ISSN: 0973-7510
E-ISSN: 2581-690X
Development of industrialization is directly proportional to the development of nations. Rising industrializations also increase the pollutions. One of them is poultry industry which discharges a huge amount of keratinous waste. The present study focused on the selection of feather degrading bacteria from Barshi region of Maharashtra. Bacterial strains were grown in whole feather broth medium. Out of 16 proteolytic bacterial isolates, 9 feather degrading isolates were selected from the Parande road side feather waste dumping area. Among these isolates, P3A was selected and classified on the basis of morphological and biochemical analysis. 16S rRNA studies confirmed that the isolated strain was Staphylococcus epidermidis. Staphylococcus epidermidis P3A shows 84% feather degradation and 72 U/ml maximum keratinase activity after 96 hrs. The isolate shows potential use in feather waste disposal methods which are safe and environment friendly.
Environmental pollution, Feather waste, Keratin, Keratinase, 16S rRNA
Disposal of environmental waste is becoming a significant issue across the world and it majorly polluted water sources and land1-4. The waste byproducts resulting from food processing are keratinous substances including animal hairs, chicken feathers, wool and horns, and vast amounts of certain byproducts are generated yearly. For example, during the year 2019 it has been approximated that over 4.7 million tons of waste feathers were generated by poultry industries alone5.
Waste material from the poultry sector which always includes excreta, wasted feed, cracked eggs, dead birds, feathers and offal may appear as significant environmental pollutants. Feathers are excellent sources of essential amino acids and could be processed into necessary products such as bio-fuel, recycled plastics, feather food and organic fertilizers6. Keratinous components are special since they are composed of essential amino acids including glycine, arginine proline, leucine, valine and threonine. Nevertheless, the robust structure, elevated keratin cohesion are especially due to disulphide bonds formed between cysteine chains within as well as among keratin polypeptide chains7 but existing feather waste management strategies such as incineration, soil filling or processing to animal food by high pressure steam cooking often need a lot of energy inputs also generates handling, storage, pollution control problems8. Development of keratin hydrolysates through physical or chemical methods usually requires a high temperature and therefore effects in deterioration of heat susceptible amino acids, such as lysine, tryptophan and methionine which reduce in the nutritional value of the hydrolysates9. Feather contains more than 90% of protein, the primary element of which is beta-keratin, insoluble and fibrous structural protein that bridges widely interconnected by disulphide bonds, this makes them very rigid to digestion by livestock, foremost to severe problem for discarding10. Keratin belongs to the class of scleroprotein is highly stable and have low rate of degradation11,12.
The Bacteria secretes enzymes which specifically breakdown the beta-keratin part of feathers13. Keratinase is an extra cellular enzyme only secreted when keratinous materials such as hairs, wool, feather, horn nails etc are present and act on the disulphide bond of keratin14,15.
In spite of their foremost ability of withstand, keratins do not mount up in environment because of the numerous microorganisms has ability to hydrolyze them. Keratinolytic activity was widely recognized for strains of the genera Bacillus as well as Streptomyces among bacteria16-18. Bacterial keratinase enzyme may have significant uses during the expansion of non-polluting processes in biotechnological processes relating to keratinous waste from the poultry and leather productions. Biodegradation of feathers using keratinase producing microorganisms attracts several researchers as it can offer an effective technique to dispose feather waste that can be combined with useful product formation19. The purpose of this study was to select novel bacteria for degradation of chicken feather from dumping sites of feather waste in Barshi region of Maharashtra.
Collection of soil samples
For the purpose of selection of feather bio-degradable micro-organisms soil samples from the depths of approximately 3.0 to 4.0 cm were collected from various chicken feather waste dumping sites.
Processing of Feathers
Waste feathers were thoroughly cleaned in potable water to eliminate the blood particles, dust and eventually washed with distilled water. Feathers were dehydrated in sunlight and after that at 60°C for 48 hrs and stored at 5°C for further study20.
Isolation of bacteria
Five-fold series dilution was prepared from soil samples, enriched in whole feather broth medium and incubated for 7 days at 37°C with 120 rpm.
Primary screening
Suspension from enrichment culture was evaluated for proteolytic activity on skim milk agar. Well-grown Clear zone forming colonies were selected for the study of keratinolytic potential.
Secondary screening
24 hrs old cultures were prepared from the promising isolates selected in primary screening and inoculated in sterile whole feather broth Medium composed as (gm/l): Feathers-5.0, K2HPO4-0.3, KH2PO4-0.4, NaCl-0.5 and pH-7.5. The flasks were incubated on shaker for 7days at 37°C.
Identification of isolates
The promising strains were identified in accordance with the taxonomic scheme of Bergey’s manual based on their morphological, cultural and physiological characters. The tests carried out include gram-nature, motility, catalase, coagulase, oxidase, urease, indole production, production of hydrogen sulfide, methyl red, Voges-Proskauer, nitrate reduction, oxidative fermentative test 21.
Further identification was confirmed by 16S rRNA sequencing. Bacterial DNA was isolated using DNA isolation kit. Gene fragment was amplified by the PCR. A single specific PCR amplicon band was selected and unincorporated PCR primer and dNTPs from PCR products were removed by purification of the column. The PCR amplicon DNA sequencing reaction was performed with 27F/1492R primers using the BDT v3.1 Cycle sequencing kit on the ABI 3730xl Genetic Analyzer. The gene sequence was used to perform BLAST with the database of NCBI Genbank database. On the basis maximum similarity score, initial 10 sequences were selected and matched using multiple alignment software programs and the phylogenetic tree was constructed using the Neighbor-Joining method.
Determination of keratinase activity
24 hrs of old culture of isolated strain was inoculated in whole feather broth medium incubated at 37°C on shaker with 120 rpm for production of keratinase enzyme. Aliquot of the broth culture was withdrawn after 24 hrs of interval; crude enzyme was extracted by filtration and centrifugation. Keratinase activity was measured with Keratin Azure (K8500 Sigma Aldrich) as substrate with slight changes of methods described by (22, 23, 15 and 24). In brief, the reaction mixture prepared as 0.8 ml 50mM potassium phosphate buffer of pH 7.5 contain keratin azure (0.4% w/v) added with the 0.2 ml crude form of enzyme and incubated at 50°C for 1 hr with 200 rpm shaking condition. The enzyme substrate reaction was stopped by addition of 0.2 ml 10% trichloroacetic acid. Fallowed by 15 minute’s centrifugation at 15000 rpm and supernatant was utilized to measure the absorbance at 595 nm as compare with control sample. The control sample was prepared by similar procedure used for preparation of test sample except 10% trichloroacetic acid was added before crude enzyme in reaction mixture. 1 unit of enzyme activity equal to the quantity of enzyme that responsible for 0.01 absorbance’s changes at 595 nm under the defined reaction conditions.
Determination Feather degradation
Degradation of feather was determined by visual observation further percentage of feather degradation was calculated, after biodegradation process, the remaining feathers particles were accumulated from whole feather broth medium by filtering through the whatman filter paper No 3. The collected feathers particles were dehydrated at 50°C up to its weight stabilized to constant value and compared to initial weight of feather taken in preparation of whole feather broth medium. The Isolates which effectively degraded feather were selected for further analysis. The percentage of feather degradation was determined using the formula:
Feather degradation (%) = (Initial weight of feathers – Final weight of feathers)/Initial weight of feathers×100 25,26.
Isolation and identification
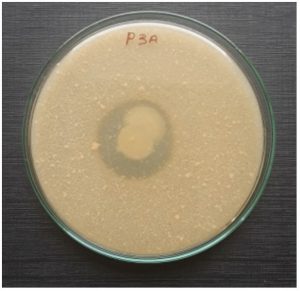
Sixteen isolates exhibited protease activity were confirmed by skim milk agar plates, which were selected from soil sample collected from the feather waste dumping yard on the side of Parande road, out of them nine isolates showed potential for degrading feathers and therefore they were used for further studies. The isolate P3A showed significant feather degradation (Fig 1) was identified by various morphological and biochemical characterizations (Table 1). The strain was Circular, Creamy white, Raised, Translucent, viscid, Gram positive, non motile. According to 16S rRNA analysis the selected bacteria was identified as Staphylococcus epidermidis and designated as Staphylococcus epidermidis P3A. DDBJ had received the isolates under the accession number LC530604. Phylogenic tree showed the detailed relationship between the isolated strain and other homologous species of the genus Staphylococcus epidermidis (Fig 3). Feather degradation process mostly carried out by the gram-positive bacteria27 also the feather-degradation by gram-negative bacteria is noted in few reports28-30. Our results of isolation of feather degrading bacteria support the results of Lin et al.31 they isolated seven proteolytic bacteria among them five bacterial strain had potential of feather degradation identified as Bacillus licheniformis. Several Bacillus sp. including Bacillus subtilis, Bacillus coagulans, Bacillus cereus and Bacillus pumilus have been recognized to be prospective sources of keratinases32. In addition to these, vast variety of keratinase producing microorganism also exists such as Nocardiopsis dassonvillei NRC2aza33, Stenotrophomonas34, Chryseobacterium sp.35, Vibrio sp. kr236 and Arthrobactercreatinolyticus KP01574437.
Table (1):
Biochemical characterizations of P3A isolate.
Sr. no. |
Types of test |
Results obtained |
|---|---|---|
1 |
Glucose |
A |
2 |
Fructose |
A |
3 |
Lactose |
A |
4 |
Maltose |
A |
5 |
Manitol |
N |
6 |
Indole production |
N |
7 |
Methyl red test |
N |
8 |
Voges Proskauer test. |
P |
9 |
Citrate utilization |
N |
10 |
Catalase activity |
P |
11 |
Starch hydrolysis test. |
N |
12 |
Hydrogen sulphide test |
P |
13 |
Urease Activity |
P |
14 |
Gelatine hydrolysis test |
N |
15 |
Oxidase |
N |
16 |
Nitrate reduction |
P |
17 |
Endospore Formation. |
N |
Note: A=acid production; N=Negative, P=Positive.
There are very few reports available on feather degradation by the staphylococcus sp. Venkata and Divakar38 isolated the Staphylococcus aureus, Bacillus cereus and Bacillus licheniformis from poultry waste dumping yard. Feather waste dumping sites are good habitat and best resource for isolation of feather degrading bacteria.
Determination of keratinase activity
Promising isolate P3A effectively degraded the feather by secreting extracellular keratinase enzyme. The maximum keratinase production recoded as 72 U/ml after 96 hrs in whole feather broth medium. The enzyme activity was decreases as per increases the incubation time after 96 hrs (Table 2). Mursheda akhter et al.39 recorded highest keratinase activity 60 U/ml after 96 hrs from the Bacillus cereus and 44 U/ml after 72 hrs from Pseudomonas sp using chicken feather. Similar work of Jahan et al.40 revealed that isolate Z4 showed highest keratinase activity 22.3 U/ml after 72 hrs and complete degradation of feather after 7 days at 37°C. Inamdar et al.41 in his study on the feather degradation reported 50 U/ml maximum keratinase activities from the Bacillus genus. Radha and Gunasekaren42 revealed that the Bacillus licheniformis MKU secreted high amount of keratinase within 48 hrs of growth. It was clear that feather functions as an enzyme inducer in the feather growth medium.
Table (2):
Keratinase production by isolate P3A in whole feather broth.
| Time (hrs) | Keratinase activity (U/ml) |
|---|---|
| 24 | 8 |
| 48 | 19 |
| 72 | 36 |
| 96 | 72 |
| 120 | 26 |
| 144 | 7 |
| Feather degradation 84% after 96 hrs | |
Percentage of feather degradation
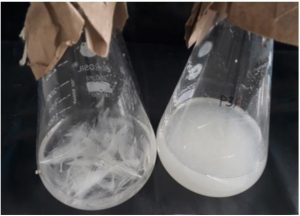
Isolate P3A exhibited the 84% feather degradation after 96 hrs in whole feather broth medium (Fig 2). Nonso et al.43 during their study obtained rate of feather degradation as 86%, 82% 69%, 66% and 62% and 69% from Keratinolytic bacteria Bacillus sp. FPF-1, Chryseobacterium sp. FPF-8, Brevibacillus sp. FSS-1, Brevibacillus sp. Nnolim-K2 and Brevibacillus sp. FPF-12 respectively. Suchitra Godbole et al.44 isolated five feather degrading bacteria identified as Aeromicrobium sp., Exiguobacter sp., Marinococcus sp., Bacillus sp. 1 and Bacillus sp. 2. and reported highest feather degradation 72.55% from Aeromicrobium sp. Mursheda akhter et al.39 revealed that complete degradation of feather was obtained within 5 days. Jendri Mamangkey et al.45 reported newly isolated strain Azotobacter chroococcum B4 capable of degrading whole feathers after 5 days. Similar work done by Mehta et al.46 they reported that entire degradation of feather waste within 7 days by the Bacillus sonorensis strain NRRLB-23154 isolated from poultry farm. While Shah and Vaidya8 recorded 70% degradation after 6 days by Acinetobacter sp. PD12 isolated from feather waste dumping area.
In the current reports we have attempted to isolate and classify a novel bacterial strain capable for degradation of keratinous wastes into soluble and useful products. Many bacterial members from the genus Bacillus47 and Streptomyces sp.48,18,49 were selected from poultry chicken feathers waste, these bacteria degrade feather keratin for utilization carbon, nitrogen and sulfur as primary source for their growth. Similar work done by Suntornsuk et al.50 they observed that the isolate Bacillus licheniformis FK14 degrade the feather within 5 days at 50°C. Giongo et al.51 reported that keratinolytic Bacillus sp. selected from Brazilian Amazon basin shows significant degradation after 72 hrs. We conclude from the results of this study that chicken feather waste dumping sites are the rich source of feather degrading bacteria. Feather degrading bacteria utilize the feather keratin protein for their growth.
From the discussion it might be concluded that a novel gram positive Staphylococcus epidermidis P3A was isolated from feather waste discarding area from Barshi town. The bacteria effectively utilize feather keratin substrate as the predominant resource of energy, nitrogen, carbon, and sulphur. The isolated strain can be applicable for reduction of environmental pollution arises due to poultry chicken feather waste.
ACKNOWLEDGMENTS
Authors sincerely thanks to Triyat Genomics Laboratory Nagpur India for 16S rRNA sequencing analysis. ARJ acknowledges to BARTI Pune for PhD fellowship for the year 2015-2017.
CONFLICT OF INTEREST
The authors declare that there is no conflict of interest.
AUTHORS’ CONTRIBUTION
ARJ: concept of the main idea, complied information from the literature, performing the experiment, drafted the manuscript. MGB: verifying the analytical methods supervised findings of this project.
FUNDING
Authors are thankful to the BARTI Pune for M.J.P. National Research Fellowship (2015-2017).
ETHICS STATEMENT
This article does not contain any studies with human participants or animals performed by any of the authors.
AVAILABILITY OF DATA
All datasets generated or analyzed during this study are provided in this manuscript. 16S rRNA gene sequences generated during this study was deposited in the DNA Data Bank of Japan under accession numbers: LC530604.
- Bhavy KS, Raji P, Selvarani AJ, Samrot AV, Javad PTM, Appalaraju VVSS. Leather processing, its effects on environment and alternatives of chrome tanning. International Journal of Advanced Research In Engineering And Technology. 2019;10:69-79.
Crossref - Samrot AV, Sahithya CS, Selvarani, AJ, Pachiyappan S, Kumar SS. Surface-Engineered Super-Paramagnetic Iron Oxide Nanoparticles for Chromium Removal. Int J Nano Med. 2019;14;8105-8119.
Crossref - Raji P, Samrot AV, Bhavya KS, Sharan M, Priya S, Paulraj P. Greener Approach for Leather Tanning Using Less Chrome with Plant Tannins and Tannins Mediated Nanoparticles. J Clust Sci. 2019;30:1533-1543.
Crossref - Samrot AV, Shobana N, Sruthi DP. Sahithya CS. Utilization of chitosan coated superparamagnetic iron oxide nanoparticles for chromium removal. Appl Water Sci. 2018;8:192.
Crossref - Li Z, Reimer C, Picard M, Mohanty AK, Misra M. Characterization of chicken feather biocarbon for use in sustainable biocomposites. Front Mater. 2020;7(3):1-12.
Crossref - Thyagarajan D, Barathi M and Sakthivadivu R. Scope of Poultry Waste Utilization. IOSR Journal of Agriculture and Veterinary Science. 2013;6(5):29-35.
Crossref - Callegaro K, Brandelli A, Daroit D J. Beyond plucking: feathers bioprocessing into valuable protein hydrolysates. Waste Manag. 2019;95:399-415.
Crossref - Shah M, Vaidya RB. A novel feather degrading Acinetobacter sp. PD 12 isolated from feather waste dumping site in Mumbai. European Academic Research. 2015;3(1):757-773.
- Martinez JPDO, Cai G, Nachtschatt M, et al. Challenges and opportunities in identifying and characterising keratinases for value-added peptide production. Catalysts. 2020;10(2);1-23.
Crossref - Arun kumar JM, Lakshmi A, Sangeetha Rani V, Sailaja B. Isolation and characterization of feather degrading bacteria from poultry waste. Journal of Research in Biology. 2012;2(7):676-682.
- Bradbury JH. Structure and chemistry of keratin fibers. Adv Protein Chem. 1973;27:111- 211.
Crossref - Karthikeyan R, Balaji S, Sehgal PK. Industrial application of keratins – A review. J Sci Ind Research. 2007;66:710-715.
- Muthusamy G, selvakumar T, Arunprakash S. Production of keratinolytic Enzyme by a Newly Isolated feather degrading Bacillus sp. From Chick Feather Waste. Internatioal Journal of Pharma and Bio Sciences. 2011;2(3):256-265.
- Gradisar J, Fridrich I, Krizal, Jerala R. Similarities and specification of fungal keratinolytic protease: comparison of keratinase pf Paccilomyces marquandi and Doratomyces microspores to some known protease. Appl Environ Microbiol. 2005;71:3420-3426.
Crossref - Cai C, Chen J, Qi J, Yin Y, Zheng X. Purification and characterization of keratinase from a new Bacillus subtilis strain. J Zhejiang Univ Sci B. 2008;9(9):713-720.
Crossref - Lin X, Inglis GD, Yanke LJ, Cheng K J. Selection and characterization of feather degrading bacteria from canola meal compost. J Ind Microbiol Biotechnol. 1999;23:149-153.
Crossref - Kim JM, Lim WJ, Suh HJ. Feather-degrading Bacillus species from poultry waste. Process Biochemistry. 2001;37:287-291.
Crossref - Bressolier P, Letourneau F, Urdaci M, Verneuil B. Purification and characterization of a keratinolytic serine proteinase from Streptomyces albidoflavus. Appl Environ Microbiol. 1999;65:2570-2576.
Crossref - Brandelli A. Bacterial Keratinases: Useful Enzymes for Bioprocessing Agroindustrial Wastes and Beyond. Food Bioprocess Technol. 2008;1:105-116.
Crossref - Kainoor PS, Naik GR. Production and characterization of feather degrading keratinase from Bacillus sp. JB 99. Indian Journal of Biotechnology. 2010;9:384-390.
- Brenner DJ, Krieg NR, Staley JT. Bergey’s manual of systematic bacteriology (2nd Ed) Part B Springer, New York. 2004;2:323-358.
- Sivakumar T, Shankar T, Thangapandian V, Ramasubramanian V. Optimization of Cultural Condition for Keratinase Production Using Bacillus cereus TS1. Insight Microbiology. 2013;3(1):1-8.
Crossref - Korkmaz H, Hur H, Dincer S. Characterization of alkaline keratinase of Bacillus licheniformis strain HK-1 from poultry waste. Annals of Microbiology. 2004;54(2):201- 211.
- Poopathi S, Thirugnanasambantham K, Mani C, Lakshmi P V, Ragul K. Purification and characterization of keratinase from feather degrading bacterium useful for mosquito control – A new report. Tropical Biomedicine. 2014;31(1):97-109.
- Shabaan MT, Attia M, El-Sabagh and Amany AM Ahmed, Isolation, Screening and Selection of Efficient Feather Degrading Bacteria. Current Science International. 2014;3(4):488-498.
- Huang Y, Busk PK, Lange L. Production and Characterization of Keratinolytic Proteases Produced by Onygena corvina. Fungal Genomics Biol. 2015;4(1):119.
Crossref - Gupta R, Ramnani P. Microbial keratinases and their perspective applications: an overview. Appl Microbiol Biotechnol. 2006;70:21-33.
Crossref - Brandelli A, Daroit DJ, Riffel A Biochemical features of microbial keratinases and their production and applications. Appl Microbiol Biotechnol. 2010;85:1735-1750.
Crossref - Tork S, Aly MM, Nawa L Biochemical and molecular characterization of a new local keratinase producing Pseudomomanas sp. MS21. Asian J Biotechnol. 2010;2(1): 1-13.
Crossref - Bach E, Daroit DJ, Correa APF, Brandelli A. Production and properties of keratinolytic proteases from three novel Gram-negative feather-degrading bacteria isolated from Brazilian soils. Biodegradation. 2011;22:1191-1201.
Crossref - Lin XD, Kelemen W, Miller ES, Shih JCH. Nucleotide sequences and expression of kerA, the gene encoding keratinolytic proteases of bacillus licheniformis PW-1. Applied Environ Microbiol. 1995;61:1469-1474.
Crossref - Femi-Ola TO, Akinbobola OS, Oluwaniyi TT. Isolation and characterization of feather degrading bacteria from poultry soil. Agriculture and Biology Journal of North America. 2015;146-154.
- Azza M Abdel-fattah A. Novel keratinase from marine Nocardiopsis dassonvillei NRC2aza exhibiting remarkable hide dehairing. Egyptian Pharmaceutical Journal. 2013;12:142-147.
Crossref - Cao Z-J, Zhang Q, Wei D-K, et al. Characterization of a novel Stenotrophomonas isolate with high keratinase activity and purification of the enzyme. J Ind Microbiol Biotechnol. 2009;36:181-8.
Crossref - Brandelli A, Riffel A. Production of an extracellular keratinase from Chryseobacterium sp. growing on raw feathers. Electronic Journal of Biotechnology. 2005;8(1):35-42.
Crossref - Grazziotin A, Pimentel FA, de Jong EV, Brandelli A. Nutritional improvement of feather protein by treatment with microbial keratinase. Anim Feed Sci Technol. 2006;126:135-144.
Crossref - Kate S, Pethe A. Study of efficiency of Keratinase production by Arthrobacter creatinolyticus KP015744 isolated from leather sample. International Journal of Advanced Research. 2014;2(11):992-999.
- Venkata NR, Divakar G. Screening and isolation of keratinase producing bacteria from poultry waste. Int J Pharm Res All Sci. 2013;2(1):70-74.
- Akhter M, Marzan LW, Akter Y, Shimizu K. Microbial Bioremediation of Feather Waste for Keratinase Production: An Outstanding Solution for Leather Dehairing in Tanneries. Microbiology Insights. 2020;13: 1-12.
Crossref - Jahan Z, Khan SN, Mozammel Hoq M.Screening of keratinolytic bacteria from Poultry Wastes Bangladesh. Journal of Scientific and Industrial Research. 2010;45(3):261-266.
Crossref - Inamdar A, Sahera N, Siddiqui R. Screening and production of extra cellular feather degrading enzyme from bacterial isolates. Indian J L Sci. 2012;1(2):19-24.
- Rhadha S, Gunasekaren P. Sustained expression of keratinase gene under PxyLA and PamyL promotores in the recombinant Bacillus megaterium MS941. Bioresour Technol. 2008;99:5528-5537.
Crossref - Nonso EN, Anthony IO, Uchechukwu UN. Bacillus sp. FPF-1 Produced Keratinase with High Potential for Chicken Feather Degradation. Molecules. 2020;25:1505; 1-16.
Crossref - Godbole S, Pattan J, Gaikwad S, Jha T. Isolation, Identification and Characterization of Keratin degrading microorganisms from Poultry soil and their Feather degradation Potential. International Journal of Environment, Agriculture and Biotechnology. 2020;2(4):2060-2068.
Crossref - Jendri M, Dwi S, Erman M and Apon Z M. Keratinase Activity of A Newly Keratinolytic Bacteria, Azotobacter chroococcum B4. 2020; J Pure Appl Microbiol. 14(2):1203-1211.
- Mehta RS, Jholapara RJ, Sawant CS. Isolation of A Novel Feather-Degrading Bacterium and Optimization of It’s of Its Cultural Conditions for Enzyme Production. Int J Pharm Pharm Sci. 2014;6(1):194-201.
- Lin X, Lee CG, Casale ES, Shih JCH. Purification and characterization of a keratinase from a feather-degrading Bacillus licheniformis strain. Appl Environ Microbiol. 1992;58:3271-3275.
Crossref - Bockle BB, Galunsky, Muller R. Characterization of a keratinolytic serine proteinase from Streptomyces pactum DSM 40530. Appl Environ Microbiol. 1995;61:3705-3710.
Crossref - Kansoh AL, Hossiny EN, Abd E K, El-Hameed. Keratinase production from feathers wastes using some local Streptomyces isolates. Aust J Basic Applied Sci. 2009;3:561-571.
- Suntornsuk W, Tongjun J, Onnim P, et al. Purification and characterization of keratinase from a thermotollerent feather-degrading bacterium. World J Microbial. Biotechnol. 2005;21:1111-1117.
Crossref - Giongo JL, lucas FS, Casarin F, Heeb P, Brandelli A. Keratinolytic Protease of Bacillus species isolated from the Amazon basin showing remarkable dehairing activity. World J Microbial Biotechnol. 2007;70:21-33.
© The Author(s) 2020. Open Access. This article is distributed under the terms of the Creative Commons Attribution 4.0 International License which permits unrestricted use, sharing, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.