ISSN: 0973-7510
E-ISSN: 2581-690X
In this study, traditional sausage samples from different provinces of Turkey (Gaziantep, Antalya, Erzurum and Kahramanmaras) were obtained and one hundred three isolates were collected. Using the (GTG)5-PCR genomic fingerprint analysis method, seven of them were observed to be different and conventional tests of these isolates were performed. Molecular identification of two isolates carrying the bacteriocin gene and having antimicrobial activity by agar disc diffusion method was performed by 16S rRNA sequence analysis. As a result, the seven isolates were identified as Aerococcus urinaeequi (EK1), Streptococcus salivarius (EK2), Leuconostoc mesenteroides (EK3), Macrococcus caseolyticus (EK4), Lactococcus garvieae (EK5), Staphylococcus saprophyticus (EK6) and Lactobacillus sakei (EK7). Among these strains, it has been determined that Ln. mesenteroides and L. sakei carried the mecentericin and sacacin genes. When antimicrobial activity against different strains was examined, inhibition formations of Ln. mesenteroides and L. sakei on Enterococcus faecalis, Shigella dysenteriae and Escherichia coli O157: H7 were observed.
Bacteriocin, (GTG)5-PCR, 16S rRNA sequence analysis, antimicrobial activity
Bacteriocins are natural antimicrobial peptides which are used as bioprotectors in the food industry to protect human health and to increase the sustainability of foods1. They are produced by both Gram positive and Gram negative bacteria. Most of the bacteriocins are 20-70 amino acid structure2. Bacteriocins generally have cationic character and easily interact with Gram-positive bacteria that contain high amounts of anionic lipids in the cell membrane. This interaction causes the formation of pores in the cell membrane. The pores in the membrane show a bactericidal effect in the cells by changing the energy state of the cell3,4.
Bacteriocins are basically divided into two main classes. Class I bacteriocins contain lantibiotics with a molecular weight less than 5 kDa and can be distinguished from class II bacteriocins due to post-translational modifications5. Lantibiotics are divided into two groups. Type A lantibiotics are positively charged, long and flexible molecules that act on the bacterial membranes by performing pore formation. The most well-known member of this group is nisin which was described in 19286. Type B lantibiotics are smaller spherical peptides and are negatively charged or uncharged. It shows antimicrobial activity by inhibiting the specific enzymes of the host cell7. Class II bacteriocins are unmodified peptides. Class IIa (or pediosin PA1-like) peptides generally contain 37-48 amino acids5. Approximately 50 different class IIa bacteriocins have been isolated from fermented meat, fermented vegetables, dairy products, smoked salmon and the human gastrointestinal tract. Class IIb bacteriocins need two complementary peptides to show antimicrobial activity. Although some peptides of this class may exhibit antimicrobial activity separately, the addition of the complementary agent greatly enhances the activity. Class IIb bacteriocins contain amphiphilic and hydrophobic regions and are mostly cationic8. Apart from these two classes, there are class III and class IV. Class III contains heat-sensitive enzymes which degrade the cell wall9. Colicin produced by Escherichia coli is an example of class III bacteriocins, while helveticin M, helveticin J and enterolisin A produced by Lactobacillus crispatus, L. helveticus and Enterococcus faecalis are also examples of class III bacteriocins. Helveticin M has recently been characterized and found to disrupt the cell wall of Gram-positive bacteria and the outer membrane of Gram-negative bacteria. In addition, it has been stated that class III bacteriocin is effective against both Gram-positive and Gram-negative bacteria10. Class IV bacteriocins contain spherical, heat-sensitive, helix and post-translationally modified proteins consisting of 35-70 amino acids9.
Genes related to bacteriocin biosynthesis are usually found collectively and are encoded in plasmids, chromosomes or transposons. Bacteriocins are synthesized as biologically inactive prepeptides, usually containing a N-terminal leader peptide bound to the C-terminal propeptide11,12.
Bacteriocin-producing organisms are resistant to the bacteriocins they produce because they have specific immune proteins. Two separate bacteriological immune systems have been identified in the bacteriocin-producing cell which would be a special ABC carrier system consisting of immune protein and two or three subunits. These two immune systems work together to protect the bacteriocin-producing cells from their bacteriocin5.
The aim of the study was isolation and identification of lactic acid bacteria from traditional sausage samples collected from different locations of Turkey. Then, determination of whether the bacteria have bacteriocin gene or not and detection of antimicrobial effect of these isolates were aimed.
Isolation and molecular characterization of lactic acid bacteria
Lactic acid bacteria (LAB) were isolated from Turkish traditional sausages. The samples were collected from different provinces of Turkey; Gaziantep, Kayseri, Erzurum and Kahramanmaras. Each sample (25 gr) was mixed with 225 mL physiological water (0.9% NaCl) and homogenized in stomacher. Samples were diluted (10-1-10-7) and each dilution (100 µL) were spread onto MRS and M17 agar plates and incubated at 37°C for 48 h. First of all, the growing colonies were spread on M17 and MRS agar to obtain pure cultures and were eliminated by morphology differences on plates. The pure, single and different colonies were stored in the Tryptic Soy Broth with 15% glycerol content at -86°C for further studies13.
The total genomic DNA of each test strain was isolated according to the procedure of Promega™ Wizard® Genomic DNA Purification Kit. Afterwards, test isolates were subjected to rep-PCR [(GTG)5-PCR] with the special primer of (GTG)5 elements to obtain genomic fingerprinting. The 16S rRNA gene regions of the isolates which would be considered to be different according to the rep-PCR analysis were amplified with universal primers by PCR and were subjected to the cloning procedure13-17.
Conventional identification
The isolates which were selected according to genomic fingerprinting were subjected to conventional tests for pH, temperature and salt (NaCl) requirements in growth media. The pH, temperature and NaCl requirements for bacterial growth were measured in M17 and MRS broth by following optical density at 600 nm wavelength with spectrophotometer by the method of Prescott et al.18. Firstly, M17 and MRS broth media were prepared and, then a full loop of the test isolate on M17 and MRS agar was transferred to broth media and incubated at different temperatures (10-45°C) for 48 h. To measure the response to pH changes during growth, M17 and MRS broth media were prepared and pH of broth media was adjusted to different points at pH 3-11 range, before autoclaving. Then, a full loop of the test isolate on M17 and MRS agar was transferred to broth media and incubated at 37°C for 48 h. The salt (NaCl) requirement for growth was also tested in M17 and MRS broth medium containing 2-12 (w/v). For this, M17 and MRS broth media were prepared and salt at determined concentrations were added to broth media, before autoclaving. Then, a full loop of the test isolate on M17 and MRS agar was transferred to broth media and incubated at 37°C for 48 h19.
Colony morphology, Gram staining, motility, the presence of catalase and oxidase reactions and gas productions were also investigated18,20,21.
Determination of bacteriocin genes
The selected test isolates were evaluated according to bacteriocin genes with multiplex PCR reactions. 75 ng of purified DNA (2 µL) was used as the template in 30 µL total reaction mixture. Twenty-eight microliters of the reaction cocktail were prepared as follows: PCR Buffer (10X) 3 µL, MgCl2 1.2 µL (25mM), dNTPs (10 mM) 0.6 µL, bovine serum albumin 1.2 µL (20 mg/mL), primers (5 µM) 1 µL, Taq polymerase (250 U) 0.3 µL and water 13.7 µL. Primers were given in Table 1. A negative control (no DNA) was included in each PCR assay. PCR reactions were performed with a 96-well ProFlexTM PCR System, using the following conditions: an initial denaturation at 95°C for 5 min; 30 cycles consisting of 94°C for 0.5 min, annealing at 55°C for 1 min, extension at 72°C for 2 min and a final polymerization at 72°C for 5 min before cooling at 4°C22.
Table (1):
Primers used in multiplex-PCR reaction for determination of bacteriocin genes.
Gene |
Primers |
|---|---|
Garvicin A(lgnA) |
lgnA–F: 5’-ATTTAATACGGACGGTATTGAT-3’ |
lgnA–R: 5’-GGAGTAAAAAGATGGAAAACAA-3’ |
|
Mesentericin |
MESY-F: 5’-AGTCTGTGGAAGCATATCAGCA-3’ |
MESY-R: 5’-TACCAAAATCCATTTCCACCAT-3’ |
|
Sacacin |
Sak A-F: 5’-ACAGAATTACAAACAATTACCGGC-3’ |
Sak A-R: 5’-CATTCCAGCTAAACCACTAGCC-3’ |
|
Salivaricin |
Salivaricin-F: 5’-GTAGAAAATATTTACTACATACT-3’ |
Salivaricin-R: 5’-GTTAAAGTATTCGTAAAACTGATG-3’ |
For the multiplex-PCR reactions, products (2 µL) were mixed with 1.0 µL gel loading
buffer (6X) and subjected to agarose (1% w/v) gel electrophoresis in Tris–Acetate–EDTA (TAE) buffer at 90 V and for 120 min. After separation, the fragments were stained with ethidium bromide solution (2 ml Etbr/100 ml 1X TAE buffer). The amplified DNA product was monitored using the Quantum Vilber Lourmat Gel Documentation System (Australia).
Agar disk diffusion method
In the agar disc diffusion test, pathogenic strains (Enterococcus faecalis ATCC 29212, Escherichia coli O157:H7 ATCC 35150, Shigella dysenteriae ATCC 13313, Pseudomonas aeruginosa ATCC 27853, Klebsiella pneumonia ATCC 13883, Enterobacter cloacae ATCC 13047, Staphylococcus aureus ATCC 29213) were inoculated on the Tryptic Soy Agar medium using the sterile swab. 100 µL of the test isolate, grown in M17 and MRS broth, were absorbed on discs with a diameter of 6 mm. Then, these discs were placed on the petri dishes where pathogenic bacteria were inoculated and incubated at 37°C for 24 h. At the end of the period, the petri dishes were removed to check if the inhibition zone was formed and the diameter of the zone was measured23,24.
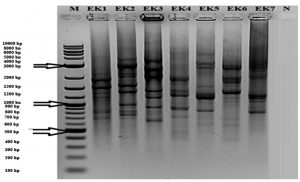
Traditional sausage samples from different locations of Turkey were collected. Isolation of lactic acid bacteria were carried out and one hundred three isolates were selected according to colony differentiation on petri plates. (GTG)5-PCR was performed to show distinction at species level (Fig.1) and according to differences, seven different isolates were decided to determine with 16S rRNA sequencing.
Fig.1. (GTG)5 profile generated with the GTG5 primer. Lanes: 1) Aerococcus urinaeequi; 2) Streptococcus salivarius subsp. salivarius; 3) Macrococcus caseolyticus subsp. hominis; 4) Leuconostoc mesenteroides subsp. dextranicum; 5) Lactococcus garvieae subsp. garvieae; 6) Staphylococcus saprophyticus subsp. saprophyticus; 7) Lactobacillus sakei subsp. sakei; N) Negative Control; M) Molecular Marker (10 kb).
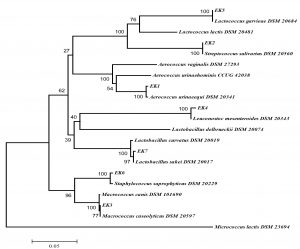
16S rRNA sequences of the seven isolates were amplified and cloned into E. coli JM101 strain. The obtained sequence was compared those in GenBank and EzTaxon. As given in Table 2; EK1, EK2, EK3, EK4, EK5, EK6 and EK7 were similar to Aerococcus urinaeequi, Streptococcus salivarius, Macrococcus caseolyticus, Leuconostoc mesenteroides, Lactococcus garvieae, Staphylococcus saprophyticus and Lactobacillus sakei, respectively. Also, phylogenetic analysis by using neighbour-joining method and data from 16S rRNA sequencing was carried out and given in Fig.2. As a result of the 16S rRNA gene sequences of the isolates, very close similarity rates (>98%) were obtained, as specified before in different experiments. Madigan and Martinko25 reported that isolates having sequence similarity over 97% might belong to the same species.
Fig. 2. Phylogenetic tree (based on neighbor-joining method) of lactic acid bacteria created with the data from 16S rRNA gene analysis results (Bootstrap value was 1000 repeats and no values below 50% were shown on nodes. Scale bar represented 0.5% deviation. Micrococcus lactis was used as outer group).
Table (2):
Comparison of 16S rRNA gene sequences of the test isolates with those in GenBank.
Isolate |
Species |
Similarity (%) |
Accession number |
|---|---|---|---|
EK1 |
Aerococcus urinaeequi |
99.86 |
MN045012 |
EK2 |
Streptococcus salivarius subsp. salivarius |
99.86 |
MN045013 |
EK3 |
Macrococcus caseolyticus subsp. hominis |
99.32 |
MN045011 |
EK4 |
Leuconostoc mesenteroides subsp. dextranicum |
99.72 |
MN045173 |
EK5 |
Lactococcus garvieae subsp. garvieae |
99.17 |
MN094108 |
EK6 |
Staphylococcus saprophyticus subsp. saprophyticus |
99.93 |
MN045172 |
EK7 |
Lactobacillus sakei subsp. sakei |
99.93 |
MN045009 |
These isolates were selected according to genetic polymorphism of (GTG)5 region. Svec et al.26 reported that (GTG)5-PCR was highly effective in the identification of lactobacilli isolated from food samples, it enabled the rapid and reliable identification of lactobacilli and other lactic acid bacteria, which were important in the food fermentation industries.
Table (3):
Morphological and biochemical properties of of LAB.
Isolate |
Morphology |
Gram reaction |
Motility |
Oxidase |
Catalase |
Gas production |
|---|---|---|---|---|---|---|
EK1 |
smooth/coccus |
+ |
– |
– |
– |
Homofermentative |
EK2 |
smooth/coccus |
+ |
– |
– |
– |
Homofermentative |
EK3 |
smooth/coccus |
+ |
– |
+ |
+ |
Homofermentative |
EK4 |
smooth/coccus |
+ |
– |
– |
– |
Homofermentative |
EK5 |
smooth/coccus |
+ |
– |
– |
– |
Homofermentative |
EK6 |
smooth/coccus |
+ |
– |
– |
+ |
Homofermentative |
EK7 |
smooth/bacilli |
+ |
– |
– |
– |
Homofermentative |
As a result of conventional identification (Table 3), all the isolates were smooth and coccus (except EK7). All of them were Gram positive, non-motile, oxidase (except EK3) and catalase negative (except EK3 and EK6) and homofermentative (except EK4). To determine physicochemical requirements of the isolates, they were grown in different culture conditions (Table 4). The isolates EK1, EK3 and EK6 were grown optimally at between 15-37°C, EK4 and EK7 were at between 25-37°C, EK2 were at between 35-37°C and EK5 were at between 15-40°C. The tolerance of pH changes was also measured. EK2, EK4, EK6 and EK7 could show tolerance between pH 5-7, while EK1, EK3 and EK5 could survive at between pH 7-9, pH 5-9 and pH 7-9. respectively. Salt requirements of isolates were also different. EK1, EK2, EK4 and EK7 grown at between 2-4% NaCl while EK3, EK5 and EK6 could grow at between 4-10%, 4-6% and 4-8% NaCl, respectively.
Table (4):
Temperature, pH and salt requirements of of LAB.
Isolate |
Temperature (°C) |
pH |
Salt concentration (%) |
|---|---|---|---|
EK1 |
15-37 |
7-9 |
2-4 |
EK2 |
35-37 |
5-7 |
2-4 |
EK3 |
15-37 |
5-9 |
4-10 |
EK4 |
25-37 |
5-7 |
2-4 |
EK5 |
15-40 |
7-9 |
4-6 |
EK6 |
15-37 |
5-7 |
4-8 |
EK7 |
25-37 |
5-7 |
2-4 |
Karani et al.27 revealed that M. caseolyticus strain developed in the presence of salt as high as 8%. Diaz et al.15 stated that halophilic or halo-tolerance microorganisms tolerated high salt concentrations and this was making them very important in remediation. Considering this information, it was concluded that M. caseolyticus, which was not normally LAB, could be widely used in biotechnological processes because of maintaining vital functions even in high salt concentrations. Ramirez-Chavarin et al.28 reported that thermotolerant lactic acid bacteria became dominant flora in foods cooked like sausage and act as bioprotective agents. Varsha and Nampoothiri29 found in their study that the bacteria L. garvieae showed growth at 42°C. The optimum pH range of S. saprophyticus was 5-6.830 and Ln. mesenteroides’ optimum pH was 5.531. Optimum pH range was 8.5-9 for A. urinaeequi32, 6-7 for S. salivarius33, 6 for L. sakei34 and 9.6 for L. garvieae35. Karani et al.27 found that the optimum pH range for M. caseolyticus was 6-11. Dhakar and Pandey36 emphasized that bacteria, archaea and eukaryotic organisms that had the potential to develop in a wide pH range could be used in industrially important bioprocesses.
Determination of bacteriocin presence
The potentials of isolates to produce bacteriocin were analyzed using specific PCR. Mesentericin and sacacin genes were determined in two isolates, EK4 (Ln. mesenteroides subsp. dextranicum) and EK7 (L. sakei subsp. sakei) (data not shown), respectively.
Todorov and Dicks37 were reported presence of mesentericin in Ln. mesenteroides subsp. dextranicum ST99 from Boza samples. de Paula et al.38 were isolated Ln. mesenteroides SJRP55 from cheese samples and purified bacteriocins which were identical to mesentericin Y105 and mesentericin B105. Hechard et al.39 purified and characterized mesentericin Y105 from L. mesenteroides.
Sawa et al.40 isolated L. sakei D98 from rice malt and purified three novel bacteriocin belonging to class IIa and class IId. Moretro et al.41 optimized sacacin P production from L. sakei CCUG 42687 in a completely defined medium. Jiang et al.42 defined a novel sacacin from L. sakei LSJ61.
Antimicrobial effect of LAB on pathogenic strains
When the effects of the test isolates against different pathogenic microorganisms were investigated by agar disc diffusion method (Table 5), it was observed that, the isolates EK4 (Ln. mesenteroides subsp. dextranicum) had antimicrobial effect on E. faecalis ATCC 29212 and S. dysenteriae ATCC 13313; another isolate, EK7 (L. sakei subsp. sakei) was effective on E. faecalis ATCC 29212 and E. coli O157: H7 ATCC 35150.
Table (5):
Antimicrobial effect of test isolates against pathogenic microorganisms.
Isolate |
Species |
E. faecalis |
E.coli O157:H7 |
S. dysenteriae |
P. aeruginosa |
K. pneumoniae |
E. cloacae |
S. aureus |
|---|---|---|---|---|---|---|---|---|
EK1 |
Aerococcus urinaeequi |
– |
– |
– |
– |
– |
– |
– |
EK2 |
Streptococcus salivarius subsp. salivarius |
– |
– |
– |
– |
– |
– |
– |
EK3 |
Macrococcus caseolyticus subsp. hominis |
– |
– |
– |
– |
– |
– |
– |
EK4 |
Leuconostoc mesenteroides subsp. dextranicum |
+ |
– |
+ |
– |
– |
– |
– |
EK5 |
Lactococcus garvieae subsp. garvieae |
– |
– |
– |
– |
– |
– |
– |
EK6 |
Staphylococcus saprophyticus subsp. saprophyticus |
– |
– |
– |
– |
– |
– |
– |
EK7 |
Lactobacillus sakei subsp. sakei |
+ |
+ |
– |
– |
– |
– |
– |
Mesentericins from Ln. mesenteroides SJRP55 showed antimicrobial effect against to E. faecalis ATCC 19443, Enterococcus sp. and E. faecium38. Mesentericin from Ln. mesenteroides subsp. dextranicum ST99 was also active on E. faecalis. Bacteriocin containing supernatants of isolates from fresh fruit and vegetables had antimicrobial effect against different LAB and non-LAB43.
Sacacin-like bacteriocins from L. sakei D98 had antimicrobial activity on E. faecalis JCM 580340. A novel bacteriocin from L. sakei LSJ61 showed antimicrobial effect on E. coli ECX442. Another novel bacteriocin from L. sakei C2, isolated from traditional Chinese fermented cabbage had a broad antimicrobial spectrum and showed inhibitory effect on S. flexneri CMCC 51606 and E. coli ATCC 2592244. Bacteriocin from L. sakei ST154Ch was active on E. faecalis, E. faecium and E. coli, while bacteriocins from L. sakei ST22Ch and L. sakei ST153Ch had antimicrobial effect on E. faecium and E. coli45.
Reliable foods would be crucial for human growth and development. In order for food to be reliable, it is necessary to avoid process applications and to use natural additives. Bacteriocins, one of these natural additives, play an important role in food preservation by inhibiting both pathogenic and spoilage microorganisms. In this study, traditional sausage samples demonstrated a diverse of microorganisms and introduced bacteriocin production ability. It could be concluded that fermented foods had a good potential for bacteriocin producing microorganisms and these bacteriocins would be a decent source for food hygiene problems. In addition, (GTG)5-PCR, which is one of the rep-PCR methods, exhibited the distinction between food-borne test strains at species and subspecies level.
ACKNOWLEDGMENTS
None.
CONFLICT OF INTEREST
The authors declare that there is no conflict of interest.
AUTHORS’ CONTRIBUTION
EA collected the sausage samples, EA and SA carried out the study, BG, HO and MAO participated in design to draft, GA and AA wrote the manuscript. All the authors read and approved the final manuscript
FUNDING
None.
ETHICS STATEMENT
All text, data, figures/tables or other illustrations presented in the manuscript are completely original and does not contain or include material taken from other copyrighted sources. This article does not contain any studies about human or animal objects.
AVAILABILITY OF DATA
All datasets obtained or studied during this study are incorporated in the manuscript.
- Veskovic Moracanin S, Turubatovic L, Skrinjar M, Obradovic D. Antilisterial Activity of Bacteriocin Isolated from Leuconostoc mesenteroides ssp. mesenteroides IMAU: 10231 in the Production of Sremska Sausages: Lactic Acid Bacteria Isolation, Bacteriocin Identification and Meat Application Experiments. Food Technol Biotechnol. 2013;51:247-256.
- Diep DB, Straume D, Kjos M, Torres C, Nes IF. An overview of the mosaic bacteriocin pln loci from Lactobacillus plantarum. Peptides. 2009;30:1562-1574.
Crossref - Meera N, Devi MC. Partial characterization and optimization of parameters for Bacteriocin production by Probiotic Lactic acid bacteria. Journal of Microbiology and Biotechnology Research. 2012;2:357-365.
- Settanni L, Corsetti A. Application of bacteriocins in vegetable food biopreservation. Int J Food Microbiol. 2008;121:123-138.
Crossref - Martinez FAC, Balciunas EM, Converti A, Cotter PD, de Souza Oliveira RP. Bacteriocin production by Bifidobacterium spp. A review. Biotechnol Adv. 2013;31:482-488.
Crossref - Guder A, Wiedemann I, Sahl HG. Posttranslationally modified bacteriocins – the lantibiotics. Peptide Science. 2000;55:62-73.
Crossref - And HC, Hoover D. Bacteriocins and their food applications. Comprehensive Reviews in Food Science and Food Safety. 2003;2:82-100.
Crossref - Mokoena MP. Lactic acid bacteria and their bacteriocins: Classification, biosynthesis and applications against uropathogens: A mini-review. Molecules. 2017;22:1255.
- Sanchez-Hidalgo M, Montalban-Lopez M, Cebrian R, Valdivia E, Martinez-Bueno M, Maqueda M. AS-48 bacteriocin: close to perfection. Cell Mol Life Sci. 2011;68:2845-2857.
Crossref - Kumariya R, Garsa AK, Rajput Y, Sood S, Akhtar N and Patel S. Bacteriocins: Classification, synthesis, mechanism of action and resistance development in food spoilage causing bacteria. Microb Pathog. 2019;128:171-177.
Crossref - Perez RH, Zendo T, Sonomoto K. Novel bacteriocins from lactic acid bacteria (LAB): various structures and applications. Microb Cell Fact. 2014;13:S3.
Crossref - Zacharof M, Lovitt R. Bacteriocins produced by lactic acid bacteria a review article. Apcbee Procedia. 2012;2:50-56.
Crossref - Albayrak S, Genc B, Ozkan H, Taskin M, Adiguzel A. Presence of different bacterial species in thermal sources and novelty in their industrial enzyme productions. J Pure Appl Microbiol. 2019;13:1375-1387.
Crossref - Muslu S, Genc B, Adiguzel M, Albayrak S, Adiguzel A. Proteolitic, Lipolitic and Amilolitic Bacteria Reservoir of Turkey; Cold-Adaptive Bacteria in Detergent Industry. J Pure Appl Microbiol. 2020;14(1):63-72.
Crossref - Diaz MP, Boyd KG, Grigson SJ, Burgess JG. Biodegradation of crude oil across a wide range of salinities by an extremely halotolerant bacterial consortium MPD-M, immobilized onto polypropylene fibers. Biotechnol Bioeng. 2002;79:145-153.
Crossref - Altschul SF, Gish W. Local alignment statistics, in Methods in Enzymology. Academic Press. 1996;460-480.
Crossref - Benson G. Tandem repeats finder: a program to analyze DNA sequences. Nucleic Acids Res. 1999;27:573.
Crossref - Prescott LM, Harley JP, Klein DA. Laboratory exercises in microbiology. McGraw-Hill Companies, 2002
- Adiguzel G, Faiz O, Sisecioglu M, et al. A novel endo-β-1,4-xylanase from Pediococcus acidilactici GC25; purification, characterization and application in clarification of fruit juices. International Journal of Biological Macromolecules. 2019;129:571-578.
Crossref - Buck JD. Nonstaining (KOH) method for determination of gram reactions of marine bacteria. Appl Environ Microbiol. 1982;44:992-993.
Crossref - Sari B, Faiz O, Genc B, Sisecioglu M, Adiguzel A, Adiguzel G. New xylanolytic enzyme from Geobacillus galactosidasius BS61 from a geothermal resource in Turkey. International Journal of Biological Macromolecules. 2018;119:1017-1026.
Crossref - Maldonado-Barragan A, Cardenas N, Martinez B, et al. A novel class IId bacteriocin from Lactococcus garvieae that inhibits septum formation in L. garvieae strains. Appl Environ Microbiol. 2013;79;4336-4346.
Crossref - Somarathna T, Fernando W, Ranaweera K, Premakumara G, Abeysinghe T, Weerakkody N. Antimicrobial activity and phytochemical screening of Alpinia malaccensis (Ran-kiriya) against food-borne bacteria. J Appl Microbiol. 2018;125:1276-1285.
Crossref - Venkadesan D, Sumathi V. Screening of lactic acid bacteria for their antibacterial activity against milk borne pathogens. International Journal of Applied Research. 2015;1:970-973
- Madigan MT, Martinko JM. Brock biology of microorganisms. Pearson Prentice HallUpper Saddle River, NJ, 2006.
- Svec P, Vancanneyt M, Seman M, et al. Evaluation of (GTG)5-PCR for identification of Enterococcus spp. FEMS Microbiol Lett. 2005;247:59-63.
Crossref - Karani M, Shashidhar R, Kakatkar A, et al. Radiation-resistant Macrococcus caseolyticus (A) isolated from radiation-processed semidried prawns. Can J Microbiol. 2015;61:89-92.
Crossref - Ramirez-Chavarin M, Wacher C, Eslava-Campos C, Perez-Chabela M. Probiotic potential of thermotolerant lactic acid bacteria strains isolated from cooked meat products. International Food Research Journal. 2013;20:991.
- Varsha KK, Nampoothiri KM. Lactococcus garvieae subsp. bovis subsp. nov., lactic acid bacteria isolated from wild gaur (Bos gaurus) dung, and description of Lactococcus garvieae subsp. garvieae subsp. nov. Int J Syst Evol Microbiol. 2016;66:3805-3809.
Crossref - Schleifer KH, Kloos WE. Isolation and characterization of Staphylococci from human skin I. Amended descriptions of Staphylococcus epidermidis and Staphylococcus saprophyticus and descriptions of three new species: Staphylococcus cohnii, Staphylococcus haemolyticus, and Staphylococcus xylosus. Int J Syst Evol Microbiol. 1975;25: 50-61.
Crossref - Mataragas M, Metaxopoulos J, Galiotou M, Drosinos E. Influence of pH and temperature on growth and bacteriocin production by Leuconostoc mesenteroides L124 and Lactobacillus curvatus L442. Meat Science. 2003;64:265-271.
Crossref - Felis GE, Torriani S, Dellaglio F. Reclassification of Pediococcus urinaeequi (ex Mees 1934) Garvie 1988 as Aerococcus urinaeequi comb. nov. Int J Syst Evol Microbiol. 2005;55:1325-1327.
Crossref - Roger P, Delettre J, Bouix M, Beal C. Characterization of Streptococcus salivarius growth and maintenance in artificial saliva. J Appl Microbiol. 2011;111:631-641.
Crossref - Rimaux T, Riviere A, Illeghems K, Weckx S, De Vuyst L, Leroy F. Expression of the arginine deiminase pathway genes in Lactobacillus sakei is strain dependent and is affected by the environmental pH. Appl Environ Microbiol. 2012;78:4874-4883.
Crossref - Altun S, Onuk EE, Ciftci A, Buyukekiz AG, Duman M. Phenotypic, Genotypic Characterisation and Antimicrobial Susceptibility Determination of Lactococcus garvieae Strains. Kafkas University Journal of Veterinary Faculty. 2013;19(3):375-381.
- Dhakar K, Pandey A. Wide pH range tolerance in extremophiles: towards understanding an important phenomenon for future biotechnology. Appl Microbiol Biotechnol. 2016;100:2499-2510.
Crossref - Todorov SD, Dicks LM. Characterization of mesentericin ST99, a bacteriocin produced by Leuconostoc mesenteroides subsp. dextranicum ST99 isolated from boza. J Ind Microbiol Biotechnol. 2004;31:323-329.
Crossref - de Paula AT, Jeronymo-Ceneviva AB, Silva LF, et al. Leuconostoc mesenteroides SJRP55: a bacteriocinogenic strain isolated from Brazilian water buffalo mozzarella cheese. Probiotics and Antimicrobial Proteins. 2014;6:186-197.
Crossref - Hechard Y, Derijard B, Letellier F, Cenatiempo Y. Characterization and purification of mesentericin Y105, an anti-Listeria bacteriocin from Leuconostoc mesenteroides. Microbiology. 1992;138:2725-2731.
Crossref - Sawa N, Koga S, Okamura K, Ishibashi N, Zendo T, Sonomoto K. Identification and characterization of novel multiple bacteriocins produced by Lactobacillus sakei D98. J Appl Microbiol. 2013;115:61-69.
Crossref - Moretro T, Aasen I, Storro I, Axelsson L. Production of sakacin P by Lactobacillus sakei in a completely defined medium. J Appl Microbiol. 2000;88:536-545.
Crossref - Jiang J, Shi B, Zhu D, et al. Characterization of a novel bacteriocin produced by Lactobacillus sakei LSJ618 isolated from traditional Chinese fermented radish. Food Control. 2012;23(2):338-344.
Crossref - Trias R, Badosa E, Montesinos E, Baneras L. Bioprotective Leuconostoc strains against Listeria monocytogenes in fresh fruits and vegetables. Int J Food Microbiol. 2008;127:91-98.
Crossref - Gao Y, Jia S, Gao Q, Tan Z. A novel bacteriocin with a broad inhibitory spectrum produced by Lactobacillus sake C2, isolated from traditional Chinese fermented cabbage. Food Control. 2010;21:76-81.
Crossref - Todorov SD, Vaz-Velho M, De Melo Franco BDG, Holzapfel WH. Partial characterization of bacteriocins produced by three strains of Lactobacillus sakei, isolated from salpicao, a fermented meat product from North-West of Portugal. Food Control. 2013;30:111-121.
Crossref
© The Author(s) 2020. Open Access. This article is distributed under the terms of the Creative Commons Attribution 4.0 International License which permits unrestricted use, sharing, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.