ISSN: 0973-7510
E-ISSN: 2581-690X
Klebsiella pneumoniae is a common bacterial pathogen causes wide range of infections all over the world. The antimicrobial resistance of K. pneumoniae is a global concern and expresses several virulence factors contributing to the pathogenesis. The incidences of bacterial co-infection in viral pneumonia are common. Increased risk of K. pneumoniae co-infection in viral respiratory tract infection should be alerted in COVID-19 pandemic period. The study aims to detect the association between antimicrobial resistance and factors causing pathogenicity of K. pneumoniae. For the current study, 108 K. pneumoniae clinical isolates were included. Antimicrobial susceptibility test was done by Kirby-Bauer disc diffusion method according to CLSI guidelines. Virulence factors such as biofilm formation, haemagglutination, haemolysins, hypermucoviscocity, siderophore, amylase, and gelatinase production were determined by phenotypic method. In this study K. pneumoniae showed high level of antimicrobial resistance towards ampicillin (92.59%) followed by amoxicillin-clavulanic acid (67.59%) and cotrimoxazole (47,22%). An important association between biofilm formation and antimicrobial resistance was found to be statistically significant for cotrimoxazole (P-value 0.036) and amoxicillin-clavulanic acid (P-value 0.037). Other virulence factors like hypermucoviscocity, haemagglutination, amylase, and siderophore production were also showed a statistically significant relation (P-value <0.05) with antimicrobial resistance. Further molecular studies are necessary for the identification of virulence and antimicrobial resistance genes, for the effective control of drug-resistant bacteria.
Antimicrobial susceptibility, Biofilm, Klebsiella pneumoniae, Pathogenesis, Virulence factors
The burgeoning antimicrobial resistance in bacteria is a global challenge. Genus Klebsiella is one of the most important Gram-negative, non-motile, non-sporing, short stout bacilli, normally present as normal flora of the intestine of humans and animals.1 Klebsiella pneumoniae is an opportunistic pathogen causing various diseases; however, urinary tract and respiratory tract infections are repeatedly reported. It also caused intra-abdominal infection, meningitis, wound infection, pyogenic liver abscess, and septicemia.2 Rare cases of mycotic aneurysm, acute suppurative bacterial dacryoadenitis and community-acquired Klebsiella pneumoniae meningitis are reported in recent years.3-5 Many reports have been published regarding the co-infection of K. pneumoniae in association with COVID-19 cases.6-8
Six multidrug-resistant bacterial pathogens are coming under the acronym ESKAPE such as Enterococcus faecium, Staphylococcus aureus, Klebsiella pneumoniae, Acinetobacter baumanni, Pseudomonas aeruginosa and Enterobacter species, they are escaping from the bactericidal activity of antimicrobial agents and mainly responsible for hospital acquired infection. K. pneumoniae as ESKAPE pathogens are highly resistant to antimicrobial agents and a principal source of hospital acquired infection in developed and low-income countries.9,10 The pathogenesis of K. pneumoniae is because of the production of different virulence factors such as capsular polysaccharide, siderophore production, biofilm formation, hypermucoviscosity, haemagglutination, haemolysis and production of extracellular enzymes.11 These factors aid in the establishment of Klebsiella infection and also act as a barrier to antimicrobial agents. Among the virulence factors, the capsular polysaccharide is the most important; it protects the microbes by inhibiting phagocytosis.12 Due to the production of capsular polysaccharide, the colonies of K. pneumoniae becomes mucoid.
Due to the increased antimicrobial resistance and multidrug resistance, the genus is showing high morbidity and mortality. Presently, K. pneumoniae shows increased resistance to broad-spectrum antibiotics, including beta-lactam, aminoglycosides and fluoroquinolones.13 Antimicrobial resistance is mainly because of the production of enzyme, virulence factors, efflux pumps and mutation.14 The enzyme Extended-spectrum beta-lactamases (ESBLs) are present in various Gram-negative bacteria worldwide, of which the most common bacterial isolate is K. pneumoniae. ESBLs producing bacteria are resistant to penicillins, first-, second-and third-generation cephalosporins and monobactams (aztreonam), but not to cephamycin or carbapenems.15 Thus, carbapenems are found to be the most reliable drug for the treatment of ESBL-producers; however, there have been a lot of research reports on increasing resistance to carbapenem antibiotics worldwide.16 The resistance to the carbapenem is due to the acquisition of resistant gene and the presence of some virulent factors.17
As per recent researches worldwide, in majority of viral pneumonia infections, bacterial co-infections are found to be in significant proportion. K. pneumoniae is a major bacteria acting as a co-pathogen in viral respiratory tract infection, it should be alerted in this COVID-19 pandemic. Due to its pathogenic and antimicrobial resistance, K. pneumoniae is a well-established pathogen in health care system. Therefore, this study aims to detect the virulence factors and antimicrobial susceptibility of K. pneumoniae. We also focused on the association between the virulence factors and antimicrobial resistance.
The study was done at Karuna Medical College, Palakkad during 2017 January to 2018 March, to assess the prevalence of antimicrobial resistance among K. pneumoniae and its virulence factor determination. A total of 650 samples were received to microbiology laboratory, in this 556 (85.54%) samples were showed growth. Of this One hundred and eight strains (n=108) of K. pneumoniae, were identified based on the cultural and biochemical profile described by Bergey’s Manual for systematic Bacteriology.18
Antimicrobial resistance testing
The antimicrobial susceptibility test for all the isolates were performed by Kirby-Bauer disc diffusion test with the following commercially available discs (Himedia Laboratories Pvt, Ltd., Mumbai, India) such as ampicillin (10µg), ampicillin-sulbactam (10 µg /10 µg), gentamicin (10 µg), cotrimoxazole (25 µg), levofloxacin (5µg), cefoxitin (30 µg), cefotaxime (30 µg), ceftriaxone (30 µg), ceftazidime (30 µg), amoxyclav (10 µg) and amikacin (30 µg). The results were interpreted as per CLSI guidelines.19
Detection of ESBLs production
Phenotypic screening test for the detection of ESBL production by K. pneumoniae was tested with third-generation cephalosporin such as ceftriaxone (30 µg), cefotaxime (30 µg), and ceftazidime (30 µg). All the isolate indicated resistant or intermediate sensitivity to any of these three discs, were suspected as ESBLs producers and is phenotypically confirmed by a combined disc diffusion test. The isolate was inoculated on to Muller Hinton Agar (Himedia Laboratories Pvt, Ltd., Mumbai, India) and susceptibility tested with ceftazidime or cefotaxime and with or without clavulanic acid. The plates were incubated at 37°C overnight. The ESBL production was confirmed phenotypically by the difference of zone diameter more than 5mm between the discs such as cefotaxime or ceftazidime and their respective clavulanic acid disc combination.20
Virulence factors determination
Biofilm formation
Quantitative estimation of biofilm formation of the isolate was performed according to the protocol by O” Toole and Kolter (1998) and Dusane et al.21,22 The optical density was measured by 570nm Enzyme linked immunosorbent assay reader and graded the intensity of the biofilm as strong (>0.240), moderate (0.120-0.240) and weak (<0.120).
Hypermucoviscosity
K. pneumoniae hypermucoviscosity was phenotypically determined by a string test. The colonies were stretched by a wire loop, and the formation of a viscous string at a length of ≥5mm indicates hypermucoviscosity positive.
Haemagglutination
Bacterial fimbriae were detected by the clumping of erythrocyte narrated by Vagarali et al.23 Human O+ve blood was used for the test. The 3% of RBC solution is prepared in fresh saline and one drop of this suspension was added to one drop of bacterial suspension and rotates for 5 minutes at room temperature.
Haemolysin production
The isolates were streaked on the blood agar. Haemolysins are three types; Alpha, Beta and gamma.24 Alpha haemolysis is partial lysis of RBC, shown as greenish discoloration. Beta haemolysis, complete lysis of red blood cells, is clearance around the colony. Gamma haemolysis doesn’t exhibit haemolysis. It was detected after 24 hours of incubation at 37°C.25
Siderophore production
This was carried out by chromazerol sulphate agar disc diffusion assay.26 Positive results show blue to orange colour change due to the chelation of iron by siderophore.
Gelatinase production
Gelatinase production was detected by using a Nutrient gelatin agar medium. Isolates were inoculated on the test tube contain medium by a straight wire and incubate at 37°C for 48hrs. After incubation, keep the tubes at 4°C overnight. The Liquefaction of gelatin shows a positive result.27
Amylase production
Nutrient gar with 1% starch was used to detect amylase production. After 48hrs of incubation, plates flooded with an Iodine solution. The zone of clearance indicates a positive reaction.28
Statistical analysis
All the data was analyzed statistically by using SPSS version 21(IBM Corporation/Armonk, New York/USA). The association between drug resistance and the factors related to pathogenicity of K. pneumoniae isolates and ESBLs producers and virulence determinants were evaluated by the chi-square test. ‘P’ value less than or equal to 0.05 was considered as statistically significant.
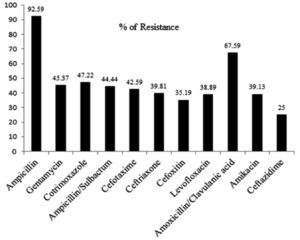
For the current study, K. pneumoniae isolates (n= 108) were collected from various clinical samples such as urine (41), sputum (39), pus (23), blood, secretion tip, anal fistula, wound swab and bronchial wash (1 each) (Table 1). Among this, more than 90% of isolates were resistant to ampicillin (92.59%; 100/108) followed by, amoxicillin-clavulanic acid (67.59%; 73/108) and Co-trimoxazole (47.22 %; 51/108) (Fig. 1). Out of total 108 isolates 30.55% of K. pneumoniae isolates were resistant to 3-4 functional group of antimicrobial agents, which is considered as MDR (multidrug resistance) (Table 2). The present study, ESBLs production was found in 49 isolates (45.37%, 49/108) and non ESBLs producers 59 (54.63%) were detected by combined disc diffusion test (Table 3).
Table (1):
Distribution of Klebsiella pneumoniae isolates in different samples.
No. |
Particulars |
No. of isolates |
|---|---|---|
1 |
Urine |
41 (37.96%) |
2 |
Sputum |
39 (36.11%) |
3 |
Pus |
23 (21.3%) |
4 |
Blood |
1 (0.93%) |
5 |
Secretion tip |
1 (0.93%) |
6 |
Anal fistula |
1 (0.93%) |
7 |
Wound swab |
1 (0.93%) |
8 |
Bronchial wash |
1 (0.93%) |
Table (2):
Incidence of MDR isolates in different sample.
No |
Sample |
No: (%) of MDR isolates |
|---|---|---|
1 |
Urine |
19/41 (46.34) |
2 |
Sputum |
6/39 (15.38) |
3 |
Pus |
5/23 (21.74) |
4 |
Blood |
0 |
5 |
Secretion tip |
1 (100) |
6 |
Anal fistula |
0 |
7 |
Wound swab |
1(100) |
8 |
Bronchial wash |
1(100) |
Table (3):
Distribution of ESBL producing K. pneumoniae in different samples.
No. |
Sample |
No. of ESBL producers (N=49; 45.37%) |
No. of Non ESBL producers (N=59; 54.63%) |
|---|---|---|---|
1 |
Urine |
26 (53.06%) |
15 (25.42%) |
2 |
Sputum |
11 (22.45%) |
28 (47.46%) |
3 |
Pus |
8 (16.33%) |
15 (25.42%) |
4 |
Blood |
0 |
01 (1.70) |
5 |
Secretion tip |
1 (2.04%) |
0 |
6 |
Anal fistula |
1 (2.04%) |
0 |
7 |
Wound Swab |
1 (2.04%) |
0 |
8 |
Bronchial wash |
1 (2.04%) |
0 |
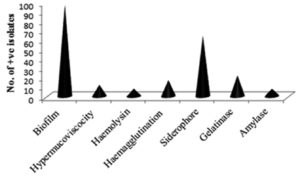
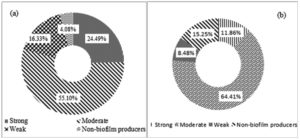
The biofilm and siderophore production was found to be the most common virulence factors among the isolates, but the incidence of Haemolysin and Amylase production was not often (Fig. 2). In the present study most of the K. pneumoniae isolates were biofilm producers. The majority of isolates produced a moderate intensity of biofilm in both ESBL producing and non-ESBLs producing K. pneumoniae and different categories of biofilm production are present in (Fig. 3). The current study reveals prevalence of haemagglutination (P-value 0.033) and siderophore production (P-value 0.001) was higher among ESBL producing K. pneumoniae (Table 4).
Table (4):
Association of ESBL producing K. pneumoniae with respect to virulence factors.
| Virulence factors | ||||||
|---|---|---|---|---|---|---|
| Hypermucoviscocity | Haemolysin | Haemagglutination | Amylase | Gelatinase | Siderophore | |
| Positive | 4.1 | 6.1 | 24.5 | 12.2 | 16.3 | 77.6 |
| Negative | 95.9 | 93.9 | 75.5 | 87.8 | 83.7 | 22.4 |
| P-value | 0.061 | 0.726 | 0.033 | 0.137 | 0.472 | 0.001 |
A significant association was noticed between antimicrobial resistance and some virulence factors. Table 5 shows the prevalence of virulence factors and antimicrobial susceptibility of K. pneumoniae. The siderophore production exhibited a statistically significant association in the antimicrobial resistance with all antimicrobial agents used in the current study (P-value <0.05). Moreover, significant associations were observed between biofilm formation and cotrimoxazole (P-value 0.036) and amoxicillin-clavulanic acid resistance (P-value 0.037); hypermucoviscosity with gentamicin (P-value 0.034) and ceftriaxone (P-value 0.026) and cefotaxime (P-value 0.044); haemagglutination (P-value 0.045) and amylase production (P-value 0.044) with cefotaxime.
Table (5):
Association between antibiotic resistance and virulence factors of K. pneumoniae.
| Virulence markers | Antibiotics | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| AMP | GEN | COT | A/S | CTX | CTR | LE | AMC | ||
| Biofilm formation | Strong | 19 | 18.4 | 23.5 | 18.8 | 26.1 | 27.9 | 26.2 | 19.2 |
| Moderate | 57 | 67.3 | 54.9 | 62.5 | 54.3 | 53.5 | 59.5 | 54.8 | |
| Weak | 13 | 6.1 | 17.6 | 14.6 | 15.2 | 14 | 11.9 | 17.8 | |
| Negative | 11 | 8.2 | 3.9 | 4.2 | 4.3 | 4.7 | 2.4 | 8.2 | |
| P-value | 0.126 | 0.309 | 0.036 | 0.298 | 0.069 | 0.065 | 0.072 | 0.037 | |
| Hypermucoviscosity | Positive | 11 | 4.1 | 9.8 | 6.2 | 4.3 | 2.3 | 7.1 | 9.6 |
| Negative | 89 | 95.9 | 90.2 | 93.8 | 95.7 | 97.7 | 92.9 | 90.4 | |
| P-value | 0.897 | 0.03 | 0.683 | 0.15 | 0.044 | 0.026 | 0.295 | 0.467 | |
| Haemolysin | Positive | 7 | 4.1 | 5.9 | 8.3 | 6.5 | 9.3 | 7.1 | 8.2 |
| Negative | 93 | 95.9 | 94.1 | 91.7 | 93.5 | 90.7 | 92.9 | 91.8 | |
| P-value | 0.568 | 0.229 | 0.567 | 0.74 | 0.762 | 0.541 | 0.933 | 0.642 | |
| Haemagglutination | Positive | 17 | 14.3 | 15.7 | 20.8 | 23.9 | 20.9 | 16.7 | 19.2 |
| Negative | 83 | 85.7 | 84.3 | 79.2 | 76.1 | 79.1 | 83.3 | 80.8 | |
| P-value | 0.204 | 0.705 | 0.988 | 0.194 | 0.045 | 0.228 | 0.833 | 0.157 | |
| Amylase | Positive | 6 | 8.2 | 11.8 | 10.4 | 13 | 9.3 | 11.9 | 9.6 |
| Negative | 94 | 91.8 | 88.2 | 89.6 | 87 | 90.7 | 88.1 | 90.4 | |
| P-value | 0.48 | 0.785 | 0.102 | 0.285 | 0.044 | 0.541 | 0.155 | 0.211 | |
| Gelatinase | Positive | 19 | 18.4 | 19.6 | 18.8 | 19.6 | 18.6 | 23.8 | 19.2 |
| Negative | 81 | 81.6 | 80.4 | 81.3 | 80.4 | 81.4 | 76.2 | 80.8 | |
| P-value | 0.211 | 0.638 | 0.852 | 0.708 | 0.858 | 0.711 | 0.479 | 0.657 | |
| Siderophore | Positive | 60 | 77.6 | 70.6 | 75 | 80.4 | 74.4 | 83.3 | 69.9 |
| Negative | 40 | 22.4 | 29.4 | 25 | 19.6 | 25.6 | 16.7 | 30.1 | |
| P-value | 0** | 0** | 0.023 | 0.003 | 0** | 0.009 | 0** | 0.001** | |
P-Value * -Significant ** highly significant
AMP- Ampicillin, GEN- Gentamicin, COT- Cotrimoxazole, A/S- Ampicillin Sulbactum, CTX- Cefotaxime, CTR- Ceftriazone, LE- Levofloxacine, AMC- Amoxicillin clavulanic acid
Klebsiella pneumoniae is one of the frequently isolated, antimicrobial resistant organisms from different clinical samples. It is also associated with hospital acquired infection and a co-pathogen in viral diseases. A study from China reported 91.9% 0f COVID-19 patient had bacterial co-infection, where it was predominant with Streptococcus pneumoniae followed by K. pneumoniae.29 In the present study, the predominant isolation of K. pneumoniae was found from urine samples followed by sputum and pus, which corroborates the observation of Phamba and Domanic.30
Antimicrobial resistance is a severe threat to the human for the successful treatment of health care infection. The current study revealed that 92.59 % (100/108), 67.59% (73/100) and 47.22% (51/108) of K. pneumoniae were resistance to ampicillin, amoxicillin-clavulanic acid and co-trimoxazole respectively. The prolonged usage of antibiotics like ampicillin and cotrimoxazole, the bacteria developed resistance against them.31 As study done by Ahanjan et al.32 reported that K. pneumoniae showed 100% resistance against cefotaxime and 47% to gentamycin. In our study, the resistance against gentamycin was 45.37% (49/108), but the cefotaxime resistance (42.59%; 46/108) was less than the mentioned study. In this survey 73/108 (67%) isolates of K. pneumoniae were resistant to amoxicillin-clavulanic acid. This report supported by Derakhshan et al.33 that found 121/200 (60.5%) of resistance against amoxicillin-clavulanic acid. Meanwhile, another study revealed that 36.69% of resistance, which is lower than our result.34
In different studies reported, the incidence of ESBL producers in India ranging from 60%-80 %.35 The incidence of ESBL producers in the present study was 45.37%, which supports the study of Wettal et al.36 Nevertheless, Thenkhiwale et al.,37 Mumtaz et al.38 and Chander and Shestha39 had reported a much lower rate of ESBL Klebsiella spp. The increased occurrence of multi drug resistant bacteria, influenced by various risk factors such as prolonged hospitalization, indiscriminate use of antibiotics and self-medication. In the present study, the rate of isolation of ESBL producers were more in urine 53.06% (26/49) followed by sputum 22.45% (11/49) and pus 16.33% (8/49). The similar results were reported by Akila et al.,40 which is dependent with the sample size.
Virulence factors are contributing to the pathogenesis and antimicrobial resistance of the bacteria. The high density of bacterial population present in biofilm contributes to antimicrobial resistance. A significant association between biofilm formation and co-trimoxazole (P-value 0.036) and amoxicillin-clavulanic (P-value 0.037) was revealed in this study. Siderophore iron uptake is one of the major virulence determinants. In the current study, siderophore production showed a significant association with antimicrobial resistance. Karam et al.41 reported a significant correlation between the presence of iron acquisition gene chuA and resistance of antibiotics such as co-trimoxazole and ceftazidime. Pramodhini et al.42 reported that other bacteria showed a significant association between certain virulence factors and antimicrobial resistance. However, they suggested that all virulence factors are may not be significantly associated with antimicrobial resistance.
The bacterial virulence and antimicrobial resistance are not independent factors, the association between these factors may play a crucial role in pathogenesis of invasive infections. The present study indicated certain virulence factors of K. pneumoniae are associated with antimicrobial resistance. The antimicrobial resistant K. pneumoniae possess virulence characteristics, which lead to complications and treatment failure. Therefore the strict monitoring of virulence factors and antimicrobial resistance can be recommended in clinical laboratories with an emphasis on surveillance and infection control activities. Furthermore, molecular studies are required for the good understanding of the genetic relation between virulence factors and drug resistance of K. pneumoniae.
ACKNOWLEDGMENTS
The authors are thankful to the Principal, Karuna Medical College, Palakkad and Head, Department of Microbiology, Karuna Medical College, Kerala, India for all the facilities provided.
CONFLICT OF INTEREST
The authors declare that there is no conflict of interest
AUTHORS’ CONTRIBUTION
All authors listed in this study have made a substantial direct and intellectual contribution to the work and approved it for publication.
FUNDING
None.
ETHICS STATEMENT
Not applicable.
AVAILABILITY OF DATA
All datasets generated during the study are included in the manuscript
- Podschun R, Ullmann U. Klebsiella spp. as nosocomial Pathogens: Epidemiology, Taxonomy, Typing Methods, and pathogenicity factors. Clin Microbiol Rev. 1998;11(4):589-603.
Crossref - Lin YT, Wang FD, Chan YJ, Fu YC, Fung CP. Clinical and microbiological characteristics of tigecycline non – susceptible Klebsiella pneumoniae bacteremia in Taiwan. BMC Infect Dis. 2014;14:1.
Crossref - Chao CM, Lee KK, Wang CS, Chen PJ, Yeh TC. Case reports a fatal case of Klebsiella pneumoniae Mycotic Aneurism. Case Rep Emerg Med. 2011;2011:498545.
Crossref - Chandravanshi SL, Mishra V. Acute suppurative bacterial dacryoadenitis: a case report. Eur J Opthalmol. 2014;24(5):790-792.
Crossref - Lee B, Yeroushalmi K, Me HM, et al. community acquired Klebsiella pneumoniae meningitis: a case report. Germ. 2018;8(2):92.
Crossref - Chen WC, Lai YC, Lin CH, et al. First COVID-19 mortality case in Taiwan with bacterial co-infection by national surveillance of critically ill patients with influenza-negative pneumonia. J Microbiol Immunol Infect. 2020;53(4):652-656.
Crossref - Arcari G, Raponi G, Sacco F, et al. Klebsiella pneumoniae infection in COVID-19 patients: a 2-month retrospective analysis in an Italian hospital. Int J Antimicrob Agents. 2021;57(1):106245.
Crossref - Dabrowska WM, Lange S, Zorena K, Dabroski S, Ozaga D, Tomaszek L. Carbapenem-resistant Klebsiella pneumoniae infection in ICU COVID-19 patients-A scoping review. J Clin Med. 2021;10(10):2067.
Crossref - Rice LB. Federal funding for the study of antimicrobial resistance in nosocomial pathogens: no ESKAPE. J Infect Dis. 2008;197(8):1079-1081.
Crossref - Navidinia M. The clinical importance of emerging ESKAPE pathogens in nosocomial infections. Archives of Advances in Biosciences. 2016;7(3):43-57.
Crossref - Fertas- Aissani R, Messai Y, Alouache S, Bakour R. Virulence profile and antibiotic susceptibility pattern of Klebsiella pneumoniae strains isolated from different clinical specimens. Pathol Biol. 2013;61(5):209-216.
Crossref - Sahly H, Podschun R, Oelschlaeger TA, et al. Capsule impedes adhesion to and invasion of epithelial cells by Klebsiella pneumoniae. Infect Immuni. 2000;68(12):6744-6749.
Crossref - Dsouza R, Pinto NA, Hwang I, et al. Panel strain of Klebsiella Pneumoniae for beta- lactam antibiotic evaluation: their phenotypic and genotypic characterization. Peer J. 2017;5:e2896.
Crossref - Papp-Wallace KM, Endimiani A, Taracila MA, Bonomo RA. Carbapenems: past, present, and future. Antimicrob Agents Chemother. 2011;55(11):4943-4960.
Crossref - Silago V, Kovacs D, Samson H, et al. Existence of Multiple ESBL Genes among phenotypically confirmed ESBL producing Klebsiella pneumoniae and Escherichia coli concurrently isolated from clinica, l colonization and contamination samples neonatal units at Bugando medical centre Mwanza, Tanzania. Antibiotics. 2021;10(5):476.
Crossref - World Health Organization. Antimicrobial resistance: Global report on surveillance 2014.
- Derakhshan S, Peerayeh SN, Bakhshi B. Association between presence of virulence genes and antibiotic resistance in clinical Klebsiella pneumoniae isolates. Lab Med. 2016;47(4):306-311.
Crossref - Garrity GM, Boone DR, Castenholz RW. Bergey’s Manual for Systematic Bacteriology, 2nd ed. Springer – verlag, New York; 2001.
- Wayne PA. CSLI. Performance standards for antimicrobial susceptibility testing twentieth informational supplement. Clinical and Laboratory Standard Institute, 2010;M100-S20:40-51 and 315.
- Wayne PA. CSLI. Performance standards for antimicrobial susceptibility testing twenty-fourth international supplements. Clinical and Laboratory Standard Institute. 2014; M100-S24.
- O’Toole GA, Kolter R. Initiation of biofilm formation in Pseudomonas fluorescens WCS365 proceeds via multiple, convergent signaling pathways: a genetic analysis. Mol Microbiol. 1998;28(3):449-461.
Crossref - Dusane DH, Rajput JK, Kumar AR. Disruption of fungal and bacterial biofilms by lauroyl glucose. Lett Appl Microbiol. 2008;47(5):374-379.
Crossref - Vagarali MA, Karadesai SG, Patil CS, Metgud SC, Mutnal MG. Haemagglutination and siderophore production as the urovirulence markers of uropathogenic Escherichia coli. Indian J Med Microbiol. 2008;26(1):68-71.
Crossref - Payment P, Coffin E, Paquette G. Blood agar to detect virulence factor in tap water heterotrophic bacteria. Appl Environ Microbiol. 1994;60(4):1179-1183.
Crossref - Gharrah MM, El-Mahdy AM, Barwa RF. Association between virulence factors in tap Extended Spectrum Beta-Lactamase producing Klebsiella pneumoniae compared to non producing isolates. Interdiscip Perspect Infect Dis. 2017;2018:1023076.
Crossref - Schwyn B, Neilands JB. Universal chemical assay for the detection and determination of siderophores. Anal Biochem. 1987;160(1):47-56.
Crossref - Mc Faddin JF. Biochemical tests for identification of medical bacteria. 2nd ed. Williams and Wilkins, Baltimore, 1980.
- Bergey DH, Holt JG. Bergey’s Manual of determinative bacteriology. 9th ed. Williams and Wilkins, Baltimore, Maryland, USA, 1994:12204.
- Zhu X, Ge Y, Wu T, et al. Co-infection with respiratory pathogen among COVID-19 cases. Virus Res. 2020;285:198005.
Crossref - Phamba SG, Dominic SRM. Prevelence of Extened spectrum beta-lactamase (ESBL) producing Klebsiella species and Escherichia coli among clinical isolates in a tertiary care hospital. Journal of International Medicine and Dentistry. 2017;4(1):6-12.
Crossref - Tabasi M, Karam MRA, Habibi M, Yekaninejad MS, Bouzari S. Phenotypic assay to determine virulence factors of uropathogenic Escherichia coli (UPEC) isolates and their correlation with antibiotic resistance pattern. Osong Public Health Res Perspect. 2015;6(4):261-268.
Crossref - Ahanjan M, Naderi F, Solimanni A. prevalence of beta-lactamases genes and antibiotic resistance pattern of Klebsiella pneumoniae isolated from teaching hospitals, sari, Iran. J Mazandaran Univ Med Sci. 2017;27(149):79-87.
- Derakhshan S, Peerayeh SN, Bakhshi B. association between prevalence of virulence genes and antibiotic resistance in clinical Klebsiella pneumoniae isolates. Science. Laboratory Medicine. 2016;47(4):306-311.
Crossref - Nirwati H, Sinanjung K, Fahrunissa F, et al. Biofilm formation and antibiotic resistance of Klebsiella pneumoniae isolated from clinical samples in tertiary care hospital, Klaten, Indonesia. BMC proceedings. 2019;13(11):20.
Crossref - Kaur J, Mahajan G, Chand K, Sheevani, Chopra S. Enhancing phenotypic detection of ESBL in AmpC producers by using Cefepime and Tazobactam. J Clin Diagn Res. 2016;10(1):DC05-DC08.
Crossref - Wattal C, Goel N, Oberoi JK, Raveendran R, Data S, Prasad KJ. Surveillance of multidrug resistant organism in tertiary care hospital in Delhi, India. J Assoc Physicians India. 2010;58:32-36.
- Tankhiwale SS, Jalgaonkar SV, Ahamad S, Hassani U. Evaluation of Extended Spectrum Beta Lactamase in urinary isolates. Indian J Med Res. 2004;120(6):553-556.
- Mumtaz S, Ahmed J, Ali L, Hamid H. Prevalence of extended spectrum beta- lactamases(ESBL) in clinical isolates from a teaching hospitalin Peshawar, Pakistan. Afr J Microbiol Res. 2011;5(19):2880-2884.
Crossref - Chander A, Shrestha CD. Prevalence of extended spectrum beta lactamase producing Escherichia coli and Klebsiella pneumoniae urinary isolates in tertiary care hospital in Kathmandu, Nepal. BMC Res Notes. 2013;6:487.
Crossref - Akila K, Nithyalakshmi J, Mohanakrishnan K, Sumathi G. Prevalence of ESBL producing Klebsiella species and their in- vitro antimicrobial susceptibility pattern in a tertiary care hospital. IOSR Journal of Dental and Medical Sciences. 2016;15(11):5-10.
- Asadi Karam MR, Habibi M, Bauzari S. Relationship between virulence factor and antimicrobial resistance among Escherichia coli isolated from urinary tract infections and commensal isolates in Tehran, Iran. O Song Public Health Research Perspectives. 2018;9(5):217-224.
Crossref - Pramodhini S, Umadevi S, Seetha KS. Detection of virulence determinants and its association with drug resistance in clinical isolates of Pseudomonas aeruginosa. Int J Res Med Sci. 2016;4(9):3917-3923.
Crossref
© The Author(s) 2022. Open Access. This article is distributed under the terms of the Creative Commons Attribution 4.0 International License which permits unrestricted use, sharing, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.