ISSN: 0973-7510
E-ISSN: 2581-690X
This study was carried out to investigate the relationship between common variants in two vitamin D pathway genes (VDR and CYP27B1) and vitamin D3 serum levels. In this study, serum vitamin D metabolite levels were measured in the blood samples of 200 patients with alopecia areata. Then, single nucleotide polymorphisms (SNPs) in VDR and CYP27B1 were analyzed using polymerase chain reaction (PCR)-sequencing. Sixty-three variations were observed in these genes (42 variations in CYP27B1 and 21 variations in VDR). A significant difference in Rs1544410 (odds ratio: 7, P < 0.0005) and rs4646536 (odds ratio: 4.043, P < 0.0005) variants was found between the patients and controls. The study showed the relationship between the two polymorphisms, Rs1544410 (odds ratio: 7, 95% CI, 1–8) and rs4646536 (odds ratio: 4.043, 95% CI, 3–14.038) on the genes VDR and CYP27B1, respectively, with increased risk of developing vitamin D3 insufficiency in the Iranian population. Therefore, SNPs in the VDR and CYP27B1 genes can be considered as prognostic biomarkers of the risk of developing vitamin D3 deficiency.
VDR, CYP27B1, VITAMIN D3, RS 1544410, SNPs
Vitamin D is a group of biologically inactive, fat-soluble prohormones with known importance in maintaining body health. There are two forms of vitamin D (calciferol): vitamin D2 (ergocalciferol) and vitamin D3 (cholecalciferol). The accepted indicator of vitamin D3 sufficiency in humans is serum 25-hydroxyvitamin D3 (25(OH)D3) levels (normal levels defined as 25(OH)D3 serum levels from 30–100 ng/mL). Currently, there is ample evidence suggesting an association of vitamin D3 deficiency (< 10 ng/mL) with various physiological disorders, especially immunological dysfunctions such as alopecia areata. The main causes of human vitamin D3 insufficiency (defined as serum 25(OH)D3 levels <30 ng/mL) include limited dietary sources, inadequate sun exposure, and presence of polymorphisms in several vitamin D genes (VDR, VDBP, CYP24A1, CYP2R1, CYP27A1, and CYP27B1)1-4.
Several studies have reported on the biological role of the vitamin D receptor (VDR) on the immunomodulatory capabilities of 1,25-dihydroxy-vitamin D3 (1,25(OH)2D3). The VDR gene is located in the human 12q12-q22 region. It has more than 470 single nucleotide polymorphisms (SNPs), some of which modulate 1,25(OH)2D3 uptake. In several experimental animal models, 1,25(OH)2D3 administration markedly prevented the development of autoimmune diseases. Therefore, it is possible that point mutations in the polymorphisms of VDR may have a pivotal role in the development of autoinflammatory and other autoimmune diseases5-8.
Studies have shown a significant association between SNPs in CYP27B1 and serum 25(OH)D3 levels (9, 10). CYP27B1 is a mitochondrial cytochrome P450. Its gene encodes 25-hydroxyvitamin D-1α-hydroxylase (1α-hydroxylase), an enzyme that catalyzes the final step of vitamin D activation to calcitriol and the 1α-hydroxylation of 25(OH)D3 to produce 1,25(OH)2D3. Furthermore, 1,25(OH)2D3 plays a significant role in immune system regulation of the human body and also has prodifferentiative and antiproliferative properties on numerous cell types11,12.
The main goal of this study was to investigate the association of common polymorphisms in the VDR and CYP27B1 genes with vitamin D3 serum levels in an Iranian population. The Consider of the examined polymorphisms as biomarkers for the exact diagnosis of vitamin D deficiency cannot be ignored.
Subject selection and sampling
Archived blood samples from 2016 to 2019 of 800 patients with alopecia areata were obtained from the blood bank of the Iranian Association of Patients with Alopecia areata. The eligibility criteria of the patients included individuals aged 25 ± 5 years living in central Tehran. In order to eliminate the effect of sunlight on the level of vitamin D3 in the blood, each year (2016, 2017, 2018 and 2019) only the blood samples of patients collected between July and September were examined. Patients who traveled outside of Tehran during the study, pregnant or lactating women, and patients with a history of taking vitamin d supplements were excluded from the study.
The levels of 25(OH)D in frozen serum were measured using an ELISA kit (EIA Kit, IDS, UK 25-OH-D). The usual quality control method was used to ensure the accuracy and precision of the tests throughout the experimental investigation. The sensitivity range of the kit was 5 nmol/L or 1.5 ng/mL, the intra-test coefficient of variation was 4.3%, and the extracorporeal coefficient of variation was 8.7%.
Questionnaires that detailed demographic data, sex, age, history of various diseases, medications, and smoking were collected from the Data Bank of the Iranian Association of Patients with Alopecia areata. Based on this, 200 of the 800 studied cases were included in the study. One hundred subjects were assigned as the patient group (50 cases with vitamin D3 insufficiency and 50 cases with vitamin D3 deficiency) and 100 individuals as the normal group (normal levels defined as 25(OH)D3 serum levels from 30–100 ng/mL) through random selection (Table 1).
Table (1):
Demographic and serum vitamin D3 variables in participant.
| Characteristics/variables | Baseline vitamin D3 status* (N=200) | ||
|---|---|---|---|
| 25(OH)D3 (ng/mL) | Deficient(N=50) patients |
Insufficient (N=50) patients |
Sufficient (N=100) Case-control |
| X≤10 | 10 <X< 30 | 30 ≤X≥100 | |
| Gender F |
84(64/12%) | 37(28/24%) | 10(7.64%) |
| M | 5(7.24%) | 47(68.12%) | 3(24.64%) |
| BMI (Kg/m2) | 20<X<27 | 20<X<27 | 20<X<27 |
| age | 25±5 years | 25±5 years | 25±5 years |
As previous studies have shown a significant association between SNPs in the CYP27B1 and VDR genes and serum 25(OH)D3 levels, we investigated SNPs in these genes using the polymerase chain reaction (PCR)-sequencing method. We also measured the association between these SNPs and changes in serum vitamin D3 metabolite levels in patients with alopecia areata.
DNA extraction and primer
Genomic DNA from the blood was isolated using the DNA extraction kit DNP (Sinaclon, Iran), and its quality and quantity were analyzed using agarose gel electrophoresis and a NanoDrop 2000 UV-Vis Spectrophotometer (Thermo Fisher Scientific), respectively.
Dedicated primers were designed using Primer 3 software to investigate important polymorphisms in the VDR (F: 5’-GTATGCCTAGACTCCACCTCC-3’. R: 5’-GGAAGAGGTCAAGGGTCAC-3’) and CYP27B1 (CYP27B1[A]: F: 5’-CCTATTCCCAAGCCCAGTC-3’. R: 5’-GGAGAGGGATTGCGTCTG-3’and CYP27B1[B]: F: 5’-CTGACCCACTTCCTGTTC-3’, R: 5’-CAGTAGAAAGGGTGCATAGG-3’) regions according to the sequencing-PCR method. These primers were synthesized by SinaClon.
Polymerase chain reaction (PCR)-sequencing
The final volume used in the PCR reactions was 25 mL, in which genomic DNA (205–250 ng) was amplified. The reaction used PCR Master Mix (2X) Sinaclon kit (0.08 units/μl Taq DNA polymerase in reaction buffer, 3 mM MgCl, 2 mM dATP, 0.4 mM dCTP, 0.4 mM dGTP, and 0.4 mM dTTP) (Cat. No.: MM2061). An advanced biometric thermocycler was used in this study. The protocol for PCR amplification is mentioned in Table 2.
Table (2):
PCR reaction protocol.
| Gene | Primer | Primer | DNA (200-250 ng) |
Master mix PCR | DEPC1 Water | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Forward | Reverse | ||||||||||
| CYP27B1(A) | F | CCTATTCCCAAGCCCAGTC | 0.5 µl | 0.5 µl | 1 µl | 10µl | 13µl | ||||
| R | GGAGAGGGATTGCGTCTG | ||||||||||
| CYP27B1(B) | F | CTGACCCACTTCCTGTTC | 0.5 µl | 0.5 µl | 1 µl | 10µl | 13µl | ||||
| R | CAGTAGAAAGGGTGCATAGG | ||||||||||
| VDR (V) | F | GTATGCCTAGACTCCACCTCC | 0.7 µl | 0.7 µl | 1 µl | 10µl | 12.6µl | ||||
| R | GGAAGAGGTCAAGGGTCAC | ||||||||||
| First denaturation | Cycle:35 | Final extension | Product size | ||||||||
| Denaturation | Annealing | Extension | |||||||||
| CYP27B1(A) | 950C – 5 min | 950C- 30 sec | 58 0C- 45sec | 720C- 30 sec | 720C – 15 min | 703pb | |||||
| CYP27B1(B) | 950C – 5 min | 950C- 30 sec | 550C- 45 sec | 720C- 30 sec | 720C – 15 min | 773pb | |||||
| VDR (V) | 950C – 5 min | 950C- 30 sec | 580C- 45 sec | 720C- 30 sec | 720C – 15 min | 774pb | |||||
1Diethyl pyro carbonate
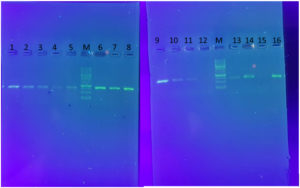
The PCR products were analyzed by electrophoresis in 1% agarose gel alongside a Thermo Scientific Gene Ruler 100 bp DNA Ladder for reference (Fig. 1).
Fig. 1. The amplified sequences of CYP27B1(B) (1,2,3,4,5), VDR (6,7,8) and CYP27B1(A) (9,10,11,13,14,16) through the sequencing- primer PCR method. M; ladder 1KB. Also, 12 and 15 are negative control.
The PCR products were sent to Rooyan Zista Gene Company for sequencing in both the forward and reverse directions. The sequencing data were analyzed using Finch TV version 1.4.0. NCBI BLAST was used as reference (Fig. 2).
Statistical analysis
Fisher’s exact test was used to evaluate the association between variables and to study the deviation from the Hardy-Weinberg equilibrium. A nominal significance level of P < 0.05 was accepted in all tests. All statistical analyses were performed using SPSS version 22.
In the present study, 100 blood samples with serum 25(OH)D3 levels under 30ng/mL and 100 blood samples from the control group with serum 25(OH)D3 levels between 30 and 100 ng/mL were genotyped for CYP27B1 and VDR region variations. The age range of all participating participants was 20 to 30 years old were reviewed. There were 69 males (24.64% normal and 75.36% patients) and 131 females (7.63% normal and 92.36% patients). The subsequent results showed 42 variations in CYP27B1 and 21 variations in VDR (Table 3). Four of these have been previously reported (Rs531190881, Rs1544410, Rs6447, and Rs4646536). Other mutations in the patients were novel mutations (5993 C=G, 6004 G>C, 6004 G=C, 6439 G=A, 6492 G=A, 6106 G>A, 6106 G=A, 6093 C=G, 6423 G=A, 6469 T=A, 6070 C=T, 6316 G=A=C=T, 6357 C=G, 7645 T>G, 7447 T=G, 7447 T>G, 7717 G=A, 7741 G=A, 7609 A=G, 7623 A=C, 7533 DEL C, 7533 C=T, 7540 G>C, 7540 G=A, 7555 G=C, 7555 G=T=C, 7565 G=C=T, 7565 G=T, 7716 G=A, 7716 G=T, 7902 G>A, 7902 G=A, 7798 T=A, 7479 C=G, 7800 G=A, 7484 C=T, 7443 INS T, 7446 T=G, 7446 INS T, 7904 G=C=T, 7904 G=T, 7948 G=A, 7823 G=A, 7724 A=G, 7843 G=A, 7553 T=C, 7842 A=T, 7936 G=T, 7879 T=C, 7878 G=C, 7941 G=A, 7643 INS T, 63459 G=A, 63459 G>A, 63474 G=T, 93959 INS G, 63814 G=C, 64086 T>G, 63472 G>T, 63472 G=T, 63454 A>C, 64066 INS A, 64073 INS G, 63481 A=T, 64118 G>A, 63565 C>A, 63558 T=A=C, 63545 G=C, 63537 G>T=C, 64076 INS C) (Table 3). Most of the variations were single nucleotide substitutions. The significant associations between the two variants Rs4646536 (CYP27B1) and Rs1544410 (VDR) with risk of developing vitamin D3 insufficiency and deficiency (P < 0.05) are shown in Table 3.
Table (3):
Frequency of SNPs in Case-control and patients.
| genotype | X < 10 patients |
k | pvalue | OR | 95% CI | 10 < X < 30 patients |
k | pvalue | OR | 95% CI | X > 30 Case-control |
||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 5993 C=G (a) | 14/28571 | 15.054a | 0.000 | 1.00 | 1.074 | 1.000 | 0 | _ | _ | _ | _ | _ | 0 |
| 6004 G>C | 7/142857 | 7.000a | 0.007 | 1.075 | 1.019 | 1.000 | 0 | _ | _ | _ | _ | _ | 0 |
| 6004 G=C | 7/142857 | 7.000a | 0.007 | 1.075 | 1.019 | 1.000 | 0 | _ | _ | _ | _ | _ | 0 |
| 6439 G=A | 7/142857 | 7.000a | 0.007 | 1.075 | 1.019 | 1.000 | 0 | _ | _ | _ | _ | _ | 0 |
| 6492 G=A | 7/142857 | 7.000a | 0.007 | 0 | 0 | 0 | 0 | 22.000a | 0.005 | 0 | 0 | 0 | 20 |
| 6106 G>A | 7/142857 | 7.000a | 0.007 | 1.075 | 1.019 | 1.000 | 0 | _ | _ | _ | _ | _ | 0 |
| 6106 G=A | 7/142857 | 7.000a | 0.007 | 1.075 | 1.019 | 1.000 | 0 | _ | _ | _ | _ | _ | 0 |
| 6093 C=G | 7/142857 | 7.000a | 0.007 | 1.075 | 1.019 | 1.000 | 0 | _ | _ | _ | _ | _ | 0 |
| 6423 G=A | 0 | _ | _ | _ | _ | _ | 12/5 | 13.000a | 0.004 | 1 | 1.065 | 1.000 | 0 |
| 6469 T=A | 0 | _ | _ | _ | _ | _ | 12/5 | 13.000a | 0.005 | 1 | 1.065 | 1.000 | 0 |
| 6070 C=T | 0 | _ | _ | _ | _ | _ | 12/5 | 13.000a | 0.005 | 1 | 1.065 | 1.000 | 0 |
| 6316 G=A=C=T | 0 | _ | _ | _ | _ | _ | 12/5 | 13.000a | 0.005 | 1 | 1.065 | 1.000 | 0 |
| 6357 C=G | 0 | _ | _ | _ | _ | _ | 12/5 | 13.000a | 0.005 | 1 | 1.065 | 1.000 | 0 |
| 7645 T>G (b) | 5/882353 | 6.000a | 0.013 | 1.064 | 1.012 | 1.000 | 0 | _ | _ | _ | 0 | ||
| 7447 T=G | 5/882353 | 6.000a | 0.013 | 1.064 | 1.012 | 1.000 | 8/333333 | 8.000a | 0.004 | 1.087 | 1.026 | 1.000 | 0 |
| 7447 T>G | 0 | _ | _ | _ | 8/333333 | 8.000a | 0.004 | 1.087 | 1.026 | 1.000 | 0 | ||
| 7717 G=A | 5/882353 | 6.000a | 0.013 | 1.064 | 1.012 | 1.000 | 0 | _ | _ | _ | _ | _ | 0 |
| 7741 G=A | 5/882353 | 6.000a | 0.013 | 1.064 | 1.012 | 1.000 | 0 | _ | _ | _ | _ | _ | 0 |
| 7609 A=G | 5/882353 | 6.000a | 0.013 | 1.064 | 1.012 | 1.000 | 0 | _ | _ | _ | _ | _ | 0 |
| 7623 A=C | 5/882353 | 6.000a | 0.013 | 1.064 | 1.012 | 1.000 | 0 | _ | _ | _ | _ | _ | 0 |
| 7989 T=C | 29/41176 | 11.000a | 0.0001 | 3 | 1 | 8.045 | 8/333333 | 0.000a | 0.0005 | 0.00 | 0.000 | 2.073 | 10 |
| 7989 T>C | 5/882353 | 6.000a | 0.013 | 1.064 | 1.012 | 1.000 | 16/66667 | 18.000a | 0.004 | 1 | 1 | 1 | 0 |
| 7989 T=G | 0 | 22.000a | 0.0005 | 0 | 0 | 0 | 0 | 22.000a | 0.005 | 0 | 0 | 0 | 20 |
| 7533 DEL C | 0 | _ | _ | _ | _ | _ | 8/333333 | 8.000a | _ | _ | 1.026 | 1.000 | 0 |
| 7533 C=T | 0 | _ | _ | _ | _ | _ | 8/333333 | 8.000a | 0.004 | 1.087 | 1.026 | 1.000 | 0 |
| 7540 G>C | 0 | _ | _ | _ | _ | _ | 8/333333 | 8.000a | 0.004 | 1.087 | 1.026 | 1.000 | 0 |
| 7540 G=A | 0 | 22.000a | 0.0005 | 0 | 0 | 0 | 0 | 22.000a | 0.005 | 0 | 0 | 0 | 20 |
| 7555 G=C | 11/76471 | 12.000a | 0.0005 | 1 | 1.057 | 1.000 | 0 | _ | _ | _ | 0 | ||
| 7555 G=T=C | 0 | _ | _ | _ | _ | _ | 8/333333 | 8.000a | 0.004 | 1.087 | 1.026 | 1.000 | 0 |
| 7565 G=C=T | 0 | _ | _ | _ | _ | _ | 8/333333 | 8.000a | 0.004 | 1.087 | 1.026 | 1.000 | 0 |
| 7565 G=T | 0 | _ | _ | _ | _ | _ | 8/333333 | 8.000a | 0.004 | 1.087 | 1.026 | 1.000 | 0 |
| 7716 G=A | 0 | _ | _ | _ | _ | _ | 8/333333 | 8.000a | 0.004 | 1.087 | 1.026 | 1.000 | 0 |
| 7716 G=T | 0 | _ | _ | _ | 8/333333 | 8.000a | 0.004 | 1.087 | 1.026 | 1.000 | 0 | ||
| 7902 G>A | 5/882353 | 6.000a | 0.013 | 1.064 | 1.012 | 1.000 | 8/333333 | 8.000a | 0.004 | 1.087 | 1.026 | 1.000 | 0 |
| 7902 G=A | 0 | _ | _ | _ | 8/333333 | 8.000a | 0.004 | 1.087 | 1.026 | 1.000 | 0 | ||
| 7479 C=G | 11/76471 | 12.000a | 0.0005 | 1 | 1.057 | 1.000 | 8/333333 | 8.000a | 0.004 | 1.087 | 1.026 | 1.000 | 0 |
| 7798 T=A | 0 | _ | _ | _ | _ | _ | 8/333333 | 8.000a | 0.004 | 1.087 | 1.026 | 1.000 | 0 |
| 7800 G=A | 0 | _ | _ | _ | _ | _ | 8/333333 | 8.000a | 0.004 | 1.087 | 1.026 | 1.000 | 0 |
| 7483 G=A | 5/882353 | 6.000a | 0.013 | 1.064 | 1.012 | 1.000 | 0 | _ | _ | _ | _ | _ | 0 |
| 7484 C=T | 5/882353 | 6.000a | 0.013 | 1.064 | 1.012 | 1.000 | 0 | _ | _ | _ | _ | _ | 0 |
| 7443 INS T | 5/882353 | 6.000a | 0.013 | 1.064 | 1.012 | 1.000 | 0 | _ | _ | _ | _ | _ | 0 |
| 7446 T=G | 5/882353 | 6.000a | 0.013 | 1.064 | 1.012 | 1.000 | 0 | _ | _ | _ | _ | _ | 0 |
| 7446 INS T | 0 | _ | _ | _ | _ | _ | 8/333333 | 8.000a | 0.004 | 1.087 | 1.026 | 1.000 | 0 |
| 7904 G=C=T | 0 | _ | _ | _ | _ | _ | 8/333333 | 8.000a | 0.004 | 1.087 | 1.026 | 1.000 | 0 |
| 7904 G=T | 5/882353 | 6.000a | 0.013 | 1.064 | 1.012 | 1.000 | 0 | _ | _ | _ | _ | _ | 0 |
| 7948 G=A | 5/882353 | 6.000a | 0.013 | 1.064 | 1.012 | 1.000 | 0 | _ | _ | _ | _ | _ | 0 |
| 7823 G=A | 5/882353 | 6.000a | 0.013 | 1.064 | 1.012 | 1.000 | 0 | _ | _ | _ | _ | _ | 0 |
| 7724 A=G | 5/882353 | 6.000a | 0.013 | 1.064 | 1.012 | 1.000 | 0 | _ | _ | _ | _ | _ | 0 |
| 7843 G=A | 5/882353 | 6.000a | 0.013 | 1.064 | 1.012 | 1.000 | 0 | _ | _ | _ | _ | _ | 0 |
| 7553 T=C | 5/882353 | 6.000a | 0.013 | 1.064 | 1.012 | 1.000 | 0 | _ | _ | _ | _ | _ | 0 |
| 7842 A=T | 5/882353 | 6.000a | 0.013 | 1.064 | 1.012 | 1.000 | 0 | _ | _ | _ | _ | _ | 0 |
| 7936 G=T | 11/76471 | 12.000a | 0.0005 | 1 | 1.057 | 1.000 | 0 | _ | _ | _ | _ | _ | 0 |
| 7879 T=C | 5/882353 | 6.000a | 0.013 | 1.064 | 1.012 | 1.000 | 0 | _ | _ | _ | _ | _ | 0 |
| 7878 G=C | 5/882353 | 6.000a | 0.013 | 1.064 | 1.012 | 1.000 | 0 | _ | _ | _ | _ | _ | 0 |
| 7941 G=A | 5/882353 | 6.000a | 0.013 | 1.064 | 1.012 | 1.000 | 0 | _ | _ | _ | _ | _ | 0 |
| 7643 INS T | 0 | _ | _ | _ | _ | _ | 8/333333 | 8.000a | 0.004 | 1.087 | 1.026 | 1.000 | 0 |
| 63459 G=A (v) | 7/692308 | 8.000a | 0.004 | 1.087 | 1.026 | 1.000 | 0 | _ | _ | _ | 0 | ||
| 63459 G>A | 0 | _ | _ | _ | _ | _ | 10 | 10.000a | 0.001 | 1 | 1.041 | 1 | 0 |
| 63474 G=T | 7/692308 | 8.000a | 0.004 | 1.087 | 1.026 | 1.000 | 0 | _ | _ | _ | 0 | ||
| 63980 G=A | 15/38462 | 16.000a | 0.0005 | 1 | 1.083 | 1.000 | 30 | 35.000a | 0.005 | 1 | 1 | 1 | 0 |
| 63980 G>A | 38/46154 | 7.000a | 0.005 | 2 | 1.000 | 4.000 | 40 | 9.000a | 0.002 | 2 | 1 | 5.020 | 20 |
| 63980 G>A=T | 7/692308 | 8.000a | 0.004 | 1.087 | 1.026 | 1.000 | 0 | _ | _ | _ | _ | _ | 0 |
| 93959 INS G | 7/692308 | 8.000a | 0.004 | 1.087 | 1.026 | 1.000 | 0 | _ | _ | _ | _ | _ | 0 |
| 63814 G=C | 7/692308 | 8.000a | 0.004 | 1.087 | 1.026 | 1.000 | 0 | _ | _ | _ | _ | _ | 0 |
| 63959 INS G | 7/692308 | 8.000a | 0.004 | 1.087 | 1.026 | 1.000 | 0 | _ | _ | _ | _ | _ | 0 |
| 64069 INS G | 7/692308 | 8.000a | 0.004 | 1.087 | 1.026 | 1.000 | 0 | _ | _ | _ | _ | _ | 0 |
| 64086 T>G | 7/692308 | 8.000a | 0.004 | 1.087 | 1.026 | 1.000 | 0 | _ | _ | _ | _ | _ | 0 |
| 63472 G>T | 7/692308 | 8.000a | 0.004 | 1.087 | 1.026 | 1.000 | 0 | _ | _ | _ | _ | _ | 0 |
| 63472 G=T | 7/692308 | 8.000a | 0.004 | 1.087 | 1.026 | 1.000 | 0 | _ | _ | _ | _ | _ | 0 |
| 63471 G=T | 7/692308 | 8.000a | 0.004 | 1.087 | 1.026 | 1.000 | 0 | _ | _ | _ | _ | _ | 0 |
| 63454 A>C | 7/692308 | 8.000a | 0.004 | 1.087 | 1.026 | 1.000 | 0 | _ | _ | _ | _ | _ | 0 |
| 64066 INS A | 7/692308 | 8.000a | 0.004 | 1.087 | 1.026 | 1.000 | 0 | _ | _ | _ | _ | _ | 0 |
| 64073 INS G | 7/692308 | 8.000a | 0.004 | 1.087 | 1.026 | 1.000 | 0 | _ | _ | _ | _ | _ | 0 |
| 63481 A=T | 7/692308 | 8.000a | 0.004 | 1.087 | 1.026 | 1.000 | 0 | _ | _ | _ | _ | _ | 0 |
| 64118 G>A | 0 | _ | _ | _ | _ | _ | 10 | 10.000a | 0.001 | 1 | 1.041 | 1 | 0 |
| 63565 C>A | 0 | _ | _ | _ | _ | _ | 10 | 10.000a | 0.001 | 1 | 1.041 | 1 | 0 |
| 63558 T=A=C | 0 | _ | _ | _ | _ | _ | 10 | 10.000a | 0.001 | 1 | 1.041 | 1 | 0 |
| 63545 G=C | 0 | _ | _ | _ | _ | _ | 10 | 10.000a | 0.001 | 1 | 1.041 | 1 | 0 |
| 63537 G>T=C | 0 | _ | _ | _ | _ | _ | 10 | 10.000a | 0.001 | 1 | 1.041 | 1 | 0 |
| 64076 INS C | 7/692308 | 8.000a | 0.004 | 1.087 | 1.026 | 1.000 | 0 | _ | _ | _ | 0 | ||
The P values were calculated from logistic regression analyses. Bold numbers mean significance association.
The Pc values were calculated using Bonferroni’s correction
OR, odds ratio; CI, confidence interval.
Several studies have shown that there is a significant relationship between serum vitamin D3 levels and the development of several diseases7,8. In addition, numerous investigations have indicated a marked association between some of the polymorphisms in VDR and CYP27B1 genes and serum levels of vitamin D31,13. Furthermore, several studies have shown that in viral diseases such as COVID-19, the viruses cause upper respiratory infection by using angiotensin inverted enzyme receptors that bind to heart and lung cells14. Vitamin D deficiency causes overexpression of angiotensin receptors on cell surfaces15. Hence, subjects with vitamin D deficiency may be more susceptible to viral invasion of the lungs. The present study is the first to assess the frequency of polymorphisms in the aforementioned genes in Iranian patients with alopecia areata living in Tehran.
Only 210 of the 800 studied blood samples showed 25(OH)D3 levels from 30–100 ng/mL, with 100 of them derived from patients living in Tehran. According to previous studies, vitamin D deficiency in the Iranian population is a serious health problem, with a prevalence of 81.3% in the urban population16. Therefore, the high prevalence of vitamin D3 insufficiency and deficiency in patients with alopecia areata reflects the pivotal role of vitamin D3 in the development of alopecia areata. The first record of the inverse correlation between vitamin D and alopecia areata severity (P < 0.001; r = −0. 409) was reported by Carman et al.17. Consistent with the previous findings, a negative association was reported between the severity of alopecia areata and vitamin D levels (P = 0.00; r = −0. 474) as assessed using the SALT score used by Unal and Gonulalan18, Bhat et al.19, and Rehman et al.20.
Given that 25(OH)D3 levels are known to be lower in the serum samples collected in winter and spring than in the samples collected in summer and fall2, samples collected between July and September were included in succeeding studies to decrease the rate of error during the investigation. Vitamin D deficiency is much more common among Iranian women than men16. Although the results have shown that women are at greater risk of developing vitamin D deficiency than men, a significant association between observed polymorphisms and gender was not found.
The BMI of all participants was between 20 and 27. Individuals outside this range were excluded. This is due to findings from previous research that have shown that people with a BMI >30 are at a higher risk of developing vitamin D deficiency than others21-24.
The CYP27B1 gene was evaluated by PCR sequencing in two regions located between 57764840 and 57764087 (exons 5–7) and between 57766295 and 57765610 (exon 2). These regions of the gene were selected because, according to previous studies, the most important polymorphisms related to vitamin D deficiency in these areas were rs118204007, rs118204009, rs118204010, rs118204011, rs28934606, rs28934607, rs4646536, rs1057520815, rs28934604, rs28934605, and rs387906260. In addition, regions 47846656–47845901 located on the VDR gene were investigated using the same method. Previous studies have shown that this area of the gene contains notable polymorphisms related to vitamin D3 deficiency, such as rs121909798, rs121909799, rs121909802, and rs1544410.
However, the study only illustrated 63 variations (42 in CYP27B1 and 21 variations in VDR) between the two genes, and risk of developing vitamin D3 insufficiency and deficiency was reported to be significantly associated with them (Rs4646536 in CYP27B1 and Rs1544410 in VDR) (P < 0.05). The CYP27B1 rs4646536 variation may play a vital role in the development of islet autoimmunity and the preclinical phase of type-1 diabetes25,26. In this investigation, the rs4646536 variation indicated a higher risk of developing vitamin D3 deficiency (odds ratio: 4, P < 0.0005). Furthermore, rs1544410 is known to be a BsmI polymorphism. Multiple studies analyzing bone mineral density (BMD) in women have reported a significant association between the A allele and increased risk of low BMD (odds ratio of A; A genotype: 2)27-29. In addition, a meta-analysis of several studies also identified a reduced risk of osteoporosis associated with the G; G genotype, with an odds ratio of 0.61, 95% CI, 0.40–0.9230. Rs1544410 is an important polymorphism identified in this study and has been shown to be most associated with risk of having vitamin D3 insufficiency (odds ratio of 9, 95% CI, 4–17, P < 0.0005) and deficiency (odds ratio of 6, 95% CI, 3–12, P < 0.0005).
In summary, our results showed a significant association between SNPs in the CYP27B1 and VDR genes and serum 25(OH)D3 levels. We also found that the Rs1544410 variation was associated with an increased risk of having vitamin D3 deficiency. Unfortunately, vitamin D3 insufficiency has dramatically increased in recent years. The pivotal role of serum 25(OH)D3 levels in health has been confirmed by several studies. Therefore, diagnosing polymorphisms of genes as biomarkers of genetic predisposition to vitamin D3 deficiency might be a potential approach. Our data confirm previous observations that A-A genotype (Rs1544410) carriers have a higher risk of having vitamin D3 deficiency than other genotypes. This information might have serious implications in choosing personalized lifestyle management strategies to prevent vitamin D insufficiency in genetically susceptible individuals. Thus, it seems that the evaluation of VDR and CYP27B1 polymorphisms could be considered a prognostic mechanism to identify subjects with high risk of developing vitamin D3 insufficiency. Future genetic studies in more extensive populations are needed to further evaluate the importance of these two genes in diseases related to vitamin D3 deficiency.
Limitations
This trial only tested participants with alopecia areata living in a specific area of Tehran. Since one of the important resources in the maintenance of vitamin D levels is sunlight, geographical regions may affect serum vitamin D levels. In addition, there was a time limitation in sample collection. Furthermore, there was a limitation in only selecting participants with specific BMIs, given that BMI could have an effect on serum vitamin D levels.
ACKNOWLEDGMENTS
This study is provided in the Research Institute of Royesh gene, Karaj, Iran. We would like to express our heartfelt thanks to Mr. Mostafa Mohaghegh, the manager of the Iranian Association of Patients with Alopecia areata for their valuable cooperation to collection of the blood sample and clinical data of patients.
CONFLICT OF INTEREST
The authors declare that there is no conflict of interest.
AUTHORS’ CONTRIBUTION
SS and MH: conceived of the presented idea and they had participated in study design and performed the statistical analysis. HS, MK, AD and MV participated in laboratory and clinical data gathering and analysis. All authors discussed the results and contributed to the final manuscript. HS, MK, AD and MV experimented. SS wrote the manuscript with support from other authors. The sample was provided by SS. All authors give final approval of the version to be submitted and any revised version, as well as all authors, participate in drafting the article or revising it critically for important intellectual content.
FUNDING
None.
ETHICS STATEMENT
This study was approved by the ethical committee of the Royesh gene, Karaj, Iran. The signed consent forms by participants in the research have been collected by the Iranian Association of Patients with Alopecia areata.
AVAILABILITY OF DATA
The datasets generated during and/or analyzed during the current study are available from the corresponding author on reasonable request.
- Barry EL, Peacock JL, Rees JR, et al. Vitamin D receptor genotype, vitamin D3 supplementation, and risk of colorectal adenomas: a randomized clinical trial. JAMA Oncol. 2017;3(5):628-635.
Crossref - Muindi JR, Adjei AA, Wu ZR, et al. Serum vitamin D metabolites in colorectal cancer patients receiving cholecalciferol supplementation: correlation with polymorphisms in the vitamin D genes. Hormones and Cancer. 2013;4(4):242-250.
Crossref - Dezhgir A, Talebzadeh T, Hosseini IA, et al. The prevalence of polymorphisms of thiopurine s-methyltransferase gene In Iranian alopecia areata patients.
- Dong LM, Ulrich CM, Hsu L, et al. Vitamin D related genes, CYP24A1 and CYP27B1, and colon cancer risk. Cancer Epidemiol Biomarkers Prevn. 2009;18(9):2540-2548.
Crossref - Khodadadi H, Khabazi A, Ghaderian SMH, et al. Molecular analysis of vitamin D receptor gene polymorphisms rs2228570 (FokI) and rs1544410 (BsmI) in patients with Behcet’s Disease. Life Science Journal. 2013;10(4):608-615.
- Miyamoto K-I, Kesterson RA, Yamamoto H, et al. Structural organization of the human vitamin D receptor chromosomal gene and its promoter. J Mol Endocrinol. 1997;11(8):1165-1179.
Crossref - Khoo A-L, Chai LY, Koenen HJ, et al. 1, 25-dihydroxyvitamin D3 modulates cytokine production induced by Candida albicans: impact of seasonal variation of immune responses. J Infect Dis. 2011;203(1):122-130.
Crossref - Adams JS, Hewison M. Unexpected actions of vitamin D: new perspectives on the regulation of innate and adaptive immunity. Nat Clin Pract Endocrinol Metab. 2008;4(2):80-90.
Crossref - McGrath JJ, Saha S, Burne TH, Eyles DW. A systematic review of the association between common single nucleotide polymorphisms and 25-hydroxyvitamin D concentrations. J Steroid Biochem Mol Biol. 2010;121(1-2):471-477.
Crossref - Bu F-X, Armas L, Lappe J, Zhou Y, Gao G, Wang H-W, et al. Comprehensive association analysis of nine candidate genes with serum 25-hydroxy vitamin D levels among healthy Caucasian subjects. Hum Genet. 2010;128(5):549-556.
Crossref - Manchanda PK, Kibler AJ, Zhang M, Ravi J, Bid HK. Vitamin D receptor as a therapeutic target for benign prostatic hyperplasia. Indian J Urol: IJU: Journal of the Urological Society of India. 2012;28(4):377-381.
Crossref - Swanson CM, Nielson CM, Shrestha S, et al. Higher 25 (OH) D2 is associated with lower 25 (OH) D3 and 1, 25 (OH)2D3. J Clin Endocrinol Metabm (JCEM). 2014;99(8):2736-2744.
Crossref - Jacobs ET, Van Pelt C, Forster RE, et al. CYP24A1 and CYP27B1 polymorphisms modulate vitamin D metabolism in colon cancer cells. Cancer Res. 2013;73(8):2563-2573.
Crossref - Menachery VD, Yount Jr BL, Debbink K, et al. A SARS-like cluster of circulating bat coronaviruses shows potential for human emergence. Nat Med. 2015;21(12):1508-1513.
Crossref - de Borst MH, Vervloet MG, ter Wee PM, Navis G. Cross talk between the renin-angiotensin-aldosterone system and vitamin D-FGF-23-klotho in chronic kidney disease. J Am Soc Nephrol. 2011;22(9):1603-1609.
Crossref - Hashemipour S, Larijani B, Adibi H, et al. Vitamin D deficiency and causative factors in the population of Tehran. BMC Public Health. 2004;4(1):38.
Crossref - Cerman AA, Solak SS, Kivanc Altunay IK. Vitamin D deficiency in alopecia areata. B J Dermatol. 2014;170(6):1299-1304.
Crossref - Unal M, Gonulalan G. Serum vitamin D level is related to disease severity in pediatric alopecia areata. Journal of Cosmetic Dermatology. 2018;17(1):101-104.
Crossref - Bhat YJ, Latif I, Malik R, et al. Vitamin D level in alopecia areata. Indian J Dermatol. 2017;62(4):407-410.
Crossref - Rehman F, Dogra N, Wani MA. Serum Vitamin D levels and Alopecia areata-A hospital based case-control study from North-India. Int J Trichology. 2019;11(2):49-57.
Crossref - Sanghera DK, Sapkota BR, Aston CE, Blackett PR. Vitamin D status, gender differences, and cardiometabolic health disparities. Ann Nutr Metab. 2017;70(2):79-87.
Crossref - Adeyemi OM, Agniel D, French AL, Tien P, Weber K, Glesby MJ, et al. Vitamin D deficiency in HIV-infected and un-infected women in the US. J Acquir Immune Defic Syndr. 2011;57(3):197-204.
Crossref - Lake JE, Adams JS. Vitamin D in HIV-infected patients. Curr HIV/AIDS Rep. 2011;8(3):133-141.
Crossref - Abd Mishani M, Hajifaraji M, Najmi M, Najmi N. The role of vitamin D in HIV disease. Iranian Journal of Nutrition Sciences & Food Technology. 2013;7(5):679-688.
- Engelman CD, Fingerlin TE, Langefeld CD, et al. Genetic and environmental determinants of 25-hydroxyvitamin D and 1, 25-dihydroxyvitamin D levels in Hispanic and African Americans. J Clin Endocrinol Metab. 2008;93(9):3381-3388.
Crossref - Frederiksen BN, Kroehl M, Fingerlin TE, et al. Association between vitamin D metabolism gene polymorphisms and risk of islet autoimmunity and progression to type 1 diabetes: the diabetes autoimmunity study in the young (DAISY). J Clin Endocrinol Metab. 2013;98(11):E1845-E1851.
Crossref - Salamone L, Ferrell R, Black D, et al. The association between vitamin D receptor gene polymorphisms and bone mineral density at the spine, hip and whole-body in premenopausal women. Osteoporosis international. 1996;6(1):63-68.
Crossref - Li Y, Xi B, Li K, Wang C. Association between vitamin D receptor gene polymorphisms and bone mineral density in Chinese women. Mol Biol Rep. 2012;39(5):5709-5717.
Crossref - Pouresmaeili F, Jamshidi J, Azargashb E, Samangouee S. Association between vitamin D receptor gene BsmI polymorphism and bone mineral density in a population of 146 Iranian women. Cell Journal (Yakhteh). 2013;15(1):75.
- Jia F, Sun R-F, Li Q-H, et al. Vitamin D receptor Bsm I polymorphism and osteoporosis risk: a meta-analysis from 26 studies. Genetic Testing and Molecular Biomarkers. 2013;17(1):30-34.
Crossref
© The Author(s) 2021. Open Access. This article is distributed under the terms of the Creative Commons Attribution 4.0 International License which permits unrestricted use, sharing, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.