Tricarboxylic acid cycle is the essential metabolic pathway for cellular biosynthetic processes e.g. gluconeogenesis, amino acid and heme biosynthesis. Exploring the functional regulatory mechanism of the tricarboxylate transporters, especially the mitochondrial inner membrane transporters of liver, has received much attention due to their implications on various metabolomic diseases. The tricarboxylic acid transporter has been purified from liver with molecular weight 32.6 kDa. Based on the amino acid sequence analysis, six hypothetical membrane-spanning alpha helices have been recognized and used for development of an initial model for protein topographical, and modelling analyses of the tricarboxylate transporter within the inner membrane. The objective of this review was to emphasize the biochemical and metabolic implications of tricarboxylic acids especially citric, aconitic and itaconic acids and their transporters on the mitochondrial membrane of liver.
Tricarboxylic acids; citrate synthase, aconitase; itaconate.
Tricarboxylic acid (TCA) cycle is the key oxidative metabolic pathway with profound roles in oxidation of acetyl-CoA, producing the reducing equivalents for the currency molecules generation ATP2. Recently, structure, function and regulation of the tricarboxylate transporter at the inner membrane of liver mitochondrial has been elucidated, these transporters have been cloned, purified and sequenced with about 32.6 kDa1. Based on the amino acid sequences of transporters, six putative membrane-spanning alpha-helices were recognized as a model for the topography of tricarboxylate transporter within the inner membrane1. TCA cycle enzymes are mainly involved in various biosynthetic processes such as gluconeogenesis, amino acid and heme biosynthesis. Saccharomyces cerevisiae has been sued as a model organism to unravel the TCA cycle enzymes functions and regulations, some of TCA cycle enzymes are not essential for cellular growth and does not implemented on assimilation of fermentable carbon sources, while, these enzymes are important for cellular growth on non-fermentable sugars particularly acetate, glycerol, lactate and/or pyruvate. Mutants defective for growth on different non-fermented sugars as source of carbon, allowed the genetic analysis of TCA cycle oxidative functions4. The 8 enzymes of TCA cycle in S. cerevisiae are encoded by about 15 genes, among these enzymes four genes CIT1, ACO1, FUM1 and MDH1 encoding citrate synthase, aconitase, fumarase, and malate dehydrogenase, respectively5. These enzymes are homo and hetero-multiple subunits encoded by multiple genes such as NAD+-dependent isocitrate dehydrogenase (NAD-IDH) that contains four Idh1p and Idh2p subunits encoded by IDH1 and IDH2 genes6, while, a-ketoglutarate dehydrogenase have 3 varied subunits encoded by KGD1, KGD2 and LPD1 genes7. Lipoamide dehydrogenase encoded by LPD1 gene is a part of pyruvate dehydrogenase and a-amino acid dehydrogenase complex, while, succinyl-CoA kinase is a hetero-dimer composed of an a- and b-subunits encoded by LSC1 and LSC2 genes8. Succinate dehydrogenase is an integral protein of membranes having 4 subunits encoded by SDH1-4 genes9, in addition, there are four genes were found to encode subunits of succinate dehydrogenase (SDH1b, YMR118c, YLR164c, YOR297c), mitochondrial citrate synthase (CIT3), aconitase (ACO2) and have been identified in Saccharomyces genome, however, their properties remains poorly characterized10. Nevertheless, isozymes such as isocitrate dehydrogenase (NADP+-dependent), malate dehydrogenase and citrate synthase were characterized in mitochondria, peroxisomes and cytosol without direct function to the TCA cycle12. These mutants are incapable of using acetate as a carbon source, were found to contain mutations in the eight TCA cycle genes12. TCA cycle is mainly implemented in initiation of peroxisomes, the growth of this organelle require the oleic acid metabolism13, and the mutants cannot use properly acetyl-CoA from the b-oxidation of oleate in developing peroxisomes14. The function of TCA cycle is strongly compromised in mutants missing TCA cycle enzymes assuming that the implementation of the entire enzyme for functional cycle15.
Mutants of succinate dehydrogenase subunits are defective in gluconeogenesis, having inability to assimilate ethanol to glucose-6-phosphate, that essential for the gene expression signal repression of gluconeogenesis, that is imperfect in glyoxylate cycle and gluconeogenic12. The a- and b-subunits of ATP-dependent succinyl-CoA ligase that encoded by LSC1 and LSC2, have recently recognized8. The carbon type expression dependent of these genes is typical of further TCA cycle genes16, both subunits of NAD-IDH are required for activity and allosteric regulation17. Moreover, NAD has been observed to conjugated with the 52 -nontranslated region of mitochondrial mRNAs18, thus might be have a regulatory function outside of its normal catalytic function in the cycle19.
In contrary to the higher solubility of unassembled Idh2p subunit, the unassembled Idh1p subunit are insoluble, the covalently modified form of this protein has been recorded with its complex of higher molecular mass as results of aberrant assembly due to the oxidation of protein19. Two sets of phenotypes idh2 mutants have been recognized, the first strain having missense alleles with active site mutation grow well on glycerol, in contrast to the poorly growth mutant idh2 nonsense that grow very poorly on YPG. Moreover, the strains of these mutations alleles naturally gather extragenic mutations that increase growth on YPG. This phenomenon has been designated as glycerol-suppressor buildup phenotype and the extragenic mutations were known as glycerol suppressors20. To understand the identity of glycerol suppression features accompanied with idh2 mutants20, a collection of 200 glycerol suppressor mutants were collected and analyzed, particularly, the mutants of CIT1 locus were the frequent suppressor mutant recognized, representing more than 20% of the entire collection. Documentation of idh2 cit double mutants in the Acn- mutant group have been observed initially21. The genetic and molecular identity of cit1 suppressors were analyzed in detail revealing that null mutations, missense and nonsense were able for enhancing the glycerol dependent on idh2 strains growth. From the chemical identity, citrate and isocitrate have not been appeared to accumulate to toxic levels in these mutants20. The phenotype suppression apparently have no effect on the covalent complex of Idh1p development in Didh2 strains, due to the appearance of the suppressor colonies in Didh1Didh2 double mutants19. The molecular expressions of various TCA cycle proteins were altered in idh2 mutants and the presence of cit1-suppressing mutants modulates these alterations supposing a connection to glycerol suppression20.
Citric acid Identity
Citric acid, an organic acid that mainly found in citrus fruits naturally. In solution, it present as citrate, a polyatomic anion ester salt, for example trisodium citrate, an ester is triethyl citrate22. Citric acid was firstly produced in England that imported Italian lemons (7-9% citric acid), it is a tribasic acid with pKa values 14.4, the solution of citric acid are buffered in pH range from 2 to 8. In biological systems around pH 7, citric acid mainly found as citrate ion and mono-hydrogen citrate ion, solution of citric acid (1 mM) gives pH of 3.2. The fruit juices pH, from oranges and lemons depends on the amount of citric acid, that being minor for higher acid concentration and vice versa24. Lemon juice as well as Aspergillus niger are the commercial source of citric acid in many countries based on glycerol fermentation through dicloroacetone as intermediates. In addition, further routes have been established from different artificial materials, but the chemical methods still the feasible and uncompetitive one. Moreover, citric acid play an significant part in metabolism of all aerobic organisms, from human, plants and microorganisms. Citromyces spp were the first organisms that reported to accumulate citric acid by growing on medium containing sugars and inorganic salts as reported by Wehmer (106). Then a plethora of microorganisms were reported as strong citric acid producers such as Aspergillus niger, A.awamori, A.nidulans, A. fonsecaeus, A. luchensis, A. phoenicus, A.wentii, A. saitoi, A. flavus, Absidia sp, Acremonium sp., Botrytis sp., Eupenicillium sp., Mucor piriformis, Penicillium janthinellum, P. restrictum, Talaromyces sp., Trichoderma viride and Ustulina vulgaris. Different strains of A. niger have the ability to grow on sugar containing medium at pH 2.0-3.0, throughout their growth, that accumulate huge quantities of citric acid which has been considered as profound millstone for citric acid industrial production15. In addition to filamentous fungi, several unicellular “yeasts” fungi particularly species belongs to the genera Hansenula, Zygosaccharomyces, Candida, Saccharomyces, Pichia, Debaromyces, Torula, Torulopsis, Kloekera and Yarrowia have the potency for citric acid production using carbohydrates and other related alkanes as carbon sources. In addition, citric acid has been produced industrially from Candida spp, such as Candia tropicalis, C. catenula, C. guilliermondii73. However, the current production of citric acid by wild isolate of A. niger was halted due to the association of high amounts of isocitrate as an undesirable byproduct with the citric acid production system, so using of mutant strains of A. niger is the alternative to overcome these challenges.
mong the different fungi, A. niger remains the industrial platform for commercial citric acid production. The nutritional requirements for citric acid production by A. niger have been optimized and properly controlled for maximum citric acid yield using commercial fermenters. Various strains of A. niger with potency to over produce citric acid regarding to diverse fermentation conditions were reported. Practically, the amount of produced citric acid is approximated by 112 g per 100 g sucrose, nevertheless, due to losses through stationary phases, the harvested citric acid from these strains usually not more than 70% of its theoretical yield on the optimum source of carbon. Although, the successful history for production of citric acid by different microbial fermentation process, however, the biochemical bases for regulating the yield of citric acid remain unclear.
Citric acid Biochemistry
Citrate is the key intermediate in TCA cycle for energy generation in all organisms from animals, plants to microorganisms. By activity of citrate synthase, oxaloacetate and acetyl CoA were condensed for citrate formation. Citrate has been considered as substrate for aconitase, that transformed into aconitic acid, generating oxaloacetate. E. coli can generate and use internal citrate as part of their TCA cycle, it lacks the enzymes required to importing it into the cell25. For citrate metabolism, firstly it should be transported from the mitochondria to the cytoplasm, then catabolized into acetyl-CoA for biosynthesis of fatty acid synthesis and oxaloacetate. Citrate is a highly modulator for allosterically regulation the activity of acetyl-CoA carboxylase, regulating the biotrans-formation of acetyl-CoA to malonyl-CoA. High concentrations of cytosolic citrate can inhibit phospho-fructokinase, pointing to great source of biosynthetic precursors, for phosphorfructo-kinase to continue for further biosynthetic process of fructose 6-phosphate and various glycolytic intermediates. Citrate is a profound constituent of bone regulating the size of apatite crystals as well as, by enhancing the inhibitory consequence of high concentrations of ATP26.
Applications
Food and drink
Citric acid has been used mainly as additive and preservative compounds in food, beverages, soft drinks, and candies. Additionally, citric acid can be implanted to ice cream as a blending agent to save fats from separating to caramel and to stop sucrose crystallization27.
Cleaning and chelating agent
Citric acid can be used as powerful chelating agent for binding with metals increasing their solubility, improving the accumulation of limescale from boilers. It can be used to improves the effectiveness of soaps and laundry detergents, and for treating of water. The mechanism of action of citric acid is by metals chelation in hard water, it lets these detergents produce foam and work better without need for water softening as well as an active ingredient in kitchen cleaning solutions, shampoo to ash out wax and hair coloring28.
Cosmetics, Cross linkers, pharmaceuticals, dietary supplements
Citric acid has been used extensively as acidulant in creams, gels, and liquids, with its alpha hydroxy acid that can be implanted as an active ingredient in chemical peels. It is frequently used as a buffer to rise the solubility of brown heroin. It is used as one of the active components in antiviral tissues production28. Practically, citric acid was implemented as chelating water hardness Ca2+ and Mg2+ ions, in contrast to phosphate, it does not contribute to the aquatic system eutrophication. Citric acid has been used efficiently in crosslink of various materials, such as ultrafine protein fibers for various medical applications, polyols for manufacturing of biodegradable films, and for eco-friendly packaging with hydroxyapatite to produce bioceramic composites tissues. Crosslinking of citric acid with the starch-glycerol films, strongly improves the lower thermal degradation, mechanical properties of these films for various biotechnological applications. In addition, an important application of citric acid as crosslinking agent has been established in 2011. Glycerol and citric acid was polymerizing to form a water-soluble resin, presenting multiple important features with rapid degradation in environment. Glycerol boiling point is 290°C, while citric acid decomposing temperature is 175°C suggesting the water is the unique molecules that liberated as steam, the resulting polymer is a 3- dimensional polyester that adheres to other materials, therefore it could be used in mixture with steel, glass, metals and other solid materials.
Aconitic acid
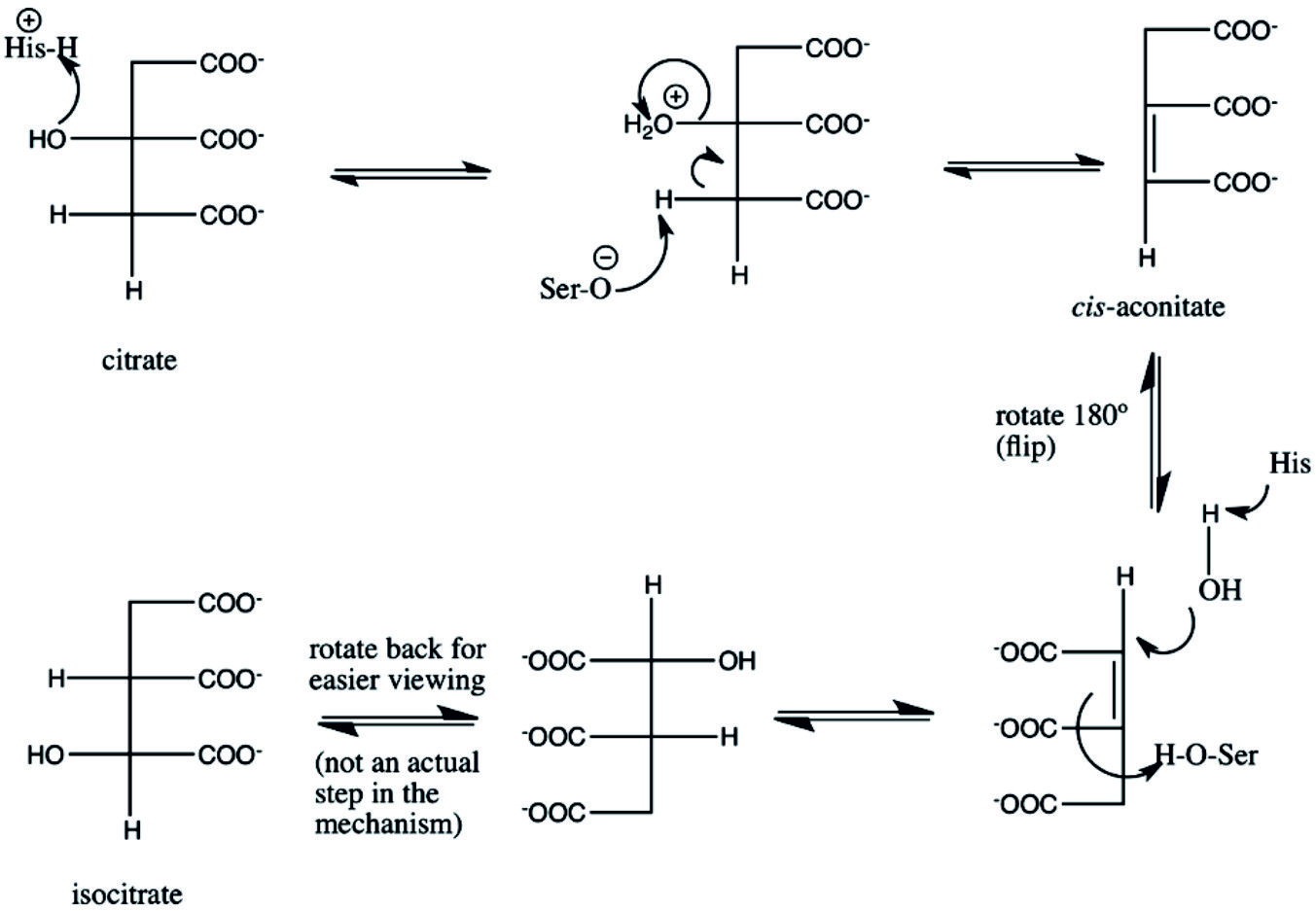
Aconitic acid, organic acid, having two isoforms, cis-aconitate and aconitic acid. Cis-aconitic acid and cis-aconitate is an conjugate base intermediate essential in the isomerization of citrate to isocitrate in the TCA cycle due to activity of aconitase. On the other hand, aconitic acid can be produced by dehydration of citric acid with sulfuric acid31. Aconitase in the TCA cycle can isomerizes citric acid to iso-citric acid via cis-aconitic acid. The mechanism of aconitase classifies it as a lyase, the relative concentrations of substrates results in it catalyzing the conversion of citrate to isocitrate31. The most common intermediates of TCA cycles produced by the dehydration of citric acid is cis-Aconitic acid, under catalysis of aconitase (aconitate hydratase; EC 4.2.1.3) that catalyze the stereo-specific isomerization of citrate to isocitrate via cis-aconitate in the TCA cycle. The mechanism of aconitase displays, it catalyzes the conversion of citrate and isocitrate into aconitate in addition to the reverse reaction, similarly to fumarase31.
Itaconic acid
Itaconic acid had been discovered as a thermal degradation product of citric acid, it is an unsaturated dicarbonic acid, that has been designated as methylene succinic acid. Owing to its chemical identity, that has one unsaturated double bond and two carboxyl groups, itaconic acid can be transformed into different valuable bio-based chemicals or materials generated from carbohydrates. The chemical and functional structure of itaconic acid allows a variety of reactions and applications and it is able to replace petrochemical industries using methacrylic or acrylic acid as precursor compound. Due to the presence of the two carboxyl groups, this acid having two pKa values, one at pKa1 3.84 and other at pKa2 5.55 at 25°C. In the aqueous solutions, the absorption of each dissociated system based on the actual pH value. Basically, the non-dissociated itaconic acid has been reported at pH values lower than pH 2, while at pH value higher than pH 7, the double dissociated itaconate has been reported. Within the pH range 2 and 7, a mixture of different dissociation forms of itaconic. Itaconic is naturally white solid compounds with higher solubility in water, ethanol, and acetone. Historically, the name itaconic acid was derived from aconitic acid, which has been as another derivative of citric acid32.
Laboratory synthesis and reactions
The first discoveries about itaconic acid root back into 1836. With distillation of citric acid, a new component as itaconic acid. The name of this acid represents an anagram of cis-aconitic acid33. This acid has been recognized from fungi which descriptively named as Aspergillus itaconicus34. Itaconic acid anhydride can be produced through dry distillation of citric acid, that undergoes hydrolysis to itaconic acid an further upon heating, itaconic anhydride can be isomerizes to citraconic acid anhydride, that subsequently can be hydrolyzed to citraconic acid35.
Compartmentalization of itaconic acid synthesis in A. terreus
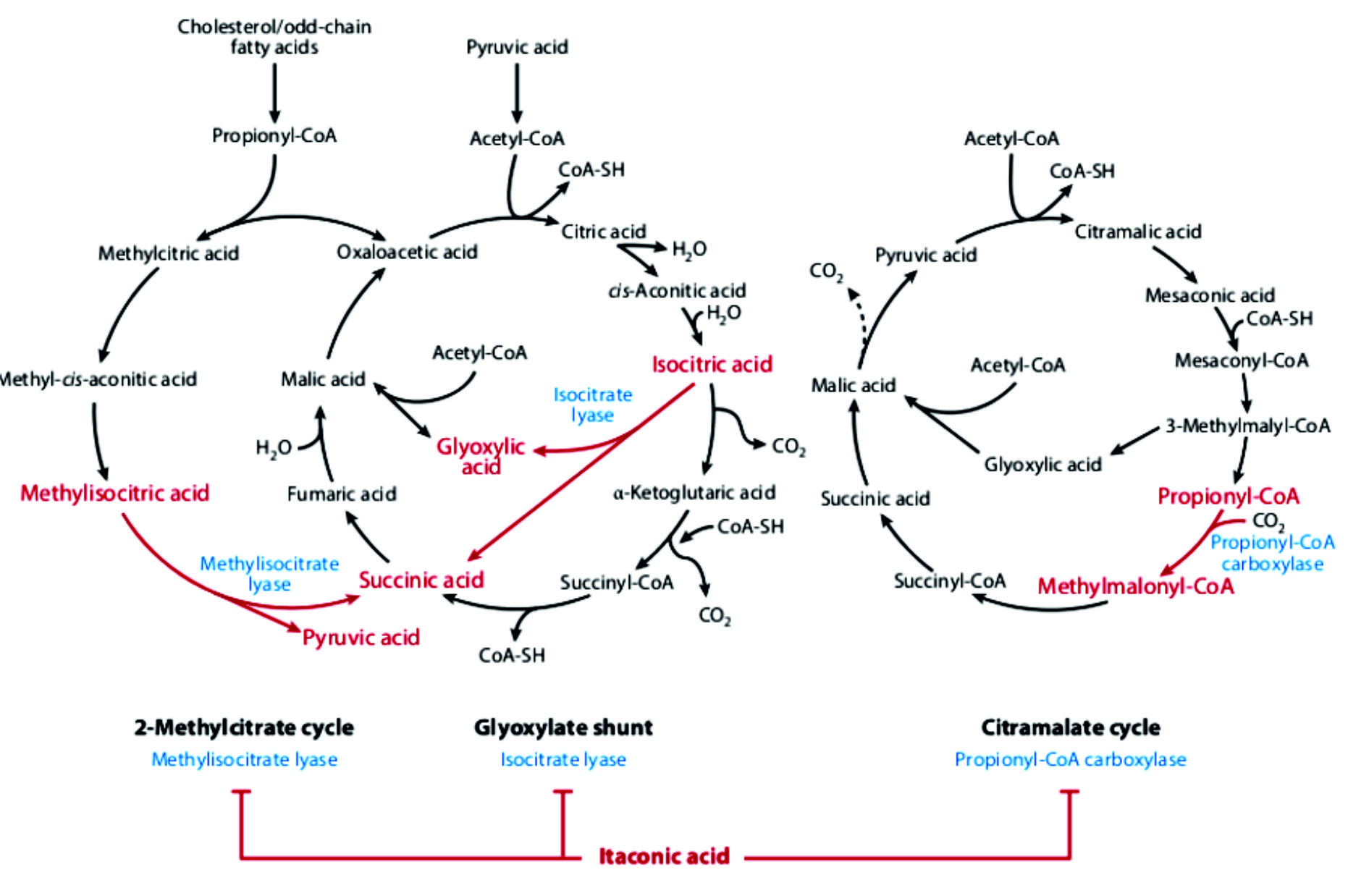
Itaconic acid is formed from the citric acid cycle transitional cis-aconitic acid by cis-aconitic acid decarboxylase, that encoded by the cad1. This pathway had been confirmed by isotope 14C and 13C labeled substrates tracing experiments36. Starting from sugar substrate, as source of carbon to produce itaconic acid, A. niger utilize glucose from the extracellular environment, and processes it via glycolysis to pyruvate in the cytoplasm. Pyruvic acid can enter the mitochondrial TCA cycle either via acetyl-CoA or malic acid. During the initial steps of TCA cycle, citrate and cis-aconitate are formed. In Aspergillus terreus, cis-aconitate decarboxylase is localized in the cytosol, and then directed to its location to produce itaconic acid. Taken it into account, transport of cis-aconitic acid from mitochodria to the cytosol is required. This relocation is done by citrate-malate antiporter37. Itaconic acid (IA) has wan significant attention over the last few years. Itaconic acid composed of two carboxylic acid functionalities and a,b-unsaturateds bond, that makes it a promising initials for a numerous chemical renovations. Itaconic acid was first manufactured in 1837 by decarboxylation of citric acid, in addition to further synthetic approaches that have been reported as platform8,9. Industrial pathways using carbohydrates have been established since the 1945, since then significant study efforts have been devoted in this field to expand the yields and practicability of this process. Currently, itaconic acid can be produced by industrially through different fermentation process of Aspergillus terreus. The overall global yield of itaconic acid has been approximated by about 80,000 tons annually with a price more than 2 US$ per kg.
Interestingly itaconic acid is regulated with a mitochondrial carrier protein, that upstream of the cadA on A. terreus genome (Li et al.38). While on the downstream of cadA gene, another transporter that designated as putative Major Facilitator Superfamily transporter. In A. niger, the expressing of cadA gene under control of various constitutive promoters with different expression strength established that the itaconic acid yield straight relates to the cadA transcript level34. It can be determined that a high transcriptional titer of this gene is indispensable for an greatest expression of these genes. A high transcriptional level of the gene is essential, because of a little enzyme stability in vivo, unlike to the instability in vitro39.
Production
The discovery of itaconic acid has been of growing interest for industry that can be used as an initial material for polymer synthesis. The structure of itaconic acid reveals its reactive methylene group which allows a self-polymerization to polyitaconic acid34. For itaconic acid production by A. terreus, glucose is metabolized into two molecules of pyruvate through EMP pathway in the cytoplasm. The moiety of pyruvate is transported to the mitochondria and decarboxylated into acetyl-CoA and / or carboxylated into oxaloacetate in the cytoplasm. The synthesized oxaloacetate is subsequently transformed into malate, that transported to the mitochondria via malate-citrate antiporter proteins. In mitochondria, a condensation of acetyl-CoA and oxaloacetate was performed into citric acid by the action of citrate synthase, that subsequently converted to citric acid by the action of cis-aconitate by aconitase. cis-Aconitate is transported to the cytosol with the using mitochondrial tricarboxylic acid transporter and serves as an ancestor for itaconic acid production via decarboxylation by cis-aconitic acid decarboxylase (CAD, EC 4.1.16). Cis-Aconitic acid has been converted into IA with crude CAD (55 kDa), that has been considered as a key enzyme on production of itaconic acid by A. terreus, as reported by Bentley and Thiessen. This cadA gene has been considered as part of the gene cluster of mitochondrial TCA transporter, membrane permease and a transcription factor that controls and regulate expression of pathway specific gene cluster. The presence of L-aspartate in producing media straight suppressed pyruvate carboxylase expression in A. terreus culture, while the limited malate flux regulated the malate/citrate antiporters subsequent increases the CAD activity to simultaneously, which in turn convert cis-aconitate into itaconic acid.
Moreover, the wide industrial applications of A. terreus for production of itaconic acid, other organisms, like Ustilaginaceae, could be also have the potentiality to form itaconic acid (Geiser et al. 2014). Affording to the present literature, triggering of itaconic acid production in Ustilago maydis is mainly dependent on ammonium concentrations (Maassen et al. 2014). For biosynthesis of itaconic acid, the pathway of U. maydis is relatively comparable to that of A. terreus. However, U. maydis can be converted to cis-aconitate in the cytosol to trans-aconitate with cytosolic aconitate-D-isomerase as hypothesized by Geiser et al. 2016 showing that U. maydis synthesized itaconate via unusual transitional trans-aconitate. In addition, aconitate-D-isomerase, trans-aconitate decarboxylase enzymes, and mitochondrial TCA transporter and itaconate transporter protein, that belongs to the Major Facilitator superfamily, that reported in the identical gene cluster for synthesis of itaconic acid (Geiser et al. 2016). Trans-aconitate is catalyzed by trans-aconitate decarboxylase (Tad1) into itaconic acid. Furthermore, Zambanini et al. (2017a) examined two of the native promoters, Ptad1 and Pmtt1 from the itaconate cluster of U. maydis. It is recognized that the activate of itaconic acid overproduction in U. maydis is ammonium dependent, and it is hypothesized that these two promoters are well suitable to persuade gene expression in responsive to the nitrogen starvation that is coupled to the itaconic acid production phase. This study delivers a new set of genetic tools foremost to further development of the organic acid synthesis using metabolic molecular manipulation techniques for Ustilago (Zambanini et al. 2017a).
Therefore, itaconic acid is a potential replacement for crude oil-based products, and it is used as a precursor for plastic polymer synthesis, resins, lattices and fibers. Toward efficient itaconicacid production certain drawbacks of using filamentous fungi should be sort out41. As mentioned above, Aspergillus niger is known to produce 200 g/L of citric acid. The only obstacle is that Aspergillus niger lacks cad1 gene. Although, after the cad1 of A. terreus engineered into A. niger, still itaconic acid titers were low compared to the synthetized citric acid. Significant increase in itaconic acid yield was detected with overexpression of mitochondrial and plasma membrane transporter42.
Itaconic acid as an antimicrobial agent
Antimicrobial activity of itaconic acid is administered through action on pathogen metabolism (Fig. 2). Modes of these actions are: inhibition of isocitrate lyase, inhibition of methylisocitrate lyase and inhibition of propionyl-CoA carboxylase44.
Itaconic acid in mammalian cells
Strelko et al.45 reported about itaconic acid as a novel mammalian metabolite that most likely plays a role in macrophages during immune responses. Sugimoto et al.46 also discovered itaconic acid in the extracellular environment of mammalian cells as a metabolite of LPS-activated macrophages, but did not discuss its biological relevance. Acod1 upregulation was observed in different type of cells: murine macrophages infected with Mycobacteria47 or Salmonella enterica48 as well as in LPS-stimulated bone marrow-derived dendritic cells48,49 performed Acod1 upregulation after being infected in vivo with Toxoplasma gondii38 and in vitro after LPS stimulation51,52,53. There are evidences about itaconic acid affecting energy metabolism, inhibiting rat liver phosphofructokinase-2, a regulatory enzyme of the glycolytic pathway. Due to the inhibition of the glycolytic pathway, itaconic acid suppress the synthesis of fatty acids from glucose.
Pathway of itaconate metabolism in murine liver mitochondria
Xiao et al.50 merged itaconic acid metabolism with the part of TCA cycle and related metabolic reactions that involve SLP. Itaconate arises from cis-aconitate, an intermediate of the aconitase reaction, but only in tissues where cis-aconitate decarboxylase is expressed. In the fungus A. terreus, CAD is an extramitochondrial protein; in mammalian cells, an iron-responsive element binding protein exhibiting aconitase activity has been found in the cytosol, however, in cells of macrophage lineage (where itaconate is formed) cis-aconitate decarboxylase associates to mitochondria34. In the mitochondrial matrix, itaconate could weakly inhibit succinate dehydrogenase in a competitive manner50. In intact liver mitochondria and in the presence of ATP and Mg2+ but absence of oxygen, itaconate became thioesterified to itaconyl-CoA which was later converted to citramalyl-CoA through methylglutaconyl-CoA hydratase (also known as methylglutaconase, MGTK). Citramalyl-CoA could be further converted to either mesaconyl-CoA by MGTK, or to acetyl-CoA and pyruvate. Mesaconyl-CoA can lose the CoASH in a reaction catalyzed by succinate-CoA ligase, forming mesaconate57.
Acknowledgements
None.
Conflicts Of Interest
The author declare that there are no conflicts of interest.
Funding
None
Data Availability
All datasets generated or analyzed during this study are included in the manuscript.
Ethics Statement
The article does not contain any studies with human participants or animals performed by the authors.
- Kaplan R.S., Mayor J.A. Structure, function and regulation of the tricarboxylate transport protein from rat liver mitochondria. J. Bioenerg. Biomembr., 1993; 25: 503-14.
- Zeng A.P., Deckwer W.D. Pathway analysis of oxygen utilization and tricarboxylic acid cycle activity in Saccharomyces cerevisiae growing on glucose. J. Biotechnol., 1994; 37: 67-77.
- Velot C., Mixon M.B., Teige M., Srere P.A. Model of a quinary structure between Krebs TCA cycle enzymes: A model for the metabolon. Biochemistry, 1997; 36: 14271-14276.
- McAlister-Henn L., Small W.C. Molecular genetics of yeast TCA cycle isozymes. Prog. Nucleic Acid Res. Mol. Biol., 1997; 57: 317-339.
- Gangloff S.P., Margueret D., Lauquin G.J.M. Molecular cloning of the yeast mitochondrial aconitase gene (ACQ1) and evidence of a synergistic regulation of expression by glucose plus glutamate. Mol. Cell. Biol., 1990; 10: 3551-3561.
- Cupp J.R., McAlister-Henn L. Cloning and characterization of the gene encoding the IDH1 subunit of NAD(+)-dependent isocitrate dehydrogenase from Saccharomyces cerevisiae. J. Biol. Chem., 1992; 267: 16417-16423.
- Repetto B., Tzagoloff A. Structure and regulation of KGD2, the structural gene for yeast dihydrolipoyl transuccinylase. Mol. Cell. Biol., 1990; 10: 4221-4232.
- Przybyla-Zawislak B., Dennis R.A., Zakharkin S.O., McCammon M.T. Genes of succinyl-CoA ligase from Saccharomyces cerevisiae. Eur. J. Biochem., 1998; 248: 736-743.
- Daignan-Fornier B., Valens M., Lemire B.D., Bolotin-Fukuhara M. Structure and regulation of SDH3, the yeast gene encoding the cytochrome b560 subunit of respiratory complex II. J. Biol. Chem., 1994; 269: 15469-15472.
- Colby G., Ishii Y., Tzagoloff A. Suppression of sdh1 mutation by the SDH1b gene of Saccharomyces cerevisiae. Yeast, 1998; 14: 1001-1006.
- van Roermund C.W.T., Hettema E.H., Ka A.J., van den Berg M., Tabak H.F. Peroxisomal beta-oxidation of polyunsaturated fatty acids in Saccharomyces cerevisiae: Isocitrate dehydrogenase provides NADPH for reduction of double bonds at even positions. EMBO J., 1998; 17: 677-687.
- Dennis R.A., Rhodey M., McCammon M.T. Yeast metabolic mutants of glucose metabolism with defects in the coordinate regulation of carbon assimilation. Arch Biochem Biophys., 1999; 12: 25-29.
- Veenhuis M., Mateblowski M., Kunau W.H., Harder W. Proliferation of microbodies in Saccharomyces cerevisiae. Yeast, 1987; 3: 77-84.
- McCammon M.T., Veenhuis M., Trapp S.B., Goodman J.M. Association of glyoxylate and beta-oxidation enzymes with peroxisomes of Saccharomyces cerevisiae. J. Bacteriol., 1990; 172: 5816-5827.
- Sumegi B., McCammon M.T., Sherry A.D., Keys D.A., McAlister-Henn L. Metabolism of [3-13C] pyruvate in TCA cycle mutants of yeast. Biochemistry, 1992; 31: 8720-8725.
- De Risi J.L., Iyer V.R., Brown P.O. Exploring the metabolic and genetic control of gene expression on a genomic scale. Science, 1997; 278: 680-686.
- Zhao W.N. and McAlister-Henn L. Affinity purification and kinetic analysis of mutant forms of yest NAD+-specific isocitrate dehydrongase. J. Biol. Chem., 1997; 272: 21811-21817.
- Elzinga S.D.J., Bednarz A.L., van Oosterum K., Dekker P.J.T., Grivell L.A. Yeast mitochondrial NAD+-dependent isocitrate dehydrogenase is an RNA-binding protein. Nucleic Acids Res., 1993; 21: 5328-5331.
- Gadde D.M., Yang E., McCammon M.T. An unassembled subunit of NAD+-dependent isocitrate dehydrogenase is insoluble and covalently modified. Arch. Biochem Biophys., 1998; 354: 102-110.
- Gadde D.M. and McCammon M.T. Mutations in the IDH2 gene encoding the catalytic subunit of yeast NAD+-dependent isocitrate dehydrongease can be suppressed by mutations in the CIT1 gene encoding citrate synthase and other genes of oxidative metabolism. Arch. Biochem. Biophys., 1997; 344: 139-149.
- McCammon M.T. Mutants of Saccharomycesce-revisiae with defects in acetate metabolism: Isolation and characterizationof Can mutants. Genetics, 1996; 144: 57-69.
- Apleblat A. Citric acid. Springer. ISBN 2014978-3-319-11232-9.
- Duarte A.M.; Caixeirinho D.; Miguel M.G., Sustelo V.; Nunes C.; Fernandes M.M., Marreiros A. : Organic acids concentration in citrus juice from conventional versus organic farming. Acta Horticulturae, 2012; 933: 601–606.
- Silva A.M.N., Xiaole H. and Robert C. Determination of the pKa value of the hydroxyl group in the a-hydroxycarboxylates citrate, malate and lactate by 13C NMR: implications for metal coordination in biological systems. Biometals, 2009; 22: 771–778.
- Powell A. Generations of bacteria, plus freezer, yield startling results. Phys. Org., 2014:
- Hu Y.-Y., Rawal A., Schmidt-Rohr K. Strongly bound citrate stabilizes the apatite nanocrystals in bone. Proceedings of the National Academy of Sciences, 2010; 107: 22425–22429.
- Greenfield H., Southgate D.A.T. Food Composition Data: Production, Management and Use. Rome: FAO; 2003; 146.
- Garden J., Roberts K., Taylor A., and Robinson D. Evaluation of the Provision of Single Use Citric Acid Sachets to Injecting Drug Users. Scottish Center for Infection and Environmental Health, 2003.
- Anchell S : The Darkroom Cookbook: 3rd Edition (Paperback). Focal Press. (2013)
- Zheng J., Xiao F., Qian L.M., Zhou Z.R. and Xiao QZ.: Erosion behavior of human tooth enamel in citric acid solution. Tribology International., 2009; 42: 1558–1564.
- Bruce W.F. Aconitic Acid, 1937; 17: 1.
- Beinert H., Kennedy M.C., Stout CD (Nov. Aconitase as Ironminus signSulfur Protein, Enzyme, and Iron-Regulatory Protein” (PDF). Chemical Reviews. 1996; 96: 2335–2374
- Sheldon R.A. Green and sustainable manufacture of chemicals from biomass: state of the art. Green Chem., 2014; 16: 950–963.
- Cordes T., Michelucci A., Hiller K. Itaconic acid: the surprising role of an industrial compound as a mammalian antimicrobial metabolite. Annu. Rev. Nutr., 2015; 35: 451-473 .
- Steiger M.G., Blumhoff M.L, Mattanovich D., Sauer M. Biochemistry of microbial itaconic acid production. Front Microbiol., 2013; 4: 23.
- Shriner R.L., Ford S.G. and Roll J. Itaconic anhydride and itaconic acid. Org. Synth., 1931; 11: 70.
- Bonnarme P., Gillet B., Sepulchre A.M., Role C., Beloeil J.C., Ducrocq C.: Itaconate biosynthesis in Aspergillus terreus. J. Bacteriol., 1995; 177: 3573-3578.
- Jaklitsch W.M., Kubicek C.P., Scrutton M.C.: The subcellular organization of itaconate biosynthesis in Aspergillus terreus. J. Gen. Microbiol., 1991; 137: 533-539.
- Li H., Gang Z., Yuling H., Luokun X., Jie X., Hao L., Li W., Chunsong H., Junyan L., Mingshen J., Youxin J., Feili G., Boquan J., Jinquan T. Different neurotropic pathogens elicit neurotoxic CCR9- or neurosupportive CXCR3-expressing microglia. J. Immunol., 2011; 177: 3644-3656.
- Okabe M., Lies D., Kanamasa S. Park E.Y.: Biotechnological production of itaconic acid and its biosynthesis in Aspergillus terreus. Appl Microbiol Biotechnol., 2009 84: 597-606.
- Yu C., Cao Y., Zou H., Xian M. Metabolic engineering of Escherichia coli for biotechnological production of high-value organic acids and alcohols. Appl. Microbiol. Biotechnol., 2011; 89: 573-583.
- El-Imam A.A., Du C.: Fermentative itaconic acid production. J. Biodivers . Bioprospect. Dev., 2014; 1: 1-8.
- van der Straat L., Vernooij M., Lammers M., van den Berg W., Schonewille T., Cordewener J., van der Meer I., Koops A., de Graaff L.H. Expression of the Aspergillus terreus itaconic acid biosynthesis cluster in Aspergillus niger. Microb Cell Fact 2014; 13: 11.
- Nemeth B., Doczi J., Csete D., Kacso G., Ravasz D., Adams D., Kiss G., Nagy A. Horvath G., Tretter L., Mocsai A., Csepanyi-Komi R., Iordanov I., Adam-Vizi V., Chinopoulos C. Abolition of mitochondrial substrate-level phosphorylation by itaconic acid produced by LPS-induced Irg1 expression in cells of murine macrophage lineage. The FASEB Journal., 2015; 54: 234-240
- Kumar R. Glyoxylate shunt: combating mycobacterium at forefront. Int. J. Integr. Biol., 2009; 7: 69-72 .
- Strelko C.L., Lu W., Dufort F.J., Seyfried T.N., Chiles T.C., Rabinowitz J.D., Roberts M.F. Itaconic acid is a mammalian metabolite induced during macrophage activation. J. Am. Chem. Soc., 2011; 133: 16386-16389.
- Sugimoto M., Sakagami H., Yokote Y., Onuma H., Kaneko M., Mori M., Sakaguchi Y., Soga T., Tomita M. Non-targeted metabolite profiling in activated macrophage secretion. Metabolomics, 2012; 8: 624-633.
- Basler T., Jeckstadt S., Valentin-Weigand P., Goethe R. Mycobacterium paratuberculosis, Mycobacterium smegmatis, and lipopoly-saccharide induce different transcriptional and post-transcriptional regulation of the IRG1 gene in murine macrophages. J. Leukoc. Biol., 2006; 79: 628-638
- Michelucci A., Cordes T., Ghelfi J., Pailot A., Reiling N., Goldmann O., Binz T., Wegner A., Tallam A., Rausell A, Buttini M, Linster C L, Medina E, Balling R, Hiller K. Immune-responsive gene 1 protein links metabolism to immunity by catalyzing itaconic acid production. Proc. Natl. Acad. Sci. USA, 2013; 110: 7820-7825
- Hoshino K., Kaisho T., Iwabe T., Takeuchi O., Akira S.: Differential involvement of IFN-ג in toll-like receptor-stimulated dendritic cell activation. Int. Immunol., 2002; 14: 1225-1231
- Xiao W., Wang L., Xiao R., Wu M., Tan J., He Y. Expression profile of human immune-responsive gene 1 and generation and characterization of polyclonal antiserum. Mol. Cell Biochem., 2011; 353: 177-187
- Thomas D.M., Francescutti-Verbeem D.M., Kuhn D.M. Gene expression profile of activated microglia under conditions associated with dopamine neuronal damage. FASEB J., 2006; 20: 515-517
- Preusse M., Tantawy M.A., Klawonn F., Schughart K., Pessler F. Infection- and procedure-dependent effects on pulmonary gene expression in the early phase of influenza A virus infection in mice. BMC Microbiol., 2013; 13: 293-77
- Smith J., Sadeyen J.R., Paton I.R., Hocking P.M., Salmon N., Fife M., Nair V., Burt D.W., Kaiser P. Systems analysis of immune responses in Marek’s disease virus-infected chickens identifies a gene involved in susceptibility and highlights a possible novel pathogenicity mechanism. J. Virol., 2011; 85: 11146-11158
- Sakai A., Kusumoto A., Kiso Y., Furuya E. Itaconate reduces visceral fat by inhibiting fructose 2,6-bisphosphate synthesis in rat liver. Nutrition, 2004; 20: 997-1002.
- Klement T., B chs J. Itaconic acid; a biotechnological process in change. Bioresour Technol., 2013; 135: 422-431.
- Mills E., O’Neill L.A.: Succinate: a metabolic signal in inflammation. Trends Cell Biol., 2014; 24: 313-320
- Abramov A.Y., Duchen M.R. The role of an astrocytic NADPH oxidase in the neurotoxicity of amyloid beta peptides. Philos. Trans R. Soc. Lond B. Biol. Sci., 2005; 360: 2309-2314
- Adler J., Wang S.F., Lardy H.A. The metabolism of itaconic acid by liver mitochondria. J. Biol. Chem., 1957; 229: 865-879.
© The Author(s) 2019. Open Access. This article is distributed under the terms of the Creative Commons Attribution 4.0 International License which permits unrestricted use, sharing, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.