plant growth–promoting traits and were further identified using molecular identification technique such as of 16S rRNA gene sequencing (Sanger’s sequencing). 20 bacterial strains were investigated for multiple biochemical characterization such as Phosphate solubilization, production of phytohormone IAA (Indole-3 acetic acid), siderophore and hydrogen cyanide. Among 20 bacterial strains 4 bacterial strains exposed outstanding results for all biochemical activities.
The selected four strains of rhizobium spp have greater ability of solubilizing phosphorus in vitro. The Rhizobium leguminosarum-26N revealed maximum phosphate solubilization index (3.7cm) followed by Rhizobium spp LN3 (2.9), Rhizobium spp LCG6 (2.1), and Rhizobium pusense LCG5 (1.9). The study of Moon et al.,31 and Gupta et al.,10 also reported that phosphate solubilizing bacteria are more commonly found in the rhizospheric soil as compared to bulk soil and directly support plant growth. Furthermore, growth promoting bacteria have ability to produce organic acid and phosphatases which help in phosphate solubilization. In this study, bacterial phosphate solubilization results are consistent with the earlier research where they found that the application of growth promoting bacteria has the ability to solubilize inorganic phosphate and could improve the quantity of effective phosphate which enhanced the growth and development of the Avena sativa.32
In the same way, another direct effect of growth promoting rhizobacteria on plants is the production of Indole acetic acid (IAA) which is important phytohormone and work as signal molecule in the regulation of plant growth and in root elongation. In the current study, more than 75% bacterial isolates were positive for IAA production without adding L-tryptophan amino acid in YEM (Yeast extract mannitol) broth while upon the addition of L-tryptophan in the growth media, 90% bacteria exposing IAA-producing ability in broth. Among four selected strains of PGPR’s, R. leguminosarum (26N strain) produced greater quantity of IAA in vitro and greater quantity of IAA directly enhanced root length and lateral root branching.33
The selected rhizobacterial strains 26N and LCG5 strongly produced HCN while other two strains produced HCN moderately in vitro. Hydrogen cyanide is a type of volatile organic compounds (VOCs) which is toxic for phytopathogenic fungi via blocked electron transport system of fungus and also provides protection from various biotic stresses. As the plants exposed to phytopathogen grow weak in their defence system hence shielding them by cyanide producing bacteria which can be helpful in overcoming the chances of plant pathogenic infection in host.34 Furthermore, earlier study it has been reported that Pseudomonas and Bacillus spp bacteria produced HCN along with various lytic enzyme which directly inhibited the growth of phytopathogens, including Phytophthora spp and Fusarium spp.35,36
Siderophore is a low molecular weight iron chelating compound (1Kd) which inhibits the proliferation of phytopathogens by utilizing sequestered Fe3+ in the rhizosphere.37 In this study all the selected strains of rhizobium spp. showed the production of siderophores. Siderophores are the key compounds produced by diverse group of antagonistic-PGPR’s such as Bacillus subtilis, B. circulance, B. coagulanse, B. licheniformis, Pseudomonas fluroscence and P. koreensis furthermore, these siderophore producing bacteria inhibit the growth of Cephalosporium maydis and protect maize crop by fungal wilt.38 Another important trait of Rhizobium spp. which was detected in most of the bacterial strains was the synthesis of auxin. Among four strains, 26N strain showed high level of IAA production which directly induced plant growth as they increased the root length along with lateral root branching.39 Selected strains of growth promoting Rhizobium enhanced root length along with shoot length upto 2 fold when compared to control, beside IAA production 26N bacterial strain was found to be more dominating over the other bacterial strains due to high efficiency of phosphate solubilization. In Earlier studies it has been described that inoculation of growth promoting Rhizobium spp. significantly improved the root and shoot weight and total dry biomass of rice, soybean and maize crop.40-42
Bioprimed seed with selected strains of bacteria improved root and shoot length upto 2 fold when compared to untreated plant (control). Increased root length by growth promoting bacteria favoured plant growth by exploring a greater volume of soil thereby increasing nutrient obtainability. Furthermore, the test strains of Rhizobium were extremely promising in promoting root growth during early stage of seed germination and plant growth, thereby improving vigour index. The germination percentage of lentil seed almost got increased by 1.6 fold by selected PGPR’S when compared to control. These results are also supported by researcher, who described that use of phosphorous solubilization and siderophore producing bacteria enhanced shoots fresh biomass, fresh root biomass, shoot length, root length, shoot dry biomass, root dry biomass as well as total dry biomass of crop.43
Except few bacterial strains all strains had greater efficiency of producing lytic enzymes like amylase, cellulase and chitinase. Catalase and oxidase enzymes reduce the oxidative damage in crop while a-amylase enzyme in the aleurone layer of seed helps in hydrolyzing the endospermic starch into sugar molecules, which provide energy for vigour index parameter.44-37 Similar results were found by Mumtaz et al.45 who reported positive results for catalase and protease activity of Bacillus strain. He also reported the cellulose degradation ability of Bacillus strains and these enzymes were comprehensively reported for their effectiveness in tolerating stress.
The present research explores the significance of isolation, screening and biochemical characterization of growth promoting Rhizobium spp. under in vitro conditions for multiple growth promoting traits and their evaluation under controlled conditions in a pot experiment with lentil. This led to the selection of effective bacterial strains which, as a result of their multiple PGPR activities, could be effective in improving vigour index, seed germination, fresh root and shoot weight of lentil crop and maintenance of soil fertility without adding chemical fertilizers. On the basis of biochemical characterization of PGPR’s along with vigour index and seed germination percentage 26N strain (Rhizobium leguminosarum) performed better growth promotion activity under pot experiment.
ACKNOWLEDGMENTS
The authors would like to thank their supervisor Professor R.K. Singh for providing assistance on different methods.
CONFLICT OF INTEREST
The authors declare that there is no conflict of interest.
AUTHORS’ CONTRIBUTION
All authors listed have made a substantial, direct and intellectual contribution to the work, and approved it for publication.
FUNDING
This study was supported by Rajiv Gandhi National Fellowship under UGC scheme, Delhi, India with grant number F1-17.1/2017-18/RGNF-2017-18-SC-CHH-34920/(SAIII/Website).
DATA AVAILABILITY
The datasets generated and/or analysed during the current study are available from the corresponding author on reasonable request.
ETHICS STATEMENT
Not applicable.
- Bahadur I, Maurya BR, Meena VS, Saha M, Kumar A, Aeron A. Mineral release dynamics of tricalcium phosphate and waste muscovite by mineral-solubilizing rhizobacteria isolated from indo-gangetic plain of India. Geomicrobiol. 2017;34(5):454-466.
Crossref - Dinca LC, Grenni P, Onet C, Onet A. Fertilization and Soil Microbial Community: A Review. Appl Sci. 2022;12(3):1198.
Crossref - Iqbal S, Riaz M, Murtaza G, et al. Chemical Fertilizers, Formulation, and Their Influence on Soil Health. In: Hakeem KR, Dar GH, Mehmood MA, Bhat RA. (eds). Microbiota and Biofertilizers. Springer, Cham. 2021.
Crossref - Saha SP,Mazumdar D. Potential of Plant Growth Promoting Rhizobacteria for Enhancement of Plant Growth and Its Role in Improving Soil Health Under Abiotic Stress. In: Roy S, Mathur P, Chakraborty AP, Saha SP. (eds) Plant Stress: Challenges and Management in the New Decade. Advances in Science, Technology & Innovation. 2022; Springer, Cham.
Crossref - He Y, Pantigoso HA, Wu Z, Vivanco JM. Co-inoculation of Bacillus sp. and Pseudomonas putida at different development stages acts as a biostimulant to promote growth, yield and nutrient uptake of tomato. J Appl Microbiol. 2019;127(1):196-207.
Crossref - Etesami H, Adl SM. Plant growth-promoting rhizobacteria (PGPR) and their action mechanisms in availability of nutrients to plants. Phyto-Microbiome in Stress Regulation. 2020:147-203.
Crossref - Mc Steen P. Auxin and monocot development. Cold Spring Harb Perspect Biol. 2010;2(3):a001479.
Crossref - Hansen BL, de Cassia Pessotti R, Fischer MS, Collins A, El-Hifnawi L, Liu MD. Cooperation, competition, and specialized metabolism in a simplified root nodule microbiome. Mbio. 2020; 11(4):e01917-20.
Crossref - Young JPW, Moeskjaer S, Afonin A, et al. Defining the Rhizobium leguminosarum species complex. Genes. 2021;12:(1):111.
Crossref - Gupta SK, Singh RK, Patel AK, Banjare U. Role of Growth-Promoting Bacteria as Biocontrol Agent Against Root Knot Nematode of Tomato. Biosci Biotechnol Res Com. 2021;14(4):1508-1513.
Crossref - Goswami D, Thakker JN, Dhandhukia PC. Portraying mechanics of plant growth promoting rhizobacteria (PGPR): a review. Cogent Food Agric. 2016;2(1):1127500.
Crossref - Tadesse T, Leggesse T, Mulugeta M, Sefera G. Correlation and path coefficient analysis of yield and yield components in lentil (Lens culinaris Medik.) germplasm in the highlands of Bale, Ethiopia. International Journal of Biodiversity and Conservation. 2014;6:(1):115-120.
Crossref - Karim MH, Alizade H, Majnoun HN, Peyghambari S. Effects of herbicides and handweeding in control of weed in winter and spring sown lentil (Lens culinaris). Iranian Journal of Crop Sciences. 2004;6:(1):68-79.
- Tayade R, Kulkarni KP, Jo H, Song JT, Lee JD. Insight into the prospects for the improvement of seed starch in legume-a review. Front Plant Sci. 2019;10:1213.
Crossref - Pikovskaya RI. Mobilization of phosphorous in soil in connection with vital activity of some microbial species. Mikrobiologiya. 1948;17:362-370.
- Bric JM, Bostock RM, Silverstone SE. Rapid in-situ assay for indole acetic acid production by bacteria immobilized on nitrocellulose membrane. Appl Environ Microbiol. 1991;57:(2):535-538.
Crossref - Baker AW, Schippers B. Microbial cyanide production in the rhizosphere in relation to potato yield reduction and Pseudomonas SPP-mediated plant growth-stimulation. Soil Biol Biochem. 1987;19:(4):451-457.
Crossref - Schwyn B, Neilands JB. Universal chemical assay for the detection and determination of siderophore. Anal Biochem. 1987;160(1):47-56.
Crossref - Payne SM. Detection, isolation, and characterization of siderophores. Methods Enzymol. 1994;235:329-344.
Crossref - Somasegaran P, Hoben HJ. Handbook for Rhizobia. Methods in Legume-Rhizobium Technology. Heidelberg, NY: Springer. 1994.
Crossref - Fadden MC. Biochemical tests for identification of medical bacteria. Williams and Wilkins, Baltimore. USA. 1980:51-54.
- Collins CH, Lyne PM, Grange JM, Falkinham JO. Collins and Liyne’s Microbiological Methods, Butterworth-Heinemann, UK. 1995;7:117.
- Hendricks CW, Doyle JD, Hugley B. A new solid medium for enumerating cellulose-utilizing bacteria in soil. Appl Environ Microbiol. 1995;61(5):2016-2019.
Crossref - Wiwat C, Siwayaprahm P, Bhumiratana A. Purification and characterization of chitinase from Bacillus circulans No. 4.1. Curr Microbiol. 1999;39:(3):134-140.
Crossref - Hayward AC. A method for characterizing Pseudomonas solanacearum. Nature. 1960;186:405.
Crossref - ISTA. Proceedings of the International Seed Testing Association, International Rules for Seed Testing. Seed Sci. Technol. 1993; 21: 25–30.
- Rodriguez PA, Rothballer M, Chowdhury SP, Nussbaumer T, Gutjahr C, Falter-Braun P. Systems biology of plant-microbiome interactions. Mole Plant. 2019;12(6):804-821.
Crossref - Schlaeppi K, Bulgarelli D. The plant microbiome at work. Mol Plant-Microbe Interact. 2015;28(3):212-217.
Crossref - Hakim S, Naqqash T, Nawaz MS, et al. Rhizosphere Engineering With Plant Growth-Promoting Microorganisms for Agriculture and Ecological Sustainability. Front Sustain Food Syst. 2021;5:617157.
Crossref - Khan N, Ali S, Shahid MA, Mustafa A, Sayyed R, Cura JA. Insights into the interactions among roots, rhizosphere, and rhizobacteria for improving plant growth and tolerance to abiotic stresses: a review. Cells. 2021;10(6):1551.
Crossref - Moon YS, Ali S. Isolation and identification of multi-trait plant growth-promoting rhizobacteria from coastal sand dune plant species of Pohang beach. Folia Microbiol. 2022;67(3):523-533.
Crossref - Gupta SK, Prasad JK, Raghuwanshi R. Characterizing rhizospheric plant growth promoting bacteria for their effects on oat Avena sativa. Int J Pharma Bio Sci. 2017;8:(4):(B)142-151.
Crossref - Dey R, Raghuwanshi R. Comprehensive assessment of growth parameters for screening endophytic bacterial strains in Solanum lycopesicum (Tomato). Heliyon. 2020;6(10):e05325.
Crossref - Sehrawat A, Satyavir S, Sindhu, Glick GR. Hydrogen cyanide production by soil bacteria: Biological control of pests and promotion of plant growth in sustainable agriculture. Pedosphere. 2022;32:(1):15-38.
Crossref - Ali S, Hameed S, Shahid M, Iqbal M, Lazarovit G, Imran A. Functional characterization of potential PGPR exhibiting broad-spectrum antifungal activity. Microbiol Res. 2020;232:126389.
Crossref - Valencia-Hernandez JA, Solano-Alvarez N, Feregrino-Perez AA. In vitro and in vivo antimicrobial activity of a synthetic capsaicinoid oleoresin against Fusarium oxysporum, Phytophthora capsici, Clavibacter michiganensis and Pseudomonas syringae. J Plant Pathol. 2022;104(6):699-710.
Crossref - Sahebani N, Gholamrezaee N. The biocontrol potential of Pseudomonas fluorescens CHA0 against root knot nematode Meloidogyne javanica is dependent on the plant species. Biological Control. 2021;152:104445.
Crossref - Ghazy N, El-Nahrawy S, Siderophore production by Bacillus subtilis MF497446 and Pseudomonas koreensis MG209738 and their efficacy in controlling Cephalosporium maydis in maize plant. Arch Microbiol. 2021;203(3):1195-1209.
Crossref - Kishor JK, Gupta SK, Raghuwanshi R. Screening Multifunctional Plant Growth Promoting Rhizobacteria Strains for Enhancing Seed Germination in Wheat (Triticum aestivum L.). Int J Agric Res. 2017;12:(2):64-72.
Crossref - Razafintsalama H, Sauvadet M, Trap J, Autfray P, Ripoche A, Becquer T. Legume Nitrogen Fixation and Symbioses in Low-Inputs Rainfed Rice Rotations. Sustainability. 2021;13(22):12349.
Crossref - Garcia J, Schmidt JE, Amelie GM, Gaudin CM. Impact of an antarctic rhizobacterium on root traits and productivity of soybean (Glycine max L.). J Plant Nutr. 2021;44:12:1818-1825.
Crossref - Nyoki D, Ndakidemi PA. Yield Response of Intercropped Soybean and Maize Under Rhizobia (Bradyrhizobium japonicum) Inoculation and P and K Fertilization. Commun Soil Sci Plant Anal. 2018:49(10):1168-1185.
Crossref - Sijilmassi B, Filali-Maltouf A, Fahde S, et al. In-Vitro Plant Growth Promotion of Rhizobium Strains Isolated from Lentil Root Nodules under Abiotic Stresses. Agronomy. 2020;10(7):1006.
Crossref - Lee YS, Nguyen XH, Naing KW, Park YS, Kim KY. Role of lytic Enzymes secreted by Lysobacter capsici YS1215 in the control of root-knot nematode of tomato plants. Indian J Microbiol. 2015;55(1):74-80.
Crossref - Mumtaz MZ, Ahmad M Jamil M Hussain T Zinc solubilizing Bacillus spp. potential candidates for biofortification in maize. Microbiol Res. 2017;202:51-60.
Crossref
© The Author(s) 2023. Open Access. This article is distributed under the terms of the Creative Commons Attribution 4.0 International License which permits unrestricted use, sharing, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.