ISSN: 0973-7510
E-ISSN: 2581-690X
Neonatal Bacteremia is the presence of live bacteria circulating in the blood of neonates. The bloodstream infections are important cause of neonatal mortality and morbidity. This study was done to know about the bacterial agents causing neonatal Bacteremia and their antibiotic sensitivity pattern. Blood cultures were done on clinically suspected bloodstream infections in neonates for a period of 18 months. 200 neonatal blood samples were cultured by conventional method. Bacterial isolates were identified and their antibiotic susceptibility testing were done. Out of 200 blood samples 88 samples (44%) showed positive blood culture. Blood culture was positive in 51 males (57.95%) and 37 females (42.1%). Among the positive blood culture samples, 66 bacterial isolates were Gram positive (75%) and 22 isolates were Gram negative (25%). Staphylococcus aureus (54.5%) was the most common bacteria isolated followed by coagulase negative Staphylococcus species(20.5%).Among the Gram negative organisms, E.coli was isolated in 13.6% of positive blood culture samples followed by Klebsiella pneumonia (9.1%) and Pseudomonas sp.(2.3%). Gram positive organisms were highly resistant to oxacillin (88.8-89.61%). They were sensitive to Linezolid (93.75-94.5%), Teicoplanin (83.4-87.5%), Vancomycin(81.25-83.33%) and Amikacin(81.25-83.4%). Gram negative organisms were mostly resistant to third generation cephalosporins and gentamycin. They were mostly sensitive to amikacin and piperacillin. Prompt treatment of positive blood culture neonates with appropriate antibiotics can prevent the complications and can reduce the mortality.
Bacteremia, coagulase negative Staphylococcus, blood culture, Antibiotic susceptibility testing, multidrug resistance.
First 28 days of life after birth is called the neonatal period and bloodstream infections occurring in this period can lead to significant morbidity and mortality in neonates. Bacteremia in neonates is associated with fever, refusal of feeds, tachycardia, tachypnoea, convulsions, bulging fontanelle, hypoglycemia and reduced movements. Bacteremia occurring in early neonatal period may be due to maternal factors like early rupture of membranes or intrapartum infections. Bacteremia occurring in late neonatal period may be community acquired or hospital acquired infections1. Neonatal bacteremia is commonly caused by Group B Streptococci, E.coli, Staphylococcus aureus, CONS, Listeria monocytogenes, Klebsiella sp. and other Gram negative bacteria. Bacteremia in late neonatal period is commonly associated with Staphylococcus sp. and Gram negative bacteria like E.coli and Klebsiella sp. These organisms are prevalent in the hospital environment and contact with colonized persons of Staphylococcus sp. may be the source of infection. Neonates with assisted ventilation may be of risk with Pseudomonas infections2. MRSA and ESBL producers and multidrug resistant Gram negative organisms are to be considered in the treatment of neonatal bacteremia. This study was conducted to know about the bacteriological agents causing neonatal bacteremia and their susceptibility pattern. This will help in the treatment of neonatal bloodstream infections which is one of the leading cause of neonatal morbidity and mortality3.
Study period
18 months from Jan 2016 to Jun 2017.
Type of study
cross sectional prospective study
Sample size
200 neonatal blood culture samples
Blood culture samples.
Total |
culture positive |
culture negative |
|---|---|---|
N(%) |
N(%) |
|
200 |
88 (44%) |
112 (66%) |
Inclusion criteria
Neonates with features suggestive of bloodstream infections associated with systemic inflammatory response with no localized focus of infection.
Exclusion criteria
Neonates who had received prior antibiotics and birth weight less than 1000 gms.
Neonatal risk factors
Low birth weight, prematurity, meconium aspiration, birth asphyxia, indwelling intravenous canula for more than 10 days and continuous positive airway pressure.
Maternal risk factors
Early rupture of membranes, genitourinary infections, recurrent abortions and amniocentesis.
Procedure
After proper hand washing, the skin over the venepuncture site is disinfected with povidone solution and 70% ethyl alcohol. The flip-off cap in the neonatal blood culture bottle is removed and disinfected with spirit. With strict aseptic precautions 2 ml of blood is collected from the neonates and with the same syringe the blood is transferred into the blood culture bottle with 20 ml of BHI broth4,5. The blood culture bottle is immediately transported to the Microbiology laboratory and it should be never refrigerated. It is incubated at 37 degree C and daily examined for the presence of bacterial growth. Bacterial growth is indicated by the presence of turbidity in the broth and hemolysis. Subcultures are made from the broth daily for first three days and then alternate days upto seven days. Subcultures are made in blood agar and MacConkey agar. Bacterial isolates are identified by colony characteristics, Gram staining, growth on special media and biochemical properties like catalase and oxidase test, coagulase test, motility, IMViC test, urease test, TSI agar test. Antibiotic susceptibility testing is done by Kirby Bauer method with specific antibiotic discs. Bacterial inoculum is compared with 0.5 McFarland standard and lawn culture is streaked on Muller Hinton agar6,7. The results are interpreted as per CLSI guidelines. ATCC strains are used as standard strains.
Culture positive organisms.
Total |
males |
females |
|---|---|---|
N (%) |
N (%) |
|
88 |
51(57.95%) |
37(22.1%) |
Culture positive organisms.
Total |
Gram positive |
Gram negative |
|---|---|---|
N(%) |
N(%) |
|
88 |
66 (75%) |
22 (25%) |
Quality control
Uninoculated blood culture bottle is also incubated and looked for any bacterial growth.
Data analysis
Chi-square test was used. P value of 0.05 or less than that was considered as statistically significant.
Antibiotic discs used
Gram positive organisms
Oxacillin (5µg), erythromycin (15 µg), vancomycin (30µg), linezolid (30µg), gentamycin (10µg), amikacin (30µg), ciprofloxacin (5µg), clindamycin (10µg), chloramphenicol (30µg) and teicoplanin (30µg).
Gram positive organisms.
Total |
Staphylococcus aureus |
CONS |
|---|---|---|
N(%) |
N(%) |
|
66 |
48(72.73%) |
18(37.5%) |
Table (1):
Drug sensitivity pattern for Gram positive organisms.
Drug |
Staphylococcus aureus (48) N (%) |
CONS (18) N (%) |
|---|---|---|
Oxacillin |
5 (10.41) |
2 (11.2) |
Erythromycin |
4 (8.3) |
2 (11.2) |
Vancomycin |
39 (81.25) |
15 (83.33) |
Linezolid |
45 (93.75) |
17 (94.5) |
Gentamycin |
15 (31.25) |
4 (32.3) |
Amikacin |
39 (81.25) |
15 (83.4) |
Ciprofloxacin |
25 (52.1) |
10 (65.6) |
Clindamycin |
39 (81.25) |
15 (83.4) |
Chloramphenicol |
25 (52.1) |
10 (55.55) |
Teicoplanin |
42 (87.5) |
15 (83.4) |
Gram negative organism
ciprofloxacin (5µg), gentamicin (10µg), amikacin (30µg), Cefotaxime (30µg), Cefoperazone (75µg), trimethoprim-sulfamethoxazole (1.25/23.75 µg),nalidixic acid
(30µg), Ampicillin (10µg), Piperacillin (100µg) and chloramphenicol (30µg).
Gram negative organisms.
Total |
E. coli |
Klebsiella sp. |
Pseudomons sp. |
N(%) |
N(%) |
N(%) |
|
22 |
12 (54.5%) |
8 (36.36%) |
2 (9.09%) |
Table (2):
Drug resistance pattern for Gram positive organisms.
Drug |
Staphylococcus aureus (48) N (%) |
CONS(18) N(%) |
|---|---|---|
Oxacillin |
43(89.6) |
16(88.88) |
Erythromycin |
44(91.66) |
16(88.88) |
Vancomycin |
9(37.5) |
3(16.7) |
Linezolid |
3(6.25) |
1(5.55) |
Gentamycin |
33(68.75) |
14(77.77) |
Amikacin |
9(18.75) |
3(16.66) |
Ciprofloxacin |
23(47.9) |
8(44.44) |
Clindamycin |
9(18.75) |
3(16.66) |
Chloramphenicol |
23(47.92) |
8(16.6) |
Teicoplanin |
6(12.5) |
3(16.6) |
Out of 200 neonatal blood culture samples, 88 samples (44%) showed positive blood culture. In the positive blood culture samples, 51 were males (57.95%) and 37 (42.05%) were females8.
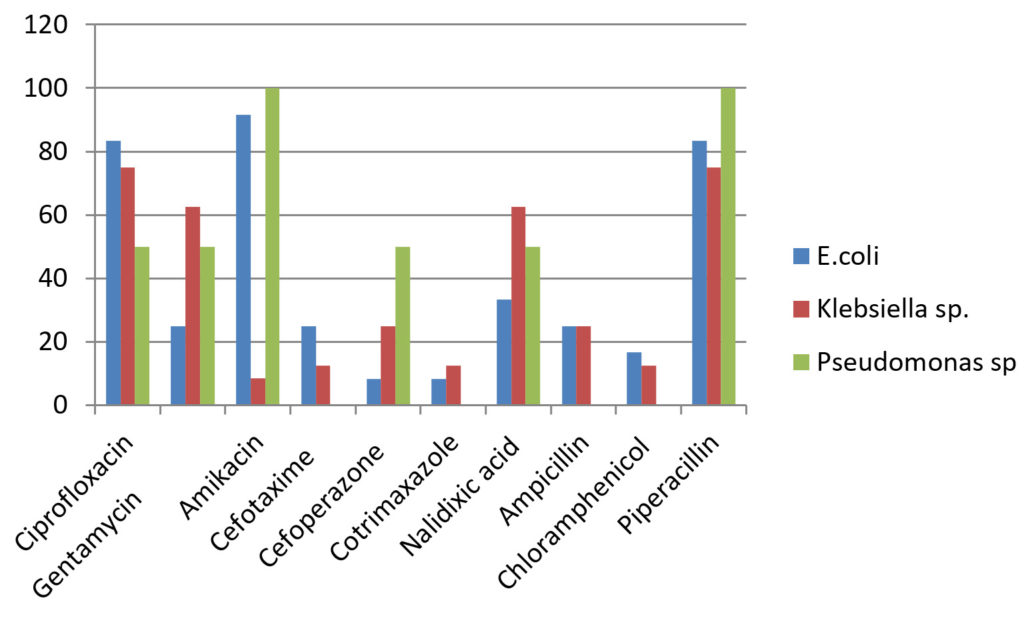
Table (3):
Drug sensitivity pattern for Gram negative organisms.
Drug |
E.coli (12) N (%) |
Klebsiella sp.(8) N (%) |
Pseudomonas sp.(2) N (%) |
|---|---|---|---|
Ciprofloxacin |
10(83.3) |
6(75) |
1(50) |
Gentamycin |
3(25) |
5(62.5) |
1(50) |
Amikacin |
11(91.66) |
7(87.5) |
2(100) |
Cefotaxime |
3(25) |
1 (12.5) |
(0) |
Cefoperazone |
1(8.33) |
2(25) |
1(50) |
Cotrimaxazole |
1(8.33) |
1(12.5) |
(0) |
Nalidixic acid |
4(33.33) |
5(62.5) |
1(50) |
Ampicillin |
3(25) |
2(25) |
(0) |
Chloramphenicol |
2(16.66) |
1(12.5) |
(0) |
Piperacillin |
10(83.33) |
6(75) |
2(100) |
Neonates presented with fever, respiratory distress, tachycardia, refusal of feeds and jaundice.
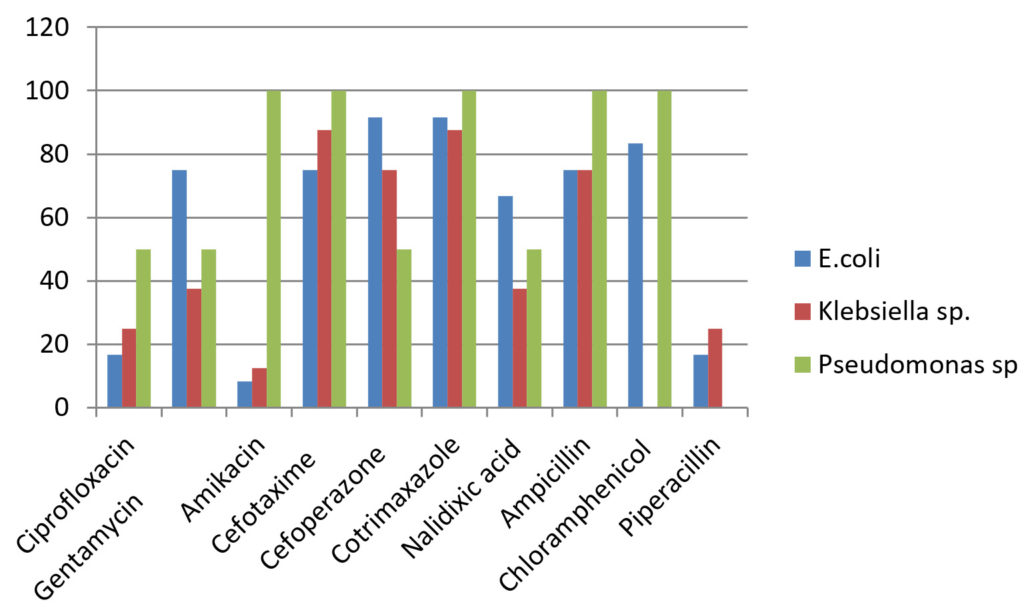
Table (4):
Drug resistance pattern of Gram negative organisms.
Drug |
E.coli (12) N (%) |
Klebsiella sp.(8) N (%) |
Pseudomonas sp.(2) N (%) |
|---|---|---|---|
Ciprofloxacin |
2(16.6) |
2(25) |
1(50) |
Gentamycin |
9(75) |
3(37.5) |
1(50) |
Amikacin |
1(8.33) |
1(12.5) |
– (0) |
Cefotaxime |
9(75) |
7(87.5) |
2(100) |
Cefoperazone |
11(91.66) |
6(75) |
1(50) |
Cotrimaxazole |
11(91.66) |
7(87.5) |
2(100) |
Nalidixic acid |
8(66.66) |
3(37.5) |
1(50) |
Ampicillin |
9(75) |
6(75) |
2(100) |
Chloramphenicol |
10(83.33) |
7(87.5) |
2(100) |
Piperacillin |
2(16.66) |
2(25) |
(0) |
In the positive blood culture samples, 66 (75%) were Gram positive organisms and 22 (25%) were Gram negative organisms. Among the Gram positive organisms, Staphylococcus aureus were isolated in 48 samples (72.72%) and CONS were isolated in 18 samples (27.27%). Among the Gram negative organisms, E. coli were isolated in 12 samples (54.55%) and Klebsiella sp. were isolated in 8 samples (36.36%) and Pseudomonas sp. were isolated in 2 samples (9.1%). CONS were considered as pathogenic organism if there was history of prior antibiotic administration, the body temperature of the neonate was one degree more than or less than 37 degree or prolonged intravenous catheterization9,10.
Neonatal bacteremia is difficult to diagnose clinically as the presentation is mostly nonspecific. Blood culture remains the gold standard method for the lab diagnosis.
The common neonatal risk factors in this study were prematurity, low birth weight and prolonged intravenous catheterisation. Immature immune system may be the reason for the occurrence of bacteremia in premature and low birth weight neonates. Maternal risk factors include genitourinary infections and premature rupture of membranes11.
Bloodstream infections in early neonatal period may be of maternal origin like early rupture of membranes or genitourinary infections. Bacteremia in late neonatal period is mostly community acquired or hospital acquired. Neonates with prolonged stay at NICU are exposed to the hospital flora. Skin and mucosal carriers of hospital personnel and baby handlers may transmit the infections to the neonates. Staphylococcus epidermidis may cause bloodstream infections because of biofilm formation in intravenous catheters. Skin damage occurring during catheter insertion inhibits TLR2-dependent pro-inflammatory cytokine production and this will allow S. epidermidis to adhere, proliferate and migrate along catheters with minimum interference from host defenses12. This is common in preterm neonates. Lipoteichoic acid and lipid S present in Staphylococcus cell wall has got antiphagocytic properties. This may be the reason for high occurrence of Gram positive organisms in neonatal bloodstream infections13.
Neonates presented with fever, respiratory distress, refusal of feeds and jaundice.
In this study, out of 200 neonatal blood culture samples, 88 (44%) were blood culture positive. This percentage was comparable to other studies like Roy et al and Kayange et al.
In the present study 47% were low birth weight neonates and 35% were preterm neonates in the positive blood culture samples. This may be because of the immature immune system in these neonates14.
In the positive blood culture samples, the percentage of males (57.95%) was higher than females (42.05%).This was comparable to other studies like Begum et al and Sreshtha et al. This may be due to X linked gene regulation of gamma globulin synthesis. The figures 1,2,3 and 4 shows the comparison of different types of drugs with gram positive and negative organism with graphical representation.
The tabular 1,2,3 and 4 are shows the isolation rate of Gram positive organisms (75%) was higher than Gram negative organisms (25%). This is in concordance with other studies like Ballot et al ,Kaufman and Fairchild and Hoogen et al. Among the Gram positive organisms, Staphylococcus aureus was the predominant isolate (72.73%) followed by CONS(27.3%).This may be due to colonization of skin and mucosa in hospital personnel’s and improper hand washing techniques. Among the Gram negative organisms, the isolation rate of E. coli was 54.5% followed by Klebsiella sp.(36.36%) and Pseudomonas sp. was isolated in 9.09%15,16.
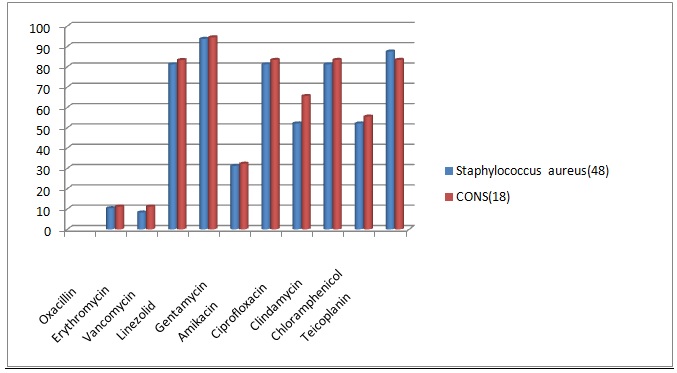
Fig. 1. Comparison of Drug sensitivity pattern for Gram positive organisms
Most of the Gram positive organisms showed oxacillin resistance. Staphylococcus aureus showed 89.6% and CONS showed 88.88% oxacillin resistance. They also showed high resistance to erythromycin, gentamycin, ciprofloxacin and chlramphenicol. Vancomycin resistance rate was 18.7% for Staphylococcus aureus and 16.7% for CONS. In this study, Staphylococcus aureus was sensitive to clindamycin in 81.25% and amikacin in 81.25%.It showed the highest sensitivity towards teicoplanin and linezolid. CONS also showed similar sensitivity pattern17.
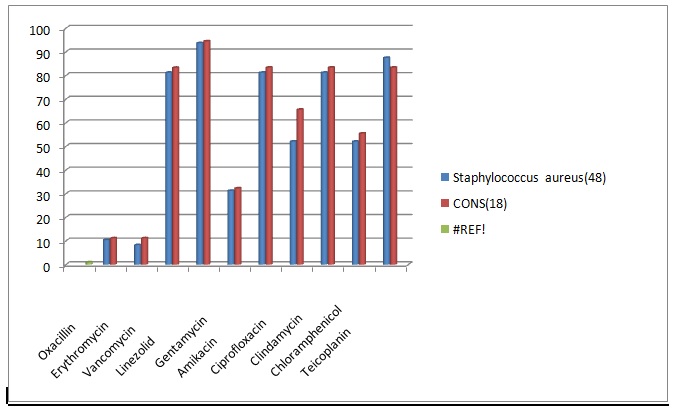
Fig. 2. Comprison of Drug resistance pattern for Gram positive organisms
Gram negative isolates like E.coli and Klebsiella sp. showed higher resistance rate towards gentamycin (37.5-75%), third generation cephalosporins (75-95.66%), nalidixic acid(37.5-75%) and chloramphenicol(83.33-87.5%)18.The sensitive rate of E.coli for amikacin was 91.66% and for piperacillin was 83.33%. Klebsiella sp. showed higher sensitivity to amikacin (87.5%). Pseudomonas sp. was resistant to gentamycin (50%), ciprofloxacin 50%), cefaperazone (50%) and cefataxime (100%). It showed 100% sensitivity towards piperacillin. The higher resistance rate of organisms may be due to injudicious use of antibiotic drugs and bacterial mechanisms of drug resistance19.
In this study Gram positive organisms were the predominant isolates in blood stream infections in neonates. Staphylococcus aureus and CONS showed higher resistance rates towards oxacllin. Gram negative organisms showed higher resistant rate towards third generation cephalosporins and gentamycin. Gram negative organisms were more sensitive towards amikacin and piperacillin. Proper hand washing and treatment of Staphylococcal carriers will reduce the incidence of blood stream infections in neonates. Proper selection of antibiotic drugs after blood culture and antibiotic susceptibility testing should be implemented and infection control policy should be formed to reduce bacteremia in neonates.
- Sankar MJ, Agarwal R, Deorari AK, Paul VK. Sepsis in the newborn. Indian J Pediatr. 2008; 75(3):261–266. doi: 10.1007/s12098-008-0056-z.
- Lever A, Mackenzie I. Sepsis: definition, epidemiology, and diagnosis. Br med J. 2007; 3(7):879. doi: 10.1136/bmj.39346.495880.AE
- Shaha C, Dey S, Shabuj H, Chisti J, Mannan M, Jashimuddin M, et al. Neonatal sepsis review. Bangladesh J Child Health. 2012; 36(2):82–89. doi: 10.3329/bjch.v36i2.13084.
- Zakariya B, Bhat V, Harish N, Babu T, Joseph M. Neonatal sepsis in a tertiary care hospital in South India: bacteriological profile and antibiotic sensitivity pattern. Indian J Pediatr. 2011;78(4):413–417. doi: 10.1007/s12098-010-0314-8. 5. Tallur S, Kasturi A, Nadgir S, Krishna B. Clinico-bacteriological study of neonatal septicemia in Hubli. Indian J Pediatr. 2000; 67(3):169–174. doi: 10.1007/BF02723654.
- Karunasekera K, Pathirana D. A preliminary study on neonatal septicaemia in a tertiary referral hospital paediatric unit. Ceylon Med J. 1999; 44(2):81–86.
- Shah BA, Padbury JF. Neonatal sepsis: An old problem with new insights. Virulence. 2014; 5(1):170-78. doi:10.4161/viru.26906.
- Simonsen KA, Anderson-Berry AL, Delair SF, Davies HD. Early-onset neonatal sepsis. Clin Microbiol rev. 2014; 27(1):21–47. doi: 10.1128/CMR.00031-13.
- Shaw G, Sim K, Randell P, Cox J, McClure Z, Li M-S, et al. Late-Onset Bloodstream Infection and Perturbed Maturation of the Gastrointestinal Microbiota in Premature Infants. PloS one. 2015; 10(7):e0132923.
- Shah GS, Budhathoki S, Das BK, Mandal RN. Risk factors in early neonatal sepsis. Kathmandu Univ med J. 2006; 4(2):187–191.
- Jyothi P, Basavaraj C, Basavaraj P. Bacteriological profile of neonatal septicemia and antibiotic susceptibility pattern of the isolates. J Nat sc Biol med. 2013; 4(2):306. doi: 10.4103/0976-9668.116981.
- Clinical and Laboratory Standards Institute (CLSI). Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically: approved standard-Tenth Edition. CLSI document M07-A10. Wayne: CLSI; 2015.
- Shehab E, El-Sokkary A, Bassiouny R, Hassan R. Epidemiology of neonatal sepsis and implicated pathogens: a study from Egypt. Biomed res Int. 2015; 509484(10):4.
- Khanal R, Manandhar S, Acharya G. Bacteriological profile of neonatal sepsis in a tertiary level Hospital of Nepal. J Nepal Paediatr soc. 2015; 34(3):175–180. doi: 10.3126/jnps.v34i3.9183.
- Gebrehiwot A, Lakew W, Moges F, Anagaw B, Yismaw G, Unakal C. Bacterial profile and drug susceptibility pattern of neonatal sepsis in Gondar University hospital, Gondar Northwest Ethiopia. Der Pharmacia Lettre. 2012; 4(6):1811–1816.
- Khan M, Khan A, Shah F, Munir A. Neonatal sepsis: a study of causative pathogens and their antimicrobial sensitivity pattern at tertiary hospital. Gomal J Med Sci. 2012; 10(2):244–7.
- Jimenez N, Tedeschi S, Saye EJ, McKenna BD, Langdon W, Wright JP, et al. Relationship between maternal and neonatal Staphylococcus aureuscolonization. Pediatrics. 2012;129(5):1252–1259. doi: 10.1542/peds.2011-2308. 18.Aamir M, Ali E, Hamouda M, Mourad F. Prevalence of multi drug resistant bacteria causing late-onset neonatal sepsis. Int J Curr Microbiol app Sci. 2015; 4(5):172–190.
- Kayange N, Kamugisha E, Mwizamholya DL, Jeremiah S, Mshana SE. Predictors of positive blood culture and deaths among neonates with suspected neonatal sepsis in a tertiary hospital, Mwanza-Tanzania. BMC pediatrics. 2010; 10(1):1. doi: 10.1186/1471-2431-10-39.
- Aamir M, Ali E, Hamouda M, Mourad F. Prevalence of multi drug resistant bacteria causing late-onset neonatal sepsis. Int J Curr Microbiol app Sci. 2015;
- Kayange N, Kamugisha E, Mwizamholya DL, Jeremiah S, Mshana SE. Predictors of positive blood culture and deaths among neonates with suspected neonatal sepsis in a tertiary hospital, Mwanza-Tanzania. BMC pediatrics. 2010; 10(1):1. doi: 10.1186/1471-2431-10-39.
© The Author(s) 2017. Open Access. This article is distributed under the terms of the Creative Commons Attribution 4.0 International License which permits unrestricted use, sharing, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.