ISSN: 0973-7510
E-ISSN: 2581-690X
To investigate the association of both tsst-1 and pvl with mecA genes in clinical Staphylococcus aureus (S. aureus) strains obtained from a tertiary care hospital, Mansoura-Egypt. Besides, relating these genes to antibiotic resistance patterns in such isolates. A prospective study was performed over clinical samples obtained from patients showing evidence of infection at Mansoura University Children Hospital, Egypt. Staphylococcus aureus isolates were obtained to test their antibiotic sensitivity pattern by disk diffusion method, E-test strips besides oxacillin agar screen test followed by screening such isolates for mecA, pvl and tsst-1 genes by polymerase chain reaction. Out of 180 isolated S. aureus strains, 88 isolates (48.9%) were methicillin resistant. The tsst-1 gene was positive in 65.9% of methicillin resistant S. aureus (MRSA) and 55.4% of methicillin sensitive S. aureus (MSSA) strains. Carriage of mecA and tsst-1 genes was not significantly associated (P=0.69). The pvl gene was positive in 23.9% of MRSA and 9.8% of MSSA isolates showing a significant association with mecA gene (P=0.04*). The mecA gene was significantly associated with multidrug resistant isolates (p= 0.041*). There was no significant association between antibiotic resistance and the presence of tsst-1 or pvl genes. The tsst-1 gene had a high prevalence among S. aureus isolates and it was similarly distributed in both MRSA and MSSA groups. The pvl gene was significantly associated with MRSA group. Antibiotic resistance pattern of S. aureus was not significantly associated to the presence of tsst-1 or pvl genes in any way.
Staphylococcus aureus, MRSA, mecA gene, pvl gene, tsst-1 gene, PCR
Staphylococcus aureus (S. aureus) is a major bacterial human pathogen held responsible for both community-acquired as well as hospital-acquired related infections1, that might cause a wide variety of clinical manifestations including sepsis, respiratory tract, surgical site and urinary tract infections2. Staphylococcus aureus infection remains a tough challenge to manage due to the emergence of multi-drug resistant strains that led to drag its clinical importance under the spot light over the past decades3. Methicillin resistant S. aureus (MRSA) strains show resistance to a large group of beta-lactam antibiotics including penicillins and cephalosporins4. The MRSA strains produce an altered penicillin-binding protein-2, referred to as PBP2a encoded by mecA gene5, such protein has more resistance to beta-lactam antibiotics when compared to the original PBP26.
Many virulence factors are responsible for the unique pathogenicity of S. aureus such as toxins, enzymes and adhesion factors. The toxins produced by S. aureus include Panton-Valentine leukocidin (PVL), hemolysin, toxic shock syndrome toxin-1 (TSST-1), exfoliative toxins and staphylococcal enterotoxins7. The PVL is a pore forming leukotoxin encoded by pvl gene and has the ability to target and kill host leukocytes through perforation of plasma membrane together with intracellular organelle membranes. The PVL producing S. aureus strains can cause various infections ranging from as simple as infections of skin and soft tissue up to serious life endangering conditions such as severe necrotic pneumonia8,9.
Nevertheless, another toxin known as TSST-1 encoded by tsst-1 gene is also produced by S. aureus strains10, and its expression can lead to an acute life threatening toxic shock syndrome (TSS) that can present itself with rash, fever and multi-organ dysfunction11. A strong relation was found between the TSS and tampon use by women due to toxin released by colonizing S. aureus strains in the vagina. However, the TSS can also occur in other different conditions such as surgical site complications, skin and respiratory tract infections12. Having said that, the secretion of TSST-1 into blood also increases the rate of TSS-like neonatal exanthematous lesion, sudden infant death syndrome and Kawasaki syndrome in pediatric age group11. The improper timing of prescribing the necessary antimicrobial treatment for patients with TSS may lead to fatal shock within hours after the onset of clinical presentations12. Therefore, to improve the outcome of such patients it is extremely important to have sufficient data about the tsst-1 gene prevalence and its relation to antibiotic susceptibility pattern.
In literature, many studies have reported an alarming increase in the MRSA strains positive for tsst-1 and pvl genes7,13. Similarly, previous studies conducted in Egypt detected tsst-1 and pvl genes among MRSA strains14,15. Toxins encoded by tsst-1 and pvl genes can cause severe infections such as TSS and necrotic pneumonia. Treatment of such infections is even more challenging when caused by MRSA because of the mecA gene- associated antimicrobial resistance. Therefore, investigating the prevalence of these toxins-encoding genes among MRSA isolates in different geographical locations is necessary for proper treatment and control of these infections. Moreover, these data will enable us to set up a proper antibiotic policy to improve the outcome of these cases. In our study, we investigated the association of both tsst-1 and pvl with mecA genes among clinical S. aureus isolates obtained from a tertiary health care center, Mansoura-Egypt. In addition, the relation of these genes to antibiotic resistance patterns was evaluated.
This prospective study was undertaken at the Medical Microbiology and Immunology Department, Faculty of Medicine, Mansoura University-Egypt. The protocol was revised and proved to be fulfilling ethical criteria by our institutional review board.
Sample Collection
Various clinical samples were collected from inpatients and outpatients showing evidence of infection at Mansoura University Children Hospital, Mansoura-Egypt from January to October 2018. Suggestive signs and symptoms differed according to the site of infection such as fever, dysuria, suprapubic tenderness, dyspnea, tachypnea, purulent sputum, wound pain or tenderness, purulent wound drainage, purulent eye secretions, etc. Samples included blood, urine, endotracheal aspirates, wound drainages, sputum, eye secretions and abscesses. Enrolled samples were sent to the Medical Microbiology and Immunology laboratory for processing.
Microbiological Processing
All samples were processed by standard microbiological techniques. Resulting S. aureus isolates were fully identified by colonial morphology, Gram-staining and biochemical testing including catalase, coagulase and mannitol fermentation. API Staph (bioMerieux, France) was also performed in order to confirm the identification of S. aureus isolates.
Antimicrobial Susceptibility Testing
Antimicrobial susceptibility pattern of S. aureus isolates was tested by disk diffusion method on Mueller-Hinton agar (MHA) using the following antibiotic disks (Oxoid, UK): penicillin (10 units), cefazolin (30mg), ciprofloxacin (5mg), erythromycin (15mg), clindamycin (2mg), Trimethoprim-sulfamethoxazole (1.25/23.75mg), gentamicin (10mg), linezolid (30mg) and tetracycline (30mg). Disk diffusion procedure was carried out according to the guidelines of clinical and laboratory standard institute (CLSI)16. Determination of the minimum inhibitory concentration for vancomycin was achieved by E-test strips (bioMerieux, France) on MHA plates following the manufacturer’s guidelines. The breakpoints for resistance as implemented by the CLSI were used for results interpretation16.
Detection of Methicillin Resistance
Methicillin resistance in previously identified S. aureus isolates was phenotypically detected by cefoxitin disk diffusion and oxacillin agar screen tests following the CLSI guidelines. Cefoxitin disk diffusion test was conducted by standard disk diffusion procedure using cefoxitin (30mg) as a surrogate for oxacillin16. As recommended by the CLSI, oxacillin agar screen test was conducted using MHA containing 4% NaCl and 6mg/mL oxacillin. A half McFarland suspension was prepared from the S. aureus isolates. Then, a sterile swab was dipped in the prepared bacterial suspension. After expressing the excess liquid, an entire quadrant of the MHA plate was streaked to be incubated at 35°C for 24 hours and then examined with transmitted light. Growth of > 1 colony on this culture medium indicated methicillin resistance16. Staphylococcus aureus ATCC 29213 was used as negative control (methicillin sensitive) while S. aureus ATCC 33591 was used as a positive control (methicillin resistant). Isolates were then kept in tryptic soy broth supplemented by 30% glycerol at -70°C until further molecular testing. Being resistant to at least 3 or more antibiotic classes in addition to beta-lactams, define multidrug resistance17.
DNA Extraction and Polymerase Chain Reaction
Extraction of Genomic DNA was conducted by using an available commercial Qia -AmpDNA Mini Kit (QIAGEN-UK) through applying the manufacturer’s guidelines. For detection of mecA, pvl and tsst-1 genes among the isolated S. aureus strains, polymerase chain reaction (PCR) was performed using specific primers as shown in Table 1 18. The PCR was performed in a final 50mL volume reaction containing Taq PCR Master Mix (QIAGEN-UK), template DNA and specific primers.
Table (1):
Primers used in the study
| Gene | Primer | Sequence (5’→ 3’) | Product size (bp) |
|---|---|---|---|
| mecA | F | AGAAGATGGTATGTGGAAGTTAG | 583 |
| R | ATGTATGTGCGATTGTATTGC | ||
| pvl | F | GGAAACATTTATTCTGGCTATAC | 502 |
| R | CTGGATTGAAGTTACCTCTGG | ||
| tsst-1 | F | TTATCGTAAGCCCTTTGTTG | 398 |
| R | TAAAGGTAGTTCTATTGGAGTAGG |
PCR identification of the mecA gene was performed with the thermo-cycling conditions set at 5 min at 94°C for initial denaturation, followed by 40 amplification cycles: 94°C for 30s, 57°C for 45 s, 72°C for 30 s then final extension at 72°C for 5 min 18. Multiplex PCR for detection of pvl and tsst-1 genes was performed with the following cycling settings: 5 min at 94°C for initial denaturation followed by 40 cycles of amplification of successive steps at 94°C for 40 s, 60°C for 40 s and 72°C for 1 min with a final extension at 72°C for 5 min 18. To determine the presence of the desired genes, PCR products were visualized by electrophoresis in agarose gel stained with ethidium bromide followed by UV lights examination. Reference strains of S. aureus ATCC 33591, S. aureus ATCC 49775 and S. aureus ATCC BAA-2094 were used as positive control for mecA, pvl and tsst-1 genes respectively while distilled water was used as the negative control.
Statistical Analysis
Chi-square test was applied to compare variables and the results were presented as percentages (%). When P value was <0.05 the difference was statistically significant. We used SPSS 22 for Windows (SPSS Inc., Chicago, IL, USA).
A total of 180 S. aureus strains were isolated from the processed 375 clinical samples (48%) [88 Male (48.9%) and 92 Females (52.1%)], the mean± SD age was 6.1 ±2.8 years. Out of the S. aureus isolates, 75 (41.7%), 43 (23.9%) and 19 (10.6%) were recovered from blood, urine and endotracheal aspirates respectively as demonstrated in Table 2. Phenotypic evaluation of methicillin resistance by cefoxitin disk diffusion and oxacillin agar screen tests revealed that 88 out of 180 (48.9%) S. aureus isolates were methicillin resistant. The mecA gene was further identified in all of these isolates that confirmed their primary identification as MRSA by phenotypic methods.
Table (2):
Different sources of Staphylococcus aureus isolates
Sample |
Number of isolates (%) |
|---|---|
Blood |
75 (41.7%) |
Urine |
43 (23.9%) |
Endotracheal aspirates |
19 (10.6%) |
Wound drainages |
13 (7.2%) |
Sputum |
12 (6.7%) |
Eye secretions |
11 (6.1%) |
Abscesses |
7 (3.9%) |
Total |
180 (100%) |
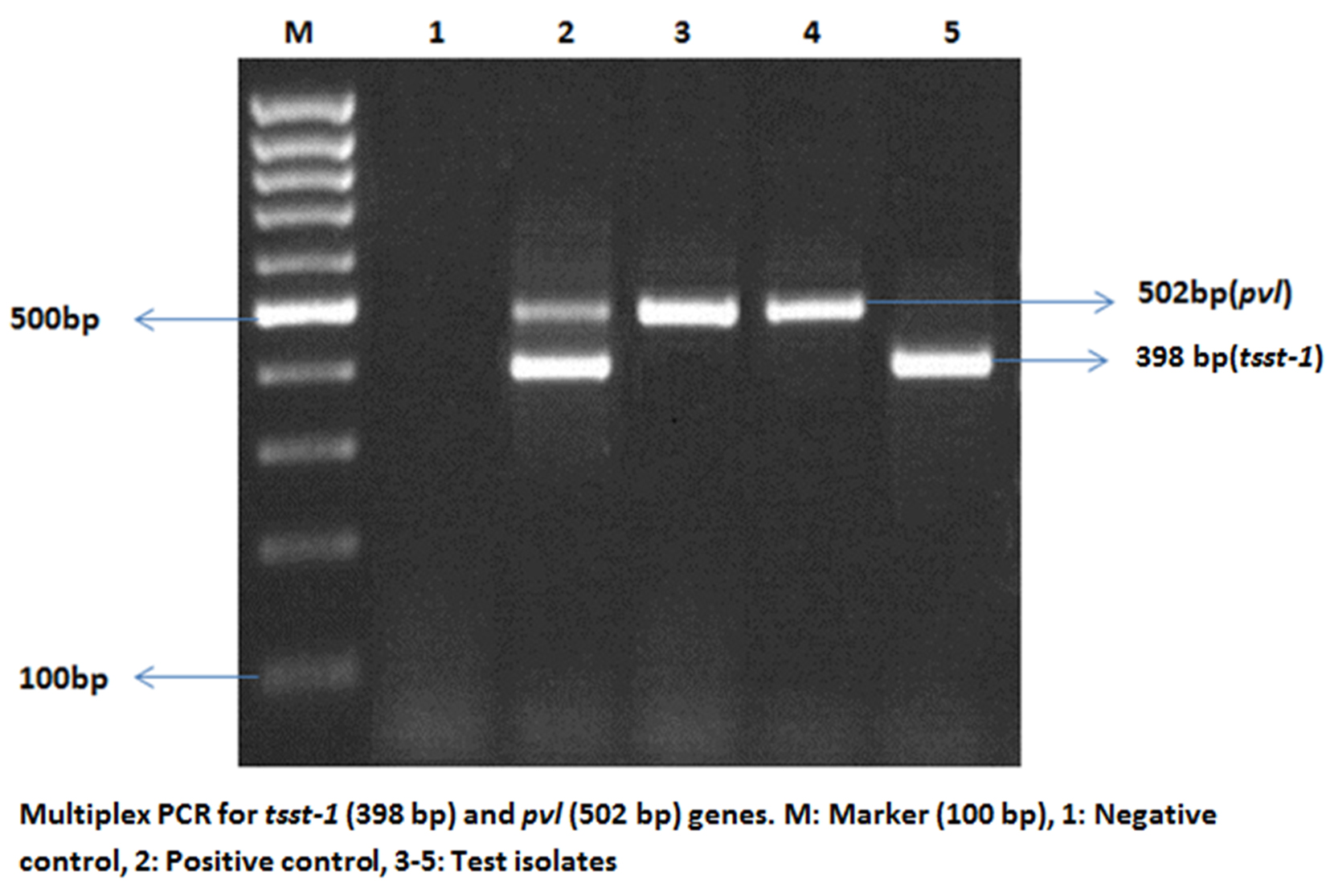
In the current study, we determined the distribution of mecA, tsst-1 and pvl genes among the recovered S. aureus strains by PCR. Out of the 180 isolated S. aureus strains, 88 (48.9%), 109 (60.6%) and 30 (16.7%) isolates had the mecA, tsst-1 and pvl genes respectively (Table 3). Fig. 1 demonstrated a representative example of multiplex PCR for detection of pvl and tsst-1 genes.
Fig. 1. Multiplex PCR for pvl (502 bp) and tsst-1 (398 bp) genes. M: Marker (100 bp); 1: Negative control, 2-3: Test isolates, 4: Positive control for pvl gene, 5: Positive control for tsst-1 gene
Table (3):
Distribution of mecA , tsst-1 and pvl genes among Staphylococcus aureus isolates
Gene |
Staphylococcus aureus Isolates (No=180) |
|---|---|
mecA positive |
88 (48.9%) |
mecA negative |
92 (51.1%) |
tsst-1 positive |
109 (60.6%) |
tsst-1 negative |
71 (39.4%) |
pvl positive |
30 (16.7%) |
pvl negative |
150 (83.3%) |
Values are expressed as No (%)
Out of 88 isolated MRSA strains, 37 (42.0%) were positive only for the tsst-1 gene and 21 (23.9%) were positive for both tsst-1 and pvl genes. Among the 92 methicillin sensitive S. aureus (MSSA) isolates,44 (47.8%) were positive only for the tsst-1 gene, 2 (2.2%) were positive only for the pvl gene and 7 (7.6%) were positive for both genes as demonstrated in Table 4. Totally, the tsst-1 gene was detected in 65.9% (58/88) of MRSA and 55.4% (51/92) of MSSA strains. No significant association was found between the carriage of mecA and tsst-1 genes (P=0.69). Regarding the pvl gene, it was found in 23.9% (21/88) of MRSA and 9.8% (9/92) of MSSA isolates. The pvl gene was significantly associated with the presence of mecA gene (P=0.04).
Table (4):
Distribution of tsst-1 and pvl genes among methicillin resistant and methicillin sensitive Staphylococcus aureus isolates
Gene |
MRSA isolates (mecA positive) |
MSSA isolates (mecA negative) |
Staphylococcus aureus Isolates |
|---|---|---|---|
Only tsst-1 positive |
37 (42.0%) |
44 (47.8%) |
81 (45.0%) |
Only pvl positive |
0 |
2 (2.2%) |
2 (1.1%) |
Both tsst-1 and pvl positive |
21 (23.9%) |
7 (7.6%) |
28 (15.6%) |
Both tsst-1 and pvl negative |
30 (34.1%) |
39 (42.4%) |
69 (38.3%) |
Total |
88 (100%) |
92 (100%) |
180 (100%) |
Values are expressed as No (%)
MRSA= Methicillin resistant Staphylococcus aureus
MSSA= Methicillin sensitive Staphylococcus aureus
Among the 88 mecA gene positive S. aureus isolates, 39(44.3%), 20(22.7%) and 8(9.1%) were recovered from blood, urine and endotracheal aspirates respectively. Of the 109 tsst-1 gene positive S. aureus strains, 41 (37.6%) were isolated from blood and 29 (26.6%) from urine. As regard the 30 pvl positive S. aureus isolates, 11(36.7%) and 7 (23.3%) were obtained from blood and urine respectively. No significant association was found between the source of clinical sample and the presence of mecA, tsst-1 or pvl genes (Table 5).
Table (5):
Distribution of mecA, tsst-1 and pvl genes among Staphylococcus aureus isolates recovered from different clinical samples.
|
Sample |
mecA gene | tsst-1 gene | pvl gene | ||||||
|---|---|---|---|---|---|---|---|---|---|
| Positive | Negative | P value | Positive | Negative | P value | Positive | Negative | P value | |
| Blood | 39 (44.3) | 36 (39.1) | 0.72 | 41 (37.6) | 34 (47.9) | 0.56 | 11 (36.7) | 64 (42.7) | 0.68 |
| Urine | 20 (22.7) | 23 (25.0) | 0.88 | 29 (26.6) | 14 (19.7) | 0.68 | 7 (23.3) | 36 (24.0) | 0.89 |
| Endotracheal aspirates | 8 (9.1) | 11 (12.0) | 0.49 | 13 (11.9) | 6 (8.5) | 0.72 | 4 (13.3) | 15 (10.0) | 0.69 |
| Wound drainages | 6 (6.8) | 7 (7.6) | 0.76 | 9 (8.3) | 4 (5.6) | 0.64 | 3 (10.0) | 10 (6.7) | 0.43 |
| Sputum | 5 (5.7) | 7 (7.6) | 0.66 | 7 (6.4) | 5 (7.0) | 0.65 | 2 (6.7) | 10 (6.7) | 1.0 |
| Eye secretions | 6 (6.8) | 5 (5.4) | 0.59 | 6 (5.5) | 5 (7.0) | 0.57 | 2 (6.7) | 9 (6.0) | 0.89 |
| Abscesses | 4 (4.5) | 3 (3.3) | 0.77 | 4 (3.7) | 3 (4.2) | 0.69 | 1 (3.3) | 6 (4.0) | 0.71 |
| Total | 88 | 92 | 109 | 71 | 30 | 150 | |||
Values are expressed as No (%)
In the present study, we determined the antibiotic resistance patterns of the recovered S. aureus isolates. Of the 180 S. aureus isolates, 108 (60.0%) were resistant to penicillin, 105 (58.3%) to cefazolin, 93 (51.7%) to tetracycline, 79 (43.9%) to ciprofloxacin, 63 (35.0%) to erythromycin, 59 (32.8%) to gentamicin, 62 (34.4%) to clindamycin and 45 (25.0%) to trimethoprim-sulfamethoxazole. The highest resistance observed was to penicillin while all isolates were sensitive to vancomycin and linezolid. Antibiotic resistance patterns in relation to the existence or absence of mecA, tsst-1 and pvl genes were given in Table 6.
Resistance patterns within MRSA strains was found to penicillin (100%), cefazolin (100%) followed by tetracycline (65.9%), erythromycin (48.9%), clindamycin (48.9%), ciprofloxacin (47.7%), gentamicin (45.5%) and trimethoprim-sulfamethoxazole (34.1%). Apart from ciprofloxacin, linezolid and vancomycin, resistance to evaluated antibiotics were significantly higher with the presence of mecA gene. Furthermore, the existence of mecA gene was significantly associated with multidrug resistant isolates (P= 0.041). No significant association was detected between antibiotic resistance and the presence of tsst-1 or pvl genes (Table 6).
Table (6):
Relation of mecA, tsst-1 and pvl genes to antibiotic resistance patterns of Staphylococcus aureus isolates.
|
Antibiotic resistance
|
mecAgene | tsst-1 gene | pvl gene | ||||||
|---|---|---|---|---|---|---|---|---|---|
| Positive | Negative | P value | Positive | Negative | P value | Positive | Negative | P value | |
| Penicillin | 88 (100) | 20 (21.7) | 0.012* | 68 (62.4) | 40 (56.3) | 0.60 | 21 (70.0) | 87 (58.0) | 0.59 |
| Cefazolin | 88 (100) | 17 (18.5) | 0.001* | 63 (57.8) | 42 (59.2) | 0.88 | 21 (70.0) | 84 (56.0) | 0.48 |
| Gentamicin | 40 (45.5) | 19 (20.7) | 0.048* | 41 (37.6) | 18 (25.4) | 0.08 | 9 (30.0) | 50 (33.3) | 0.66 |
| Erythromycin | 43 (48.9) | 20 (21.7) | 0.039* | 41 (37.6) | 22 (31.0) | 0.49 | 12 (40.0) | 51 (34.0) | 0.58 |
| Tetracycline | 58 (65.9) | 35 (38.0) | 0.04* | 55 (50.5) | 38 (53.5) | 0.51 | 14 (46.7) | 79 (52.7) | 0.7 |
| Ciprofloxacin | 42 (47.7) | 37 (40.2) | 0.62 | 52 (47.7) | 27 (38.0) | 0.22 | 16 (53.3) | 63 (42.0) | 0.42 |
| Clindamycin | 43 (48.9) | 19 (20.7) | 0.046* | 40 (36.7) | 22 (31.0) | 0.47 | 11 (36.7) | 51 (34.0) | 0.79 |
|
Trimethoprim-sulfamethoxa zole |
30 (34.1) | 15 (16.3) | 0.044* | 30 (27.5) | 15 (21.1) | 0.37 | 8 (26.7) | 37 (24.7) | 0.65 |
| Linezolid | 0 | 0 | – | 0 | 0 | – | 0 | 0 | – |
| Vancomycin | 0 | 0 | – | 0 | 0 | – | 0 | 0 | – |
| Total | 88 | 92 | 109 | 71 | 30 | 150 | |||
Values are expressed as No (%)
*Statistically significant
Staphylococcus aureus is increasingly recognized globally as a common and serious cause of various community and healthcare associated infections with a significant increase in not only the morbidity but also in mortality. They are known producers of different types of toxins and other virulence factors that shape up their clinical significance. Molecular studies have shown that some MRSA strains that carry tsst-1 and pvl genes7 have the ability to produce toxins encoded by these genes responsible for severe infections such as TSS and necrotic pneumonia with a mortality rate up to 75%19. However, treating such infections is considered a challenge to physicians in view of their antimicrobial resistant properties. Therefore, having knowledge about virulence factors of S. aureus and local prevalence of MRSA is beneficial for control and treatment of S. aureus infections.
In our study, out of the 180 S. aureus strains investigated, a predominance of 88 (48.9%) were found to be MRSA, similar to the results reported by Barakat and his colleagues who conducted a study in Egypt with a 45.3% of S. aureus isolates were MRSA20. Nevertheless, the reported prevalence of MRSA has varied widely dependent on location and the socioeconomic status of the region. The MRSA prevalence was found to be as low as 34% in European countries21,22, 18% in Russia23 and 41.6% in USA24. On the other hand, studies from developing countries have reported MRSA prevalence of as high as 72% in Lebanon25, 87.6% in Iran26 and 80% in China27 which is considerably higher than our findings. We have reported that the source of 44.3% of MRSA isolates was the blood, going through the same stream with other studies who reported that MRSA strains were responsible for majority of bloodstream infections among hospitalized patients28,29.
In the current study, phenotypic detection of methicillin resistance correlated well with genotypic detection of mecA gene. However, Motamedi and his colleagues have reported that only 30% of the phenotypically identified as MRSA isolates had the mecA gene30, that was attributed to different mechanisms of methicillin resistance other than mecA gene. Nevertheless, our results have shown that methicillin resistance is primarily mediated by mecA gene among the studied population.
In our study, the multiplex PCR has detected tsst-1 gene in 60.6% of the recovered S. aureus isolates, while Xie and his colleagues have reported that tsst-1 gene prevalence of 48% which was lower than our findings31. However, other studies have reported lower prevalence of tsst-1 gene as less as 20%24, 32-35. This variation could be attributed to the geographical location and also to different methodology encountered in these studies as their detection of the tsst-1 gene was carried out at the protein level only32,33. The expression of tsst-1 gene does not grant the production of the toxin. Therefore, despite the presence of the gene, the toxin might not been expressed among the tested group which could explain the differences in the reported tsst-1 gene prevalence.
We reported that the tsst-1 gene existed in 65.9% of MRSA and 55.4% of MSSA strains. The presence of tsst-1 gene was not significantly associated with either MRSA or MSSA groups. This was concordant with reports from Zarei Koosha and his colleagues who detected tsst-1 gene among 69.8% of MRSA isolates26. Other studies have also reported that up to 58% of MRSA strains were tsst-1 gene positive33,36,37. Similar to our findings, the prevalence of tsst-1 gene was reported up to 56% among the MSSA isolates in other studies24-26, 38.
In our study, we found that 16.7% of isolated S. aureus strains were positive for the pvl gene, putting in consideration that in previous studies, the prevalence of pvl gene has shown a wide variation ranged from 2% up to 38.9% 39,40. Moreover, we have detected the pvl gene in 23.9% of MRSA and 9.8% of MSSA isolates. This was similar to the findings of Goudarzi and his colleagues, who reported the prevalence of pvl gene of 21.4% within MRSA isolates37, and Abiri and his colleagues who detected pvl gene among 21.5% of mecA positive MRSA strains7. We found that pvl gene was significantly prevalent in MRSA group (P=0.04). Nevertheless, these findings were contrary to what been reported by Motamedi and his colleagues, that mecA and pvl genes did not show any association30.
In our study, antimicrobial susceptibility patterns of S. aureus isolates have shown higher rate of resistance to many antimicrobials that are routinely prescribed on daily bases in clinical practice for case of staphylococcal infections with the highest resistance observed toward penicillin as previously reported in other studies23,24. Such findings highlight the urge for the availability of an accurate and geographically targeted data about local antibiotic susceptibility patterns for appropriate antibiotic selection. This measure will hinder the rapid spreading of antimicrobial resistant S. aureus strains and it will give a solution for major challenge faced by healthcare providers in their clinical practice while treating infections caused by such isolates.
We found that 35.0% of S. aureus isolates were resistant to erythromycin which was similar to previous reports from Iran (51.3%)26 and China (97.8%)33, others have found the resistance to erythromycin was as low as 0%23. On the other hand, the prevalence of tetracyclin resistant strains was 51.7% close to the 48% reported by Tokajian and his colleagues in Lebanon25. Gentamicin resistance was found to be 32.8% that is nearly similar to the reports from China (28.1%)24 and higher than the reports from Russia (19%)23. In accordance with other reports, none of our isolates demonstrated resistance to vancomycin or linezolid24,25. The reported MRSA resistance patterns in our work were in line with the findings of Goudarzi and his colleagues36.
It has been shown that S. aureus strains isolated from different locations had different virulence genetic profiles. Therefore, the variations between other studies and our findings in terms of prevalence of the studied genes and antimicrobial resistance patterns might be attributed to different geographical locations. Nevertheless, different study design, successful execution of infection control measures and implementation of properly designed antibiotic stewardship policy might offer another explanation as well. Moreover, the variations in antimicrobial resistance reported from different regions emphasize the importance of tailoring antibiotic policy based on local antibiotic sensitivity patterns and international guidelines. To the best of our knowledge, this is the first research conducted in our locality to find the association of both tsst-1 and pvl genes with mecA gene in S. aureus isolates and to study their effect over the antibiotic susceptibility pattern of such isolates.
The pvl and tsst-1 genes positive S. aureus strains could lead to severe infections with high morbidity and mortality. Therefore, frequent monitoring of these strains and their virulence factors are substantially important. We reported a high prevalence of tsst-1 gene among S. aureus isolates. The tsst-1 gene was not significantly associated with either MRSA or MSSA groups. On the other hand, the pvl gene was significantly associated with MRSA group. The isolated S. aureus strains demonstrated elevated antibiotic resistance to many of the routinely used antimicrobials and the mecA gene was significantly associated with multidrug resistant isolates. Hence, implementation of antibiotic stewardship policy based on local antibiotic resistance patterns would help the physicians in selecting the choice antibiotics for their patients.
Acknowledgments
The authors like to extend their gratitude towards the professional help and constant support of the medical and nursing staff of Mansoura University Children Hospital, Iraq.
Conflicts Of Interest
The authors declare that there are no conflicts of interest.
Authors’ Contribution
AS developed the concept and designed the study. AS and YN did sample processing. AS collected and analysed data and did the interpretation of results. YN carried out protocol development and contributed in designing and writing of the methodological part of the article. AS wrote the manuscript. AS and YN did critical revision of the article. AS reviewed and approved the manuscript.
Funding
None.
Data Availability
All datasets generated or analyzed during this study are included in the manuscript.
Ethics Statement
All procedures performed in this study were in accordance with the ethical standards of the institutional research committee and with the 1964 Helsinki declaration and its later amendments or comparable ethical standards. Informed consent was obtained from at least one of parents of participants included in the study. The study protocol has been approved by Faculty of Medicine – Mansoura University ethical committee.
- Al-KhulaifiManal M., Amin ArefNagwa M., Al Salamah A.A. Phage typing, PCR amplification for mecA gene, and antibiotic resistance patterns as epidemiologic markers in nosocomial outbreaks of methicillin resistant Staphylococcus aureus. Saudi J Biol Sci., 2009; 16(1): 37–49.
- Hu Y., Liu A., Vaudrey J., Vaiciunaite B., Moigboi C., McTavish S.M., et al. Combinations of beta-lactam or aminoglycoside antibiotics with plectasin are synergistic against methicillin-sensitive and methicillin-resistant Staphylococcus aureus. PLoS One, 2015; 10(2): e0117664 .
- Lowy F.D. Antimicrobial resistance: the example of Staphylococcus aureus. J. Clin. Invest., 2003; 111(9): 1265–73.
- Pillai M.M., Latha R., Sarkar G. Detection of methicillin resistance in Staphylococcus aureus by polymerase chain reaction and conventional methods: a comparative study. J. Lab Physicians, 2012; 4(2): 83–8.
- Nowrouzian F.L., Karami N.,Welinder-Olsson C., Ahren C. Virulence gene typing of methicillin-resistant Staphylococcus aureus as a complement in epidemiological typing. J Microbiol Methods, 2013; 93(3): 173–6.
- Ba X, Harrison E.M., Edwards G.F., Holden M.T., Larsen A.R., Petersen A., et al. Novel mutations in penicillin-binding protein genes in clinical Staphylococcus aureus isolates that are methicillin resistant on susceptibility testing, but lack the mec gene. J. Antimicrob. Chemother., 2014; 69(3): 594–7.
- Abiri P., Sepahi A.A., Goudarzi H. and Goudarzi M. Distribution of Genes Encoding Toxin, Adhesion, and Antibacterial Resistance Among Various SCCmecTypes of Methicillin-Resistant Staphylococcus aureus Isolated From Intensive Care Unit, Tehran, Iran. Jundishapur J. Microbiol. 2017; 10(12): e14477-14486.
- Los FC, Randis TM, Aroian RV, Ratner AJ. Role of pore-forming toxins in bacterial infectious diseases. Microbiol. Mol. Biol Rev., 2013; 77(2): 173–207.
- Van der Meeren B.T., Millard P.S., Scacchetti M., Hermans M.H., Hilbink M., Concelho T.B., et al. Emergence of methicillin resistance and Panton-Valentine leukocidin positivity in hospital- and community-acquired Staphylococcus aureus infections in Beira, Mozambique. Trop Med Int Health., 2014; 19(2): 169–76.
- Fraser J, Arcus V, Kong P, Baker E, Proft T. Superantigens – powerful modifiers of the immune system. Trends of Mol Med Today. 2000; 6(3):125– 32.
- Llewelyn M, Cohen J. Superantigens: microbial agents that corrupt immunity. The Lancet Infectious Diseases. 2002; 2(3): 156–62.
- Deurenberg R.H., Nieuwenhuis R.F., Driessen C., London N., Stassen F.R., van Tiel F.H., et al. The prevalence of the Staphylococcus aureus tst gene among community- and hospital-acquired strains and isolates from Wegener’s Granulomatosis patients. FEMS Microbiol Lett., 2005; 245(1): 185–9.
- Durand G, Bes M, Meugnier H, Enright MC, Forey F, Liassine N, et al. Detection of new methicillin-resistant Staphylococcus aureus clones containing the toxic shock syndrome toxin 1 gene responsible for hospital- and community-acquired infections in France. J. Clin. Microbiol. 2006; 44(3): 847–53.
- Hassan R., Barwa R., EL-Sokkary M.M., Ashraf D. Virulence Characteristics of Methicillin Resistant Staphylococcus aureus Isolated From Different Clinical Sources. N Egypt J Microbiol., 2017; 48: 14-31.
- Helal Z., Radwan S., El-Tayeb W. Involvement of PVL Producing MRSA in Pneumonia Patients, Cairo, Egypt. Archives of Clinical Microbiology, 2012; 3(6).
- Clinical and Laboratory Standards Institute. Performance Standards for Antimicrobial Susceptibility Testing.28th ed. CLSI supplement M100. Wayne, PA: Clinical and Laboratory Standards Institute; 2018.
- Goudarzi M, Seyedjavadi SS, Azad M, Goudarzi H, Azimi H. Distribution of spa types, integrons and associated gene cassettes in Staphylococcus aureus strains isolated from intensive care units of hospitals in Tehran, Iran. Arch. Clin. Infect. Dis., 2016; 11(4): 38813.
- Azimian A., Havaei S.A., Fazeli H., Naderi M., Ghazvini K., Samiee S.M., et al. Genetic characterization of a vancomycin-resistant Staphylococcus aureus isolate from the respiratory tract of a patient in a university hospital in northeastern Iran. J. Clin. Microbiol. 2012; 50(11): 3581–5.
- Yanagihara K., Kihara R., Araki N., Morinaga Y., Seki M., Izumikawa K., et al. Efficacy of linezolid against Panton-Valentine leukocidin (PVL)-positive meticillin-resistant Staphylococcus aureus (MRSA) in a mouse model of haematogenous pulmonary infection. Int. J. Antimicrob. Agents, 2009; 34(5): 477–81.
- Barakat G.I., Nabil Y.M. Correlation of mupirocin resistance with biofilm production in methicillin-resistant Staphylococcus aureus from surgical site infections in a tertiary centre, Egypt. J. Glob. Antimicrob. Resist., 2016; 4: 16-20.
- Voss A., Milatovic D., Wallrauch-Schwarz C., Rosdahl V.T., Braveny I. Methicillin-resistant Staphylococcus aureus in Europe. Eur. J. Clin. Microbiol. Infect. Dis., 1994; 13(1): 50–5.
- Gbaguidi-Haore H., Thouverez M., Couetdic G., Cholley P., Talon D., Bertrand X. Usefulness of antimicrobial resistance pattern for detecting PVL- or TSST-1-producing meticillin-resistant Staphylococcus aureus in a French university hospital. J. Med. Microbiol., 2009; 58(10): 1337–42.
- Vorobieva V., Bazhukova T., Hanssen A.M., Caugant D.A., Semenova N., Haldorsen B.C., et al. Clinical isolates of Staphylococcus aureus from the Arkhangelsk region, Russia: antimicrobial susceptibility, molecular epidemiology, and distribution of Panton- Valentine leukocidin genes. APMIS, 2008; 116(10): 877–87.
- Yu F., Li T., Huang X., Xie J., Xu Y., Tu J., et al. Virulence gene profiling and molecular characterization of hospital-acquired Staphylococcus aureus isolates associated with bloodstream infection. Diagn. Microbiol Infect. Dis., 2012; 74(4): 363–8.
- Tokajian S., Haddad D., Andraos R., Hashwa F., Araj G. Toxins and Antibiotic Resistance in Staphylococcus aureus Isolated from a Major Hospital in Lebanon. ISRN Microbiology, 2011; 2011: 1–9.
- ZareiKoosha R., Mahmoodzadeh Hosseini H., Mehdizadeh Aghdam E., Ghorbani Tajandareh S., ImaniFooladi A.A. Distribution of tsst-1 and mecA Genes in Staphylococcus aureus Isolated From Clinical Specimens. Jundishapur J. Microbiol., 2016; 9(3): e29057.
- Peng Q., Hou B., Zhou S., Huang Y., Hua D., Yao F., et al. Staphylococcal cassette chromosome mec (SCCmec) analysis and antimicrobial susceptibility profiles of methicillin-resistant Staphylococcus aureus (MRSA) isolates in a teaching hospital, Shantou, China. Afr. J. Microbiol. Res., 2010; 4(9): 844–8.
- Eftekhar F., Rezaee R., Azad M., Azimi H., Goudarzi H., Goudarzi M. Distribution of adhesion and toxin genes in Staphylococcus aureus strains recovered from hospitalized patients admitted to the ICU. Arch. Pediatr. Infect. Dis., 2016; 5(1): e39349
- Rasahidi Nezhad R., Meybodi S.M., Rezaee R., Goudarzi M., Fazeli M. Molecular characterization and resistance profile of methicillin resistant Staphylococcus aureus strains isolated from hospitalized patients in intensive care unit, Tehran-Iran. Jundishapur J. Microbiol., 2017; 10(3): 41666.
- Motamedi H., RahmatAbadi S.S., Moosavian S.M., Torabi M. The Association of Panton-Valentine leukocidin and mecA Genes in Methicillin-Resistant Staphylococcus aureus Isolates From Patients Referred to Educational Hospitals in Ahvaz, Iran. Jundishapur J. Microbiol., 2015; 8(8): e22021.
- Xie Y., He Y., Gehring A., Hu Y., Li Q., Tu S.I., et al. Genotypes and toxin gene profiles of Staphylococcus aureus clinical isolates from China. PLoS One, 2011; 6(12): e28276
- Becker K., Friedrich A.W., Lubritz G., Weilert M., Peters G., Von Eiff C. Prevalence of genes encoding pyrogenic toxin superantigens and exfoliative toxins among strains of Staphylococcus aureus isolated from blood and nasal specimens. J Clin. Microbiol., 2003; 41(4): 1434–9.
- Liu M., Liu J., Guo Y., Zhang Z. Characterization of virulence factors and genetic background of Staphylococcus aureus isolated from Peking University People’s Hospital between 2005 and 2009. Curr. Microbiol., 2010; 61(5): 435–43.
- Demir C., Aslantas O., Duran N., Ocak S., Ozer B. Investigation of toxin genes in Staphylococcus aureus strains isolated in Mustafa Kemal University Hospital. Turk J. Med. Sci., 2011; 41(2): 343–52.
- Hoseini Alfatemi S.M., Motamedifar M., Hadi N., Sedigh EbrahimSaraie H.. Analysis of Virulence Genes Among Methicillin Resistant Staphylococcus aureus (MRSA) Strains. Jundishapur J. Microbiol., 2014; 7(6): e10741.
- Goudarzi M., Seyedjavadi S.S., Nasiri M.J., Goudarzi H., SajadiNia R., Dabiri H. Molecular characteristics of methicillin-resistant Staphylococcus aureus (MRSA) strains isolated from patients with bacteremia based on MLST, SCCmec, spa, and agr locus types analysis. Microb. Pathog., 2017; 104: 328–35.
- Goudarzi M., Goudarzi H., Sa Figueiredo A.M., Udo E.E., Fazeli M., Asadzadeh M., et al. Molecular Characterization of Methicillin Resistant Staphylococcus aureus Strains Isolated from Intensive Care Units in Iran: ST22-SCCmec IV/t790 Emerges as the Major Clone. PLoS One, 2016; 11(5): 0155529.
- Jimenez J.N., Ocampo A.M., Vanegas J.M., Rodriguez E.A., Garces C.G., Patino L.A., et al. Characterisation of virulence genes in methicillin susceptible and resistant Staphylococcus aureus isolates from a paediatric population in a university hospital of Medellin, Colombia. Mem. Inst. Oswaldo Cruz, 2011; 106(8): 980–5.
- Melles D.C., Gorkink R.F., Boelens H.A., Snijders S.V., Peeters J.K., Moorhouse M.J., et al. Natural population dynamics and expansion of pathogenic clones of Staphylococcus aureus. J. Clin. Invest., 2004; 114(12): 1732–40.
- Okon K.O., Basset P., Uba A., Lin J., Oyawoye B., Shittu A.O., et al. Cooccurrence of predominant Panton-Valentine leukocidin-positive sequence type (ST) 152 and multidrug-resistant ST 241 Staphylococcus aureus clones in Nigerian hospitals. J. Clin. Microbiol., 2009; 47(9): 3000–3.
© The Author(s) 2019. Open Access. This article is distributed under the terms of the Creative Commons Attribution 4.0 International License which permits unrestricted use, sharing, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.