ISSN: 0973-7510
E-ISSN: 2581-690X
Rickettsia parkeri and Rickettsia amblyommatis are spotted fever group Rickettsia (SFGR) associated with Amblyomma ticks. R. parkeri is a recognized human pathogen that causes an eschar-associated febrile illness, while R. amblyommatis has not been confirmed as a causative agent of human disease. We hypothesized that the rate of replication is one of the factors contributing to rickettsial pathogenicity. In this study, growth and infectivity of R. parkeri and R. amblyommatis in mammalian (Vero E6) and tick-derived (ISE6) cell lines were assessed and compared over a 96-hour time course of infection using quantitative real-time polymerase chain reaction and microscopy. The pathogenic R. parkeri displayed a significantly higher level of infection in both Vero E6 and ISE6 cells than R. amblyommatis at 72 hours post-inoculation (hpi). Distinct growth profiles between rickettsial species with known and uncertain pathogenicity were identified. R. parkeri burdens were significantly greater than those of R. amblyommatis from 24 to 96 hpi. The relative fold changes of load were significantly higher in the pathogenic agent than in R. amblyommatis from 48 hpi onward and reached the maximum fold increase of ~2002- and ~296-fold in Vero E6 cells and ~1363- and ~161-fold in ISE6 cells, respectively, at 96 hpi. The results from the present study demonstrate that growth rate is associated with the pathogenicity of rickettsiae. Understanding SFGR growth characteristics in mammalian and tick cells will provide insight into rickettsial biology and pathogenesis.
Rickettsia parkeri, Rickettsia amblyommatis, Amblyomma spp., growth rate, qPCR
Rickettsia species are Gram-negative alpha Proteobacteria. Several arthropods, especially ticks and fleas, are important vectors of these obligate intracellular bacteria, as they can be maintained in arthropod populations via vertical transmission. Among the diverse genotypes of identified rickettsial species, many of them have been recognized to be agents with variability in pathogenicity to humans.1,2 Nevertheless, several species of Rickettsia with uncertain pathogenicity were recently identified and reported.2
Rickettsia parkeri and Rickettsia amblyommatis are among several species of Rickettsia associated with Amblyomma ticks in the United States.3,4 R. parkeri was first isolated in 1937 from the Gulf Coast tick, Amblyomma maculatum, and was later identified as a spotted fever group Rickettsia (SFGR).5 R. parkeri was reported in 2004 as a causative agent of spotted fever disease in a human.3 It caused an eschar-associated rash illness, however, with milder symptoms than that of the bioagent that causes Rocky Mountain spotted fever (RMSF), Rickettsia rickettsii.3,6-8 Since then, additional cases of R. parkeri rickettsiosis have been reported in many southeastern and southwestern regions of the United States.9-11 In addition to the primary vector, A. maculatum, R. parkeri has also been found in different species of ticks, including Amblyomma americanum,12-14 Amblyomma triste,15 and Ixodes scapularis.14,16 R. amblyommatis, formerly named Rickettsia amblyommii and ‘Candidatus Rickettsia amblyommii’, has been detected in A. americanum throughout the United States.17,18 This rickettsial species has been proposed as a possible cause of rickettsiosis in some patients who were initially diagnosed as having RMSF.19,20 Drexler et al.21 have also suggested that much of the seroprevalence of human SFG rickettsioses with the very low case fatality rate in the United States is likely caused by infections with mildly pathogenic or non-pathogenic rickettsial species, including R. amblyommatis. This agent is also detected in other species of ticks, comprising A. maculatum,4,14 Amblyomma cajennense complex,22 Amblyomma nr maculatum,22 and I. scapularis.14,23 The relatively high prevalence of R. amblyommatis detected in A. americanum24,25 and its existence in other tick species have been suggested as a potential risk of rickettsial infection to humans; nonetheless, no confirmed cases of human infection by R. amblyommatis have been reported to date.
Little is known about the biology and pathogenic determinants of R. parkeri and R. amblyommatis species. In vitro systems are advantageous to investigate the differences between pathogenic and non-pathogenic rickettsiae.26,27 Among available cell lines, mammalian (Vero E6) and I. scapularis-derived (ISE6) cells are commonly used for rickettsial isolation and cultivation.28,29 According to their frequent use in rickettsial research and the detection of R. parkeri and R. amblyommatis in I. scapularis, Vero E6 and ISE6 cells were thus utilized in the present study as the model cells to study growth characteristics of human pathogen R. parkeri (strain Portsmouth) and R. amblyommatis (strain Darkwater), a species with uncertain pathogenicity. The growth and infectivity of the two rickettsiae were compared using quantitative real-time polymerase chain reaction (qPCR) and microscopy.
Cell culture and rickettsiae
African green monkey kidney (Vero E6) cells were maintained in Dulbecco’s modified Eagle’s medium (Invitrogen, Carlsbad, CA) supplemented with 5% heat-inactivated fetal bovine serum (Hyclone, Logan, UT) in a humidified, 5% CO2 incubator at 34°C. ISE6 cells were maintained in L15B medium (Gibco-BRL, Carlsbad, CA) contained 10% heat-inactivated fetal bovine serum (HyClone) and 10% tryptose phosphate broth (Sigma, St. Louis, MO) at pH 6.8 in a humidified, 5% CO2 cell culture incubator at 34°C. R. parkeri (strain Portsmouth) originally isolated from a skin biopsy specimen3 and R. amblyommatis (strain Darkwater) isolated from a female A. americanum tick collected in Wakulla County, Florida in 2006 (CD Paddock, unpublished data) were used throughout this study.
Propagation and semi-purification of Rickettsia
Each rickettsial species (passage 4) were propagated using the confluent Vero E6 cells for 4 days. Heavily infected cells, as determined by Diff-Quik (Dade Behring, Deerfield, IL) staining according to the manufacturer’s protocol, were washed once with culture medium. Cells were scraped from the culture flask, collected by centrifugation at 500 × g for 8 min at 4°C, and resuspended in culture medium. Semi-purification of rickettsiae was performed as previously described30 with a slight modification. Briefly, cells were mechanically lysed by forcing 5 mL of cell suspension 4-5 times through a 27-Gauge needle attached to a 10-mL syringe. The remaining intact host cells and cell debris were removed by centrifugation at 500 × g for 10 min at 4°C followed by filtration of the obtained supernatant through a 2-µm syringe filter (Whatman, Florham Park, NJ).31 Rickettsiae in the filtrate were then collected by centrifugation at 9,500 × g for 10 min at 4°C. The pellets were washed 3 times and resuspended in culture medium. Partially purified rickettsiae were assessed the viability and counted using LIVE/DEAD BacLight bacterial viability kit (Molecular Probes, Carlsbad, CA) as previously described.32
Rickettsial infection and replication in cell culture
To compare growth and infectivity of R. parkeri and R. amblyommatis, Vero E6 and ISE6 cells (105 cells per well) were seeded separately, in triplicate, into 48-well cell culture plates (NUNC, Rochester, NY) containing 500 µL of appropriate culture medium. Replicate of eukaryotic cell cultures was also prepared by seeding cells in 48-well plates containing sterile glass coverslips for assessment of the number of Rickettsia-infected cells. After incubating the plates in a humidified 5% CO2 incubator at 34°C for 2 days, each well was gently washed twice with serum-free medium and culture cells were subsequently inoculated with either partially purified R. parkeri or R. amblyommatis with an estimated multiplicity of infection of 10 in a total volume of 300 µL. Culture plates were briefly spun at 500 × g for 1 min to assist the attachment of microorganism to the host cells. After 1 hour of incubation in a humidified 5% CO2 incubator at 34°C, culture medium was removed, and cells were gently washed 3 times with 300 µL of serum-free culture medium. The appropriate culture medium of 500 µL was then added to each well and culture plates were further incubated in a humidified 5% CO2 incubator at 34°C. At 1, 6, 12, 24, 48, 72, and 96 hours post-inoculation (hpi), cells were detached by pipetting and collected by centrifugation at 13,000 × g for 15 min at 4°C. Individual wells were observed with an inverted light microscope to ensure all cells were removed. Cell pellets were stored at -80°C until used for genomic DNA (gDNA) extraction. To estimate the percentage of Rickettsia-infected cells, coverslip cultures of eukaryotic cells at each hpi were stained with Diff-Quik staining and examined with a light microscope using the oil immersion lens. A hundred intact cells of Vero E6 and ISE6 cells were examined and only cells containing rickettsiae were considered to be infected cells. Three independent experiments for growth and infectivity assessments were performed.
DNA extraction and qPCR quantitation of rickettsial load
The gDNA was extracted from the cell pellets using the DNeasy tissue kit (Qiagen, Germantown, MD) according to the manufacturer’s recommendation and stored at -20°C until used. The total number of Rickettsia in each sample was quantified by qPCR using the primer pair RaRp17.181F and RaRp17.289R that is specific to the highly conserved 17-kDa antigen gene among the species of the genus Rickettsia.33 For relative fold change analysis of rickettsial load, the qPCR for host housekeeping genes that are frequently used as reference genes (beta-actin gene for Vero E6 cells and calreticulin gene for ISE6 cells) were performed using actinF (5′-AACCTTCCTTCCTGGGCAT-3′)/actinR (5′-TGATGCTCTCCTTCTGCAT-3′) and CRT321F/CRT452R31 primer pairs, respectively. Plasmids containing single-copy portions of Rickettsia 17-kDa antigen and beta-actin gene as well as Rickettsia 17-kDa antigen and tick calreticulin gene were constructed as described by Zanettii et al.34 and Petchampai et al.,31 respectively, and used as standard templates for quantification. A 35 µL qPCR reaction mixtures were premixed in 96-well plates using 2X iTaq SYBR Green Supermix (BioRad, Hercules, CA), 100 nM of each primer, DNase/RNase-free water, and 5 µL of either gDNA template (samples), serial 10-fold dilutions of a plasmid, or water (negative control). The mixtures were then aliquoted in triplicate 10-μL reactions on 384-well plates. The qPCR was carried out with an ABI 7900HT system (Applied Biosystems, Foster City, CA) using previously optimized conditions.34 The data were analyzed with the ABI 7900HT sequence detection system (SDS 2.2.3) software. The DNA copies per well of the Rickettsia 17-kDa antigen gene at each time point of infection in the respective cultures were calculated. Relative fold change of rickettsial load, defined as fold changes of the load relative to the first time point (1 hpi), were calculated from cycle threshold (CT) values for each sample using 2-∆∆CT method.35
Statistical Analysis
Data were reported as means ± standard errors of the means (SEM) from at least two independent experiments. T-test was used for a comparison of grouped means at each time point of infection. A P-value of ≤ 0.05 was considered significantly different.
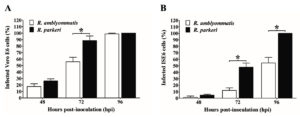
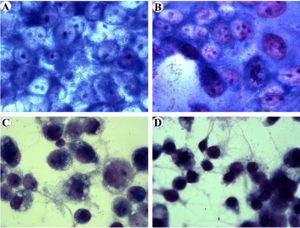
In this study, the growth and infectivity of the human pathogen R. parkeri and a Rickettsia species of undetermined pathogenicity, R. amblyommatis, were compared in Vero E6 (a model of mammalian cells) and the tick-derived ISE6 cells. It was found that rickettsiae were difficult to identify by Diff-Quik staining during the first 24 hpi. The mean percentage of infected cells at 48, 72, and 96 hpi were shown in Fig. 1. The percentage of Vero E6 and ISE6 cells infected with each rickettsial species correlated with an increased duration of infection. At 48 hpi, ~25% of Vero E6 cells were found to be infected by R. parkeri and R. amblyommatis with no significant difference (P > 0.05). The levels of infection in Vero E6 cells at 72 hpi increased ~3-fold as compared with those at 48 hpi which R. parkeri possessed significantly higher infectivity than R. amblyommatis (P < 0.05) (Fig. 1A). Although a significant difference in infectivity was not identified at 96 hpi, Vero E6 cells exposed to R. parkeri showed a greater density of rickettsiae within the cells than those of R. amblyommatis. Cell detachment was not observed, despite the level of infection being close to 100%. ISE6 cells exhibited lower percentages of rickettsial infection than Vero E6 cells. The pathogenic R. parkeri displayed a significantly higher level of infection in the ISE6 cells than R. amblyommatis at 72 and 96 hpi (P < 0.05). Moreover, infection of ISE6 cells reached nearly 100% by the pathogenic agent within 4 days and partial detachment of cells was observed in some samples. While only ~54% of ISE6 cells were infected by R. amblyommatis (Fig. 1B). At the late time point of infection (96 hpi), abundant intracellular and extracellular rickettsiae were identified in both cell lines (Fig. 2).
Fig. 1. Percentage of Vero E6 (A) and ISE6 (B) cells infected with R. parkeri and R. amblyommatis at 48, 72, and 96 hours post-inoculation. Data are presented as means ± standard errors of the means. An asterisk denotes a significant difference between species (P < 0.05; t-test).
Fig. 2. Diff-Quik stained of Vero E6 cells infected with R. pakeri (A) and R. amblyommatis (B) and ISE6 cells infected with R. parkeri (C) and R. amblyommatis (D) at 96 hours post-inoculation. Images are at a magnification of 630.
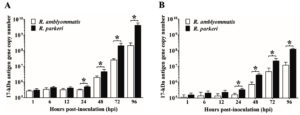
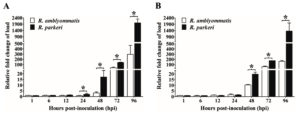
Growth of R. parkeri and R. amblyommatis in two types of eukaryotic cell lines, in terms of the total number of Rickettsia (17-kDa antigen gene copies) in each sample and the relative fold change of rickettsial load, were assessed by qPCR and compared over a 96-hour time course of infection. In Vero E6 cells, the number of inoculated rickettsiae remaining associated with the host cells at the initial time point (1 hpi) was not significantly different (P > 0.05) between the two rickettsial species and the bacterial loads did not change significantly during the first 12 hours of the experiment. After 24 hpi, Vero E6 cells displayed a continuous increase in rickettsial burdens for both species that reached the highest number at 96 hpi. Significant differences in growth between species with known and uncertain pathogenicity (P < 0.05) were identified from 24 hpi onward; such distinctions were more apparent over time of infection. The numbers of R. parkeri elevated from ~5.03×105 at 24 hpi to ~4.24×109 at 96 hpi with an increase of ~8430 times while R. amblyommatis presented only ~670 times increase (from ~3.16×105 at 24 hpi to ~2.11×108 at 96 hpi) (Fig. 3A). When the relative fold change of rickettsial load for both species was analyzed using 2-∆∆CT method, it was found that the fold change values were in good agreement with their total numbers. R. parkeri exhibited ~2-, ~17-, ~142-, and ~2002-fold increase in burden at 24, 48, 72, and 96 hpi, respectively. In contrast to R. parkeri, however, R. amblyommatis displayed significantly fewer fold changes of load (~0.9-, ~3-, ~39-, and ~296-fold at the same infection period; P < 0.05) (Fig. 4A). The results revealed that pathogenic R. parkeri propagated in Vero E6 cells faster than R. amblyommatis, a species of undetermined pathogenicity.
Fig. 3. Quantity of rickettsiae in Vero E6 (A) and ISE6 cells (B) at 1, 6, 12, 24, 48, 72, and 96 hours post-inoculation. The total number of Rickettsia (17-kDa antigen gene copy numbers) at each time point of infection was quantified by qPCR using a primer set specific to rickettsial 17-kDa antigen gene. Data are presented as means ± standard errors of the means. An asterisk denotes a significant difference between species (P < 0.05; t-test).
The growth profile of the two agents was also investigated in ISE6 cells. Comparable quantities (P > 0.05) of R. parkeri and R. amblyommatis that remained associated with ISE6 cells at 1 hpi were identified, however, with lower quantities than those of Vero E6 cells. Following the initial infection, the rickettsial burdens were relatively stable during the first 12 hours, subsequently elevated after 24 hpi, and achieved the greatest number at 96 hpi. From 24 hpi onward, loads of R. parkeri were significantly higher than those of R. amblyommatis (P < 0.05) with an increase of ~368 times (from ~3.37×105 at 24 hpi to ~1.24×108 at 96 hpi) and ~74 times (from ~1.63×105 at 24 hpi to ~1.20×107 at 96 hpi), respectively (Fig. 3B). When comparing mean 2-∆∆CT values of the two agents, it was found that R. parkeri possessed significantly greater fold changes of a burden than R. amblyommatis (~20- vs ~10-, ~176- vs ~61-, and ~1363- vs ~161-fold at 48, 72, and 96 hpi, respectively; P < 0.05) (Fig. 4B). These results were consistent with the Vero E6 cell data. Taken together, our results suggested that the human pathogenic R. parkeri grew faster than R. amblyommatis, a species of uncertain pathogenicity, in both Vero E6 and ISE6 cells.
Fig. 4. Relative fold change of rickettsial load in Vero E6 (A) and ISE6 cells (B).
The qPCR analyses for rickettsial 17-kDa antigen gene relative to host gene (beta-actin gene for Vero E6 cells or calreticulin gene for ISE6 cells) at 1, 6, 12, 24, 48, 72, and 96 hours post-inoculation (hpi) were performed. Fold changes of the load relative to the first time point (1 hpi) were calculated from cycle threshold (CT) values for each sample. Data are presented as means ± standard errors of the means. An asterisk denotes a significant difference between species (P < 0.05; t-test).
The life cycle of tick-borne SFGR involves both the tick vector and the vertebrate host. To establish an infection, rickettsiae must undertake several stages of the life cycle including adhesion to and internalization into host cells, endosomal escape into the cytoplasm, replication, and dissemination.36 In this study, the growth and infectivity of Amblyomma tick-associated SFGR, the human pathogen R. parkeri (strain Portsmouth) and R. amblyommatis (strain Darkwater), a species of uncertain pathogenicity, were compared in Vero E6 and ISE6 cells over 96 hours of infection. Quantitative analyses of R. parkeri and R. amblyommatis in both cell types showed that rickettsiae levels remained relatively constant for the first 12-24 hpi suggesting the length of the lag phase required for the bacteria to encounter the initial stages of their life cycles before beginning cell division. The lag phase duration of SFGR in cell culture system varied among species, e.g., 2-3 days for Rickettsia helvetica cultured in Vero cells,37 6-8 days for Rickettsia felis grown in S2 and C6/36 cells,38 except for Rickettsia slovaca that showed no lag phase when cultivated in Vero and L929 cells.39 Other tick-transmitted Rickettsia can grow differently in different cell types.40 The numbers, as well as fold changes in the load of the two rickettsiae, increased rapidly thereafter. Although they both entered the exponential phase of growth, however, distinct profiles for each agent were identified. The pathogenic R. parkeri possessed significantly higher numbers and fold changes of a burden than those of R. amblyommatis in both mammalian and tick cell lines indicating an association between the growth rate and rickettsial pathogenicity. In addition, the results from infectivity assessment during 72-96 hpi revealed that the levels of infection by R. parkeri were greater than those of R. amblyommatis implying the ability of the pathogenic species to disseminate more rapidly than species with uncertain pathogenicity. The incubation period of R. parkeri rickettsiosis in humans is 2-10 days8 supporting the growth and infectivity results from this study.
The distinct proliferation rate between rickettsial species with known and uncertain pathogenicity demonstrated by the current study may explain the pathogenic nature of these two Amblyomma-associated rickettsiae. R. parkeri was recognized as a human pathogen that causes remarkable clinical symptoms such as fever, headache, diffuse myalgia, macular rash, eschars-associated illnesses,3,7,8,41 while R. amblyommatis has occasionally been reported as a possible causative agent associated with rash19 and mild symptoms.42 Previous studies have demonstrated that SFGR manipulate vertebrate hosts to establish sufficient loads in the host cells for successful horizontal transfer.43,44 For other obligate intracellular bacterial pathogens, burdens of bacteria within the host have been shown to be associated with disease severity.45,46 The greater growth rate of R. parkeri in mammalian cells than that of R. amblyommatis, resulting in higher bacterial load of the pathogenic agent, may be one of the important factors contributing to increased severity of disease and transmission efficiency to tick vectors.
Tick-to-host transmission and survival in both tick and vertebrate hosts are important for the persistence of rickettsiae.47,48 To successfully alternate and survive between tick vectors to vertebrate hosts, Rickettsia pathogens require effective strategies to evade the host defense system for host cell invasion and causing disease through the obligate intracellular lifestyle. Studies in rickettsial species/strains with different levels of virulence have shown the association of bacterial loads in tick salivary glands and saliva with transmission efficiency to vertebrate hosts.49,50 Thus, the efficient proliferation of the human pathogen R. parkeri over the unknown pathogenic species, R. amblyommatis, in tick-derived ISE6 cells found in the current study supports the previous findings that pathogenicity is associated with rickettsial burden and transmission efficiency.
On the other hand, the slow growth rate would rather help promote persistent infections of non-pathogenic rickettsiae in the tick host. It was frequently found that most infected ticks harbor non-pathogenic rickettsial species.51 R. amblyommatis burdens in naturally infected A. americanum ticks were shown to be relatively stable in adult tick tissues during feeding and throughout their life cycle stages.34 Considering as an agent that causes only mild illness42 together with a slow growth characteristic found in the present study makes R. amblyommatis to be the great potential immunostimulant. R. amblyommatis has been reported to induce cross-protection against R. rickettsii in a guinea pig model.52,53 This may be due to the fact that R. amblyommatis shared immunoreactive proteins with the pathogenic rickettsial species.54 Further investigation is required to elucidate the mechanisms by which this rickettsial species, sometimes considered as tick symbiont, utilize to persist in the tick host and probably help prevent rickettsial infections. Altogether, the results from the present study demonstrate that the growth of SFGR is associated with pathogenicity. Studying SFGR growth characteristics in mammalian and tick cells facilitates a better understanding of rickettsial biology and pathogenesis leading to disease prevention and treatment.
ACKNOWLEDGMENTS
The authors would like to thank T. Kurtti and U. Munderloh (University of Minnesota, Minnesota) for providing the ISE6 cell line and members of the Macaluso laboratory for their technical assistance.
CONFLICT OF INTEREST
The authors declare that there is no conflict of interest.
AUTHORS’ CONTRIBUTION
AB and WB conceptualized the initial idea, formulated the hypothesis, designed the experiment, curated and analyzed the data. AB conducted the experiment, collected and interpreted data and drafted the manuscript. CDP and KRM supported research materials. KRM supervised the project. All authors read and approved the final manuscript for publication.
FUNDING
This work was supported by the National Institutes of Health to KRM [NIH AI077784].
ETHICS STATEMENT
This article does not contain any studies with human participants or animals performed by any of the authors.
AVAILABILITY OF DATA
All datasets generated or analyzed during this study are included in the manuscript.
- Fang R, Blanton LS, Walker DH. Rickettsiae as emerging infectious agents. Clin Lab Med. 2017;37(2):383-400.
Crossref - Blanton LS. The rickettsioses: A practical update. Infect Dis Clin North Am. 2019;33(1):213-229.
Crossref - Paddock CD, Sumner JW, Comer JA, et al. Rickettsia parkeri: a newly recognized cause of spotted fever rickettsiosis in the United States. Clin Infect Dis. 2004;38(6):805-811.
Crossref - Mendell NL, Reynolds ES, Blanton LS, et al. Detection of Rickettsiae, Borreliae, and Ehrlichiae in ticks collected from Walker County, Texas, 2017-2018. Insects. 2019;10(10):315.
Crossref - Parker RR, Kohls GM, Cox GW, Davis GE. Observations on an infectious agent from Amblyomma maculatum. Public Health Rep. 1939;54(32):1482-1484.
Crossref - Paddock CD. Rickettsia parkeri as a paradigm for multiple causes of tick-borne spotted fever in the Western Hemisphere. Ann N Y Acad Sci. 2005;1063(1):315-326.
Crossref - Whitman TJ, Richards AL, Paddock CD, et al. Rickettsia parkeri infection after tick bite, Virginia. Emerg Infect Dis. 2007;13(2):334-346.
Crossref - Paddock CD, Finley RW, Wright CS, et al. Rickettsia parkeri rickettsiosis and its clinical distinction from Rocky Mountain spotted fever. Clin Infect Dis. 2008;47(9):1188-1196.
Crossref - Paddock CD, Goddard J. The evolving medical and veterinary importance of the Gulf Coast tick (Acari: Ixodidae). J Med Entomol. 2015;52(2):230-252.
Crossref - Hecht JA, Allerdice MEJ, Karpathy SE, et al. Distribution and occurrence of Amblyomma maculatum sensu lato (Acari: Ixodidae) and Rickettsia parkeri (Rickettsiales: Rickettsiaceae), Arizona and New Mexico, 2017-2019. J Med Entomol. 2020;57(6):2030-2034.
Crossref - Yaglom HD, Casal M, Carson S, et al. Expanding recognition of Rickettsia parkeri rickettsiosis in southern Arizona, 2016-2017. Vector Borne Zoonotic Dis. 2020;20(2):82-87.
Crossref - Cohen SB, Yabsley MJ, Garrison LE, et al. Rickettsia parkeri in Amblyomma americanum ticks, Tennessee and Georgia, USA. Emerg Infect Dis. 2009;15(9):1471-1473.
Crossref - Parola P, Paddock CD, Socolovschi C, et al. Update on tick-borne rickettsioses around the world: a geographic approach. Clin Microbiol Rev. 2013;26(4):657-702.
Crossref - Lee S, Kakumanu ML, Ponnusamy L, et al. Prevalence of Rickettsiales in ticks removed from the skin of outdoor workers in North Carolina. Parasit Vectors. 2014;7:607.
Crossref - Herrick KL, Pena SA, Yaglom HD, et al. Rickettsia parkeri rickettsiosis, Arizona, USA. Emerg Infect Dis. 2016;22(5):780-785.
Crossref - Livengood J, Hutchinson ML, Thirumalapura N, Tewari D. Detection of Babesia, Borrelia, Anaplasma, and Rickettsia spp. in adult black-legged ticks (Ixodes scapularis) from Pennsylvania, United States, with a Luminex multiplex bead assay. Vector Borne Zoonotic Dis. 2020;20(6):406-411.
Crossref - Dahlgren FS, Paddock CD, Springer YP, Eisen RJ, Behravesh CB. Expanding range of Amblyomma americanum and simultaneous changes in the epidemiology of spotted fever group rickettsiosis in the United States. Am J Trop Med Hyg. 2016;94(1):35-42.
Crossref - Karpathy SE, Slater KS, Goldsmith CS, Nicholson WL, Paddock CD. Rickettsia amblyommatis sp. nov., a spotted fever group Rickettsia associated with multiple species of Amblyomma ticks in North, Central and South America. Int J Syst Evol Microbiol. 2016;66(12):5236-5243.
Crossref - Billeter SA, Blanton HL, Little SE, Levy MG, Breitschwerdt EB. Detection of Rickettsia amblyommii in association with a tick bite rash. Vector Borne Zoonotic Dis. 2007;7(4):607-610.
Crossref - Apperson CS, Engber B, Nicholson WL, et al. Tick-borne diseases in North Carolina: is “Rickettsia amblyommii” a possible cause of rickettsiosis reported as Rocky Mountain spotted fever? Vector Borne Zoonotic Dis. 2008;8(5):597-606.
Crossref - Drexler NA, Dahlgren FS, Heitman KN, Massung RF, Paddock CD, Behravesh CB. National surveillance of spotted fever group rickettsioses in the United States, 2008-2012. Am J Trop Med Hyg. 2016;94(1):26-34.
Crossref - Polsomboon S, Hoel DF, Murphy JR, et al. Molecular detection and identification of Rickettsia species in ticks (Acari: Ixodidae) collected from Belize, Central America. J Med Entomol. 2017;54(6):1718-1726.
Crossref - Trout Fryxell RT, Steelman CD, Szalanski AL, Billingsley PM, Williamson PC. Molecular detection of Rickettsia species within ticks (Acari: Ixodidae) collected from Arkansas United States. J Med Entomol. 2015;52(3):500-508.
Crossref - Jiang J, Yarina T, Miller MK, Stromdahl EY, Richards AL. Molecular detection of Rickettsia amblyommii in Amblyomma americanum parasitizing humans. Vector Borne Zoonotic Dis. 2010;10(4):329-340.
Crossref - Egizi A, Gable S, Jordan RA. Rickettsia spp. infecting lone star ticks (Amblyomma americanum) (Acari: Ixodidae) in Monmouth County, New Jersey. J Med Entomol. 2020;57(3):974-978.
Crossref - Sakamoto JM, Azad AF. Propagation of arthropod-borne Rickettsia spp. in two mosquito cell lines. Appl Environ Microbiol. 2007;73(20):6637-6643.
Crossref - Uchiyama T, Kishi M, Ogawa M. Restriction of the growth of a nonpathogenic spotted fever group Rickettsia. FEMS Immunol Med Microbiol. 2012;64(1):42-47.
Crossref - Tello-Martin R, Dzul-Rosado K, Zavala-Castro J, Lugo-Caballero C. Approaches for the successful isolation and cell culture of American Rickettsia species. J Vector Borne Dis. 2018;55(4):258-264.
Crossref - Bell-Sakyi L, Zweygarth E, Blouin EF, Gould EA, Jongejan F. Tick cell lines: tools for tick and tick-borne disease research. Trends Parasitol. 2007;23(9):450-457.
Crossref - Pornwiroon W, Bourchookarn A, Paddock CD, Macaluso KR. Proteomic analysis of Rickettsia parkeri strain Portsmouth. Infect Immun. 2009;77(12):5262-5271.
Crossref - Petchampai N, Sunyakumthorn P, Guillotte ML, et al. Molecular and functional characterization of vacuolar-ATPase from the American dog tick Dermacentor variabilis. Insect Mol Biol. 2014;23(1):42-51.
Crossref - Sunyakumthorn P, Bourchookarn A, Pornwiroon W, David C, Barker SA, Macaluso KR. Characterization and growth of polymorphic Rickettsia felis in a tick cell line. Appl Environ Microbiol. 2008;74(10):3151-3158.
Crossref - Grasperge BJ, Reif KE, Morgan TD, et al. Susceptibility of inbred mice to Rickettsia parkeri. Infect Immun. 2012;80(5):1846-1852.
Crossref - Zanettii AS, Pornwiroon W, Kearney MT, Macaluso KR. Characterization of rickettsial infection in Amblyomma americanum (Acari: Ixodidae) by quantitative real-time polymerase chain reaction. J Med Entomol. 2008;45(2):267-275.
Crossref - Schmittgen TD, Livak KJ. Analyzing real-time PCR data by the comparative CT method. Nat Protoc. 2008;3(6):1101-1108.
Crossref - Walker DH. Rickettsiae and rickettsial infections: the current state of knowledge. Clin Infect Dis. 2007;45(Suppl 1):S39-S44.
Crossref - Elfving K, Lukinius A, Nilsson K. Life cycle, growth characteristics and host cell response of Rickettsia helvetica in a Vero cell line. Exp Appl Acarol. 2012;56(2):179-187.
Crossref - Luce-Fedrow A, Macaluso KR, Richards AL. Growth of Rickettsia felis in Drosophila melanogaster S2 cells. Vector Borne Zoonotic Dis. 2014;14(2):101-110.
Crossref - Boldis V, Spitalska E. Dermacentor marginatus and Ixodes ricinus ticks versus L929 and Vero cell lines in Rickettsia slovaca life cycle evaluated by quantitative real time PCR. Exp Appl Acarol. 2010;50(4):353-359.
Crossref - Balraj P, Vestris G, Raoult D, Renesto P. Comparison of Rickettsia conorii growth within different cell lines by real-time quantitative PCR. Clin Microbiol Infect. 2009;15:294-295.
Crossref - Kaskas NM, Ledet JJ, Wong A, Muzny CA, Elopre L, Hughey L. Rickettsia parkeri: Eschar diagnosis. J Am Acad Dermatol. 2014;71(3):e87-89.
Crossref - Delisle J, Mendell NL, Stull-Lane A, Bloch KC, Bouyer DH, Moncayo AC. Human infections by multiple spotted fever group rickettsiae in Tennessee. Am J Trop Med Hyg. 2016;94(6):1212-1217.
Crossref - Souza CE, Moraes-Filho J, Ogrzewalska M, et al. Experimental infection of capybaras Hydrochoerus hydrochaeris by Rickettsia rickettsii and evaluation of the transmission of the infection to ticks Amblyomma cajennense. Vet Parasitol. 2009;161(1-2):116-121.
Crossref - Galletti MFBM, Paddock CD, Hecht JA, Biggerstaff BJ, Ritter JM, Karpathy SE. Isolate-dependent differences in clinical, pathological, and transcriptional profiles following in vitro and in vivo infections with Rickettsia rickettsii. Infect Immun. 2021;89(4):e00626-e00620.
Crossref - Groves MG, Kelly DJ. Characterization of factors determining Rickettsia tsutsugamushi pathogenicity for mice. Infect Immun. 1989;57(5):1476-1482.
Crossref - Sonthayanon P, Chierakul W, Wuthiekanun V, et al. Association of high Orientia tsutsugamushi DNA loads with disease of greater severity in adults with scrub typhus. J Clin Microbiol. 2009;47(2):430-434.
Crossref - Saito TB, Bechelli J, Smalley C, Karim S, Walker DH. Vector tick transmission model of spotted fever rickettsiosis. Am J Pathol. 2019;189(1):115-123.
Crossref - White A, Schaefer E, Thompson CW, Kribs CM, Gaff H. Dynamics of two pathogens in a single tick population. Lett Biomath. 2019;6(1):50-66.
Crossref - Ueti MW, Knowles DP, Davitt CM, Scoles GA, Baszler TV, Palmer GH. Quantitative differences in salivary pathogen load during tick transmission underlie strain-specific variation in transmission efficiency of Anaplasma marginale. Infect Immun. 2009;77(1):70-75.
Crossref - Suwanbongkot C, Langohr IM, Harris EK, Dittmar W, Christofferson RC, Macaluso KR. Spotted fever group Rickettsia infection and transmission dynamics in Amblyomma maculatum. Infect Immun. 2019;87(4):e00804-00818.
Crossref - Azad AF, Beard CB. Rickettsial pathogens and their arthropod vectors. Emerg Infect Dis. 1998;4(2):179-186.
Crossref - Blanton LS, Mendell NL, Walker DH, Bouyer DH. “Rickettsia amblyommii” induces cross protection against lethal Rocky Mountain spotted fever in a guinea pig model. Vector Borne Zoonotic Dis. 2014;14(8):557-562.
Crossref - Rivas JJ, Moreira-Soto A, Alvarado G, et al. Pathogenic potential of a Costa Rican strain of ‘Candidatus Rickettsia amblyommii‘ in guinea pigs (Cavia porcellus) and protective immunity against Rickettsia rickettsii. Ticks Tick Borne Dis. 2015;6(6):805-811.
Crossref - Pornwiroon W, Bourchookarn A, Paddock CD, Macaluso KR. Immunoproteomic profiling of Rickettsia parkeri and Rickettsia amblyommii. Ticks Tick Borne Dis. 2015;6(6):829-835.
Crossref
© The Author(s) 2022. Open Access. This article is distributed under the terms of the Creative Commons Attribution 4.0 International License which permits unrestricted use, sharing, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.