ISSN: 0973-7510
E-ISSN: 2581-690X
Sepsis, the second leading cause of death is due to infections. Intensive care units (ICUs) are having the highest burden of treating the patients with sepsis and nosocomial infections compared to other areas of hospitals. Our objective was to identify the bacteriological profile and their antibiogram of sepsis cases in all ICUs. A sum of 102 blood samples were collected from patients with clinically suspected sepsis with elevated CRP. Processed by an automated method using Bact/Alert & growth were identified by Standard guidelines. Out of 102 samples, 54 (53%) were shown positive by culture. Gram-negative bacilli were the predominant and their number were 33 (61.1% ) and the commonest organisms were from the Enterobacteriaceae family. Escherichia coli was the highest number with 15 (27.7%) followed by Klebsiella pneumoniae 10 (18.51%), & the rest were single isolates of Salmonella typhi, Proteus mirabilis and Citrobacter koseri. Nonfermenter isolated were Acinetobacter baumanii 3 (5.6%), Pseudomonas aeruginosa 2 (3.7%). The Gram-positive cocci were 17 & 32.4% of culture positivity. Coagulase-negative Staphylococcus was the highest isolated accounting for 9 (16.6%) followed by Staphylococcus aureus 6 (11.1%) and Enterococcus faecalis (3.7%). Culture positivity will be more when CRP is also included in the selection of samples for sepsis and Gram-negative bacilli are the leading cause in septicemia and organisms belonging to the Enterobacteriaceae family still dominate in septicemia infections in ICUs and a real challenge for treatment are MDRs which needs to be detected regularly by using screening tests.
Intensive Care Units, Sepsis, C-reactive protein, Bacteriological profile, Antibiogram, Resistance
Infection that triggers the inflammatory syndromic response leads to sepsis.1 It is a life threatening cause of Blood Stream Infection (BSI) with high morbidity and mortality globally2 and 2nd leading cause of death. Nosocomial infections are more prone for patients who admitted in Intensive Care Units (ICUs).3 The most common cause of infection is due to bacteria followed by fungi, virus and protozoans.4 In 2017, World Health Organization (WHO) estimated that out of 48.9 million cases, 11 million sepsis-related death occurred globally, especially in children below 5 years of age – global sepsis estimated were 20 million cases in that 2.9 million deaths occurred worldwide.5 Gram Positive Bacteria (GPB) along with Gram Negative Bacteria (GNB) developed resistance against antimicrobial agents.6 Coagulase negative Staphylococcus (CoNS) were the predominant GPB,7 Escherichia coli were the common GNB found in BSI.8 Some of the resistant pathogens were Extended Spectrum beta lactamases (ESBL) Escherichia coli, and other Enterobacteriaceae spp., metallo-beta-lactamase producing Pseudomonas aeruginosa, Methicillin resistant Staphylococcus aureus (MRSA), AmpC β-lactamases & Carbapenemase producing Klebsiella pneumoniae, Vancomycin Intermediate Staphylococcus aureus (VISA), Vancomycin Resistant Enterococcus (VRE).9 Our objective was to identify the bacteriological profile and their antibiogram of sepsis cases in all ICUs.
A cross sectional study was conducted (March 2019 – February 2020) in a tertiary care hospital which was authorized by Institutional Ethics Committee (1585/IEC/2019). Patients who were admitted in all the Intensive Care Units belonging to all age groups with clinical diagnosis of sepsis with elevated C-reactive protein (CRP) were included and those who were not willing to participate in this study were excluded. Before commencing antibiotics, whole blood (Adult 5-7 ml, Children 2-4 ml) were drawn under aseptic precautions by a trained phlebotomist as per WHO guidelines10 and it was directly inoculated into the Brain Heart Infusion broth (BHI) with an anticoagulant sodium polyanethol sulphonate (SPS) by automated method (Bact/Alert). The positive broths were subcultured onto Nutrient agar, MacConkey agar and 5% sheep blood agar and incubated for 24 hours at 37°C. A negative broth was followed up till the end of 6th day to declare no growth. Further identifications were done in growth positive organisms by Gram staining, cultural characteristics, other preliminary tests and standard biochemical tests. The antibiogram was done by Kirby Bauer’s disc diffusion method on cation adjusted Muller Hinton agar and results were interpreted with Clinical and Laboratory Standards Institute (CLSI) guidelines.11 Antibiotics tested were Amoxyclav (30 μg), Imipenem (10 μg), Ceftriaxone (30 μg), Gentamicin (10μg and 120μg), Cefoxitin (30μg), Vancomycin (30 μg), Piperacillin (100 μg), and Linezolid (30 μg), Ciprofloxacin (5 μg), Erythromycin (10 μg), Chloramphenicol (30 μg), Teicoplanin (30 μg), Colistin (10μg), Tigecycline(30μg), Piperacillin/Tazobactam (100/10 μg), Amikacin (30 μg), Clindamycin (2 μg), Ceftazidime (30 μg), PolymyxinB (10 μg), Ampicillin (30 μg), Cotrimoxazole (Trimethoprim1.25 μg/ Sulfamethoxazole 23.75 μg). Screening test for MRSA was done using Cefoxitin (30 μg) disc. For detecting ESBL production, Ceftazidime + Clavulanic acid (30/10 μg) and Ceftazidime (30 μg) were used. The results were analysed according to CLSI guidelines11 (Table: 1, Table: 2, Table: 3)
Table (1):
Number of Methicillin Sensitive / Resistant isolates among different Staphylococcus species.
| Organisms | Using Cefoxitin 30µg disk & inhibition zone diameter measurement (in mm)11 | |
|---|---|---|
| Susceptibility to Cefoxitin | Resistant to Cefoxitin | |
| Total Number of Organism (n=15) | ||
| Staphylococcus epidermidis (n=5) [CX ≥ 25mm — S, CX ≤ 24 mm — R] |
2 | 3 |
| Staphylococcus lugdunensis (n=2) [CX ≥ 22 mm — S, CX ≤ 21 mm — R] |
1 | 1 |
| Staphylococcus xylosus )(n=2) [CX ≥ 25 mm — S, CX ≤ 24 mm — R] |
2 | 0 |
| Staphylococcus aureus (n=6) [CX ≥ 22 mm — S, CX ≤ 21 mm — R] |
3 | 3 |
| Total | 8 | 7 |
Antibiogram For Cefoxitin-Among 15 Staphylococcus species, nearly 50% showed resistance to cefoxitin
Table (2):
Number of Vancomycin Sensitive Enterococcal species.
| Organisms | Using Vancomycin 30µg disk inhibition zone diameter measurement (in mm) 11 | |
|---|---|---|
| Susceptibility to Vancomycin Inhibition zone ≥ 17 mm — S, |
Resistant to Vancomycin Inhibition zone ≤ 14 mm — R |
|
| Total Number of Organism (N=2) | ||
| Enterococcus faecalis (n=2) | 2 | 0 |
Antibiogram of Vancomycin-Enterococcus faecalis showed 100% sensitivity to vancomycin
Table (3):
Gram negative bacilli profile & their Susceptibility pattern to 3rd generation cephalosporins.
| Organisms | Susceptibility to 3rd Generation Cephalosporins11 | Resistant to 3rd Generation Cephalosporins11 |
|---|---|---|
| Total No. of Organism | ||
| Klebsiella pneumoniae (n=10) | 3 | 7 |
| Escherichia coli (n=15) | 10 | 5 |
| Pseudomonas aeruginosa (n=2) | 2 | 0 |
| Acinetobacter baumanii (n=3) | 3 | 0 |
| Citrobacter koseri (n=1) | 1 | 0 |
| Salmonella typhi (n=1) | 1 | 0 |
| Proteus mirabilis (n=1) | 1 | 0 |
Antibiogram of Cephalosporin-Among 21 Gram negative isolates 12 showed resistance to Cephalosporins
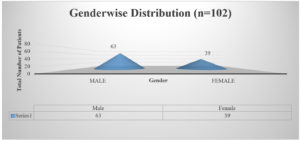
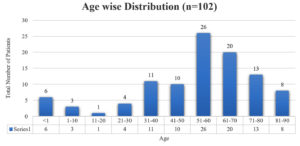
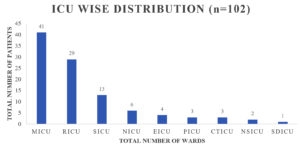
A sum of 102 samples were obtained from clinically suspected sepsis individuals having elevated CRP levels, out of which 54 (53.7%) were culture positive and 48 (46.3%) had no growth. Out of 102 samples, number of male and female were 63 (61%) and 39 (39%) respectively shown in Fig. 1, Age wise distribution shows the group belonging to 51 to 60 were more prone for sepsis as shown in Fig. 2. Among the 102 samples, majority were received from IMCU with number 41 (40%), followed by RICU which were accounted of 29 (29%). Other ICUs with more than 5% of samples were from SICU – 13 (12%) and NICU – 6 (7%). ICUs with less than 5% of samples were ER ICU with 4 (3.5%), CT ICU with 3 (3%), PICU with 3 (2.5%), SD ICU with 2 (2%), NSICU with 1 (1%) shown in Fig. 3.
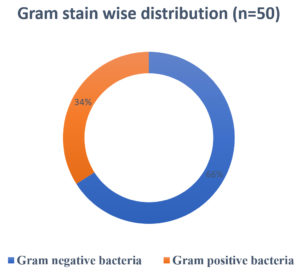
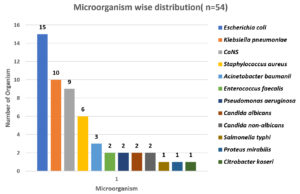
A total of 54 organisms were isolated from 102 samples. Among 54 samples, bacterial organisms were the most contributors (n=50), followed by fungus (n=4) and no growth were accounted on 48 samples. Among 54 positive samples, 23 (42.6%) were from IMCU and 17 (31.5%) from SICU followed by SICU AND NICU – 6 (11.1%) and 3 (5.6%) respectively. GNB accounted for 33 (61.1%) whereas GPC as 17 (31.5%) as shown in Fig. 4.
The predominant bacterial organism in GNB was Escherichia coli– 15 (27.7%) followed by Klebsiella pneumoniae 10 (18.51%), Acinetobacter baumanii 3 (5.6%), Pseudomonas aeruginosa (3.7%), single isolate of each Salmonella typhi, Proteus mirabilis and Citrobacter koseri (1.8%) were isolated.
In case of Gram-Positive Bacteria Coagulase negative Staphylococcus (CoNS) 9 (16.6%) was the commonest, next were Staphylococcus aureus 6 (11.1%) and Enterococcus faecalis 2 (3.7%).
Sepsis due to infection is the most common & dangerous of all the forms of nosocomial infections encountered in ICUs with sudden deaths than patients admitted to other types of units.12,13
Depending on variances in local sepsis epidemiology, the burden of sepsis may vary over time and between settings within a context. Furthermore, the prevalence of culture positive sepsis reported from several ICU in South Asia ranged from 6 to 56%, 52.9% in this study was near comparable with the study by Lisa Mellhammar et al 18 having 54%, most likely is due to discrepancies in study design and terminology.12,14-18
In our study culture positivity was 52.9% among elevated CRP Patients similar to the study of Jyoti et al study who also got 49% with CRP elevated above 6mg/dl12 n a study done by Abera Kumalo culture positivity was a 16% & even low as near 7% in recent study by Bajaj A et al.19 in may be because they did not take CRP positivity as adjunct criteria for selecting samples.20
In our study, major positive samples were from IMCU (42.6%) similarly higher percentages are seen in other studies also, a study done by Calik Z had highest percentage with 81% of his samples were from IMCU. The variations in sample percentage from different ICUs depend on the health care catering of that hospital.21
Gram negative bacteria 33 (61.1%) organisms were isolated more compared to Gram positive cocci 17 (31.5%) in our study. Similarly GNB was most commonly isolated organisms in many studies evident in Bajaj et al.18 from southern India region (65%) & from northern India one study done by Surbhikhurana et al, a 4 years study showed a percentage variations from 78% to 85%. This is well known as Gram-negative bacilli produces endotoxins that kick start the septicemia.22-25
Enterobacteriaceae was the most isolated organisms followed by non-fermenter as seen in many studies and Among Enterobacteriaceae, Escherichia coli is the commonest followed by Klebsiella pneumoniae. Similar pattern of bacteriological profile of organisms were isolated in some other studies like Alhashem F et al.25
Antimicrobial resistance to drugs such as colistin has contravened one of the final lines of defence against infections caused by multidrug-resistant bacteria.26 There were 48 samples which were culture negative accounting for 47.05 %. This may be because of empirical usage of antibiotics before the sample collection which could have made the organisms nonviable, or sepsis could be due to non-cultivable bacteria or it could be due to viral etiology which needs detection methods like PCR which has its own constrains of selection of primers & costly equipment and etc.
Culture positivity will be more when CRP is also included in the selection of samples for sepsis and Gram-negative bacilli are the leading cause in septicemia and organisms belonging to Enterobacteriaceae family still dominates in septicemic infections in ICUs and updating on local bacteriological profile regularly is necessary to select empirical treatment option depending on prevalent bacteria evident from studies like this as it is the decider many times between life & death of patients & treatment initiation awaiting culture sensitivity report and challenge for treatment are MDRs which needs to be detected regularly by using screening tests.
ACKNOWLEDGMENTS
We authors are extremely thankful to SRM Medical college Hospital and Research centre, Kattankulathur, Kanchipuram district, India for providing us all the equipment and facilities for performing this research work.
CONFLICT OF INTEREST
The authors declare that there is no conflict of interest.
AUTHORS’ CONTRIBUTION
All authors listed have made a substantial, direct and intellectual contribution to the work, and approved it for publication.
FUNDING
None.
ETHICS STATEMENT
The study is approved by Institutional ethical committee of SRM Medical College Hospital and Research Center (1585/IEC/2019).
AVAILABILITY OF DATA
All datasets generated or analysed during this study are included in the manuscript and/or the supplementary files.
- Moaveni DM. Systemic Inflammatory Response Syndrome and Sepsis in the Pregnant Patient. McEvoy MD, Furse CM, editors. Oxford Medicine Online. Oxford University Press; 2017.
Crossref - Mathur P, Varghese P, Tak V, et al. Epidemiology of Blood Stream Infections at a Level-1 Trauma Care Center of India. J Lab Physicians. 2014;6(01):022–027.
Crossref - Hazra A, Dasgupta S, Das S, Chawan NS. Nosocomial infections in the intensive care unit: Incidence, risk factors, outcome and associated pathogens in a public tertiary teaching hospital of Eastern India. Indian Journal of Critical Care Medicine. 2015;19(1):14–20.
Crossref - Nathan M. Introduction. Pathogens and Global Health. Informa UK Limited. 2012;106(8):427.
Crossref - Rudd KE, Johnson SC, Agesa KM, et al. Global, regional, and national sepsis incidence and mortality 1990-2017: analysis for the Global Burden of Disease Study. Lancet. 2020;395(10219):200-211.
Crossref - Nicolas I, Bordeau V, Bondon A, Baudy-Floc’h M, Felden B. Novel antibiotics effective against gram-positive and -negative multi-resistant bacteria with limited resistance. PLOS Biology. 2019;17(7):e3000337.
Crossref - Orsini J, Mainardi C, Muzylo E, Karki N, Cohen N, Sakoulas G. Microbiological Profile of Organisms Causing Bloodstream Infection in Critically Ill Patients. J Clin Med Res. 2012;4(6):371-377.
Crossref - Al-Otaibi FE, Bukhari EE, Badr M, Alrabiaa AA. Prevalence and risk factors of Gram-negative bacilli causing blood stream infection in patients with malignancy. Saudi Medical Journal. 2016;37(9):979–984.
Crossref - Wang TZ, Kodiyanplakkal RPL, Calfee DP. Antimicrobial resistance in nephrology. Nat Rev Nephrol. 2019;15(8):463–481.
Crossref - WHO Guidelines on Drawing Blood: World Health Organization. (2010). WHO guidelines on drawing blood: best practices in phlebotomy. World Health Organization. https://apps.who.int/iris/handle/10665/44294
- CLSI. Performance Standards for Antimicrobial Susceptibility Testing. 31st ed. CLSI supplement M100. Clinical and Laboratory Standards Institute; 2021. https://www.nih.org.pk/wp-content/uploads/2021/02/CLSI-2020.pdf
- DeNIS. Characterisation and antimicrobial resistance of sepsis pathogens in neonates born in tertiary care centres in Delhi, India: a cohort study. The Lancet Global Health. 2016;4(10):e752–e760.
Crossref - Rhee C, Jones TM, Hamad Y, et al. Prevalence, Underlying Causes, and Preventability of Sepsis-Associated Mortality in US Acute Care Hospitals. JAMA Netw Open. 2019;2(2):e187571.
Crossref - Iqbal S, Rehman FU, Ali W, Rajper SB. Pattern of bacterial isolates causing neonatal septicemia along with their pattern of antibiotic susceptibility. The Professional Medical Journal. 2020;27(04):737–741.
Crossref - Ansari S, Nepal HP, Gautam R, Shrestha S, Neopane P, Chapagain ML. Neonatal Septicemia in Nepal: Early-Onset versus Late-Onset. International Journal of Pediatrics. 2015;2015:379806.
Crossref - Shah GS, Yadav S, Thapa A, Shah L. Clinical Profile and Outcome of Neonates Admitted to Neonatal Intensive Care Unit (NICU) at a Tertiary Care Centre in Eastern Nepal. Journal of Nepal Paediatric Society. 2013;33(3):177–181.
Crossref - Monjur F, Rizwan F, Asaduzzaman M, et al. Antibiotic sensitivity pattern of causative organisms of neonatal septicemia in an urban hospital of Bangladesh. Indian J Med Sci. 2010;64(6):265-271.
Crossref - Mellhammar L, Kahn F, Whitlow C, Kander T, Christensson B, Linder A. Bacteremic sepsis leads to higher mortality when adjusting for confounders with propensity score matching. Sci Rep. 2021;11:6972.
Crossref - Bajaj A, Mishra B, Loomba PS, et al. Prevalence of Gram-negative Septicemia in a Tertiary Care Center. J Med Sci Health. 2019;5(1):36-41.
Crossref - Mama M, Abdissa A, Sewunet T. Antimicrobial susceptibility pattern of bacterial isolates from wound infection and their sensitivity to alternative topical agents at Jimma University Specialized Hospital, South-West Ethiopia. Ann Clin Microbiol Antimicrob. 2014;13:14.
Crossref - Calik Z, Acar O , Karamese M, Acar N. Analysis of Intensive Care Units (ICUs) Blood stream Infections (BSI) in Kafkas University Health Research and Application Hospital. Int J Microbiol Adv Immunol. 2015;3(1):55–59.
Crossref - Khurana S, Bhardwaj N, Kumari M, Malhotra R, Mathur P. Prevalence, etiology, and antibiotic resistance profiles of bacterial bloodstream infections in a tertiary care hospital in Northern India: A 4-year study. J Lab Physicians. 2018;10(04):426-431.
Crossref - Bendgude HH, Halgarkar CS. C-Reactive Protein (CRP) in Early Diagnosis of Neonatal Septicemia. Journal of Microbiology and Related Research. 2016;2(2):61-64
https://doi.org/10.21088/jmrr.2395.6623.2216.1. https://rfppl.co.in/view_abstract.php?jid=47&art_id=4460 - DiPiro JT. Pathophysiology and treatment of gram-negative sepsis. Am J Hosp Pharm. 1990;47(11 Suppl 3):S6-10. PMID: 2275478
- Alhashem F, Tiren-Verbeet NL, Alp E, Doganay M. Treatment of sepsis: What is the antibiotic choice in bacteremia due to carbapenem-resistant Enterobacteriaceae? World J Clin Cases. 2017;5(8):324-332.
Crossref - Liu YY, Wang Y, Walsh TR, et al. Emergence of plasmid mediated colistin resistance mechanism MCR 1 in animals and human beings in China: A microbiological and molecular biological study. Lancet Infect Dis. 2016;16(2):161-168.
Crossref
© The Author(s) 2021. Open Access. This article is distributed under the terms of the Creative Commons Attribution 4.0 International License which permits unrestricted use, sharing, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.