ISSN: 0973-7510
E-ISSN: 2581-690X
Phytopathogenic fungi pose a significant threat to agricultural sustainability, leading to huge economic losses and reducing food quality. Consequently, alternative exploration of strategies for disease management are critical, particularly utilizing naturally occurring microorganisms with antagonistic potential. In this study, bacterial isolates obtained from Jakrem and Garampani hot springs of North East India were evaluated for potential antagonism activity against four phytopathogenic fungi namely Sclerotinia sclerotiorum, Corynespora cassiicola, Fusarium oxysporum f. sp. pisi, and Colletotrichum capsici. The result recorded eight bacterial isolates with prominent antifungal activity against the test phytopathogens and their antagonistic effect was clearly visualized by scanning electron microscope analysis, which revealed mycelial deformities in the treated cultures. Crude metabolites obtained from these bacteria isolates were characterized by Gas Chromatography-Mass Spectrometry (GC-MS) and Attenuated Total Reflection (ATR) analyses. The metabolites revealed presence of several functional groups and bioactive compounds like 1,2-Benzenedicarboxylic acid, Nonanoic acid, Dibutyl phthalate, Oleic acid, Ergotamine, Citronellol, Chloroacetic acid, and Erucic acid, which were known to possess antimicrobial properties. 16S rRNA sequencing and NCBI BLAST based search for molecular identification confirmed the identity of isolates, belonging to the genera Bacillus. The study also recorded that three bacterial isolates possess the presence of surfactin and iturin antimicrobial peptides (AMP) biosynthesis gene cluster. Further, bio-formulation prepared using the three antagonistic bacterial isolates showed growth promotion in mustard seeds while inhibiting the pathogen, S. sclerotiorum in an in vitro water agar assay. The findings suggest that hot springs bacterial could be explored for bio-based agents and could serve as sustainable alternatives to synthetic agrochemicals for management and control of phytopathogenic fungi.
Hot springs, Bacillus, Biocontrol, GC-MS, Lipopeptides
In countries like India, agricultural productivity is an important critical factor for food security, as more than 60% of the population depends on agriculture for livelihood.1 Although India ranks fifth globally in agricultural production, sustaining this growth amidst the increasing population generate tremendous pressures and environmental degradation remains a significant challenge.2 While the introduction of synthetic agrochemicals during the Green Revolution dramatically increased yields, however, their long-term application has negative consequences linked with soil degradation, resistance in pathogens, and disruption of local biodiversity.3,4
Phytopathogenic fungi pose a continuous threat to crop health, reducing productivity and food quality.5 The excessive reliance on synthetic fungicides and pesticides for disease management has exacerbated issues such as pathogen resistance, making sustainable alternatives critical for future agricultural practices.6,7 Recent advances in biological control have highlighted the potential of microbial antagonists, such as Bacillus spp., as promising candidates for integrated pest management.8,9 These biocontrol agents offer a more environmentally friendly approach, leveraging natural antagonistic properties to suppress phytopathogens while promoting plant health. In this context, thermophilic bacteria from extreme environments, such as hot springs, represent a largely untapped resource with immense biotechnological potential.10 Hot springs, characterized by elevated temperatures and unique physicochemical conditions, harbour wide-range of microbiota that are better adapted to survive under harsh environments.11 These extremophilic microorganisms, especially which belonged to the genus Bacillus possess endospores which are resistant to environmental extremes, have demonstrated the ability of antibiotic production, hydrolytic enzymes and bioactive metabolites that can be harnessed for agricultural and medicinal applications.12,13 An important attribute possessed by Bacillus spp. are their capacity to form resilient endospores and produce antimicrobial metabolites providing protection against a wide array of phytopathogens, including Fusarium, Sclerotinia, and Colletotrichum.14,15
Considering the increasing global emphasis on sustainable agriculture and the limitations on used of synthetic agrochemicals, this study aims to explore the isolation of Bacillus spp. from hot springs as biocontrol agents against phytopathogenic fungi. It is generally considered that the thermophilic nature of Bacillus spp., might have the ability to produce potent metabolites that could offer an eco-friendly alternative for managing crop diseases. Therefore, the present study aims to access the antagonism potential of Bacillus isolates from Jakrem and Garampani hot springs of North East India against selected phytopathogenic fungi. Attempt was also made to characterize the metabolites of three promising isolates and their bio-formulation for management and control of phytopathogenic fungus, Sclerotinia sclerotiorum.
Sampling sites and collection of samples
Jakrem hot spring located in Meghalaya (GPS coordinates: 25°24.4632” N, 91°32.4092” E) and Garampani hot spring situated in Assam, North East India (GPS coordinates: 26°25’12.49″ N, 93°52’47.99″ E) were selected as the study sites. Portable thermometer and GPS locator system (Garnel, Model 4100) were used at each sampling site to measure the temperature and geographical positioning data of water samples. Water samples were collected in pre-sterilized 500 ml plastic containers. The containers were initially cleaned with 20% sodium hypochlorite solution and then thoroughly rinsed with sterile distilled water to remove any residual disinfectant. They were subsequently sterilized by autoclaving at 121 °C for 15 min and dried in a hot air oven at 50 °C for 1 h. To ensure further sterility, the containers were exposed to UV light for 1 h inside a laminar airflow cabinet. Samples were collected randomly in triplicates from each hot spring and transported to the laboratory for further analysis.16
Isolation of bacteria
Serial dilution technique was performed for isolating distinct isolates from water samples on three different growth media: Nutrient Agar (peptone: 5 g/l; sodium chloride: 5 g/l; HM peptone B: 1.5 g/l; yeast extract: 1.5 g/l; agar: 15 g/l), Luria-Bertani Agar (tryptone: 10 g/l; yeast extract: 5 g/l; sodium chloride: 10 g/l; agar: 15 g/l) and Reasoner’s 2A Agar (casein enzymic hydrolysate: 0.25 g/l; peptic digest of animal tissue: 0.25 g/l; casein acid hydrolysate: 0.5 g/l; yeast extract: 0.5 g/l; glucose: 0.5 g/l; starch soluble: 0.5 g/l; dipotassium phosphate: 0.03 g/l; magnesium sulphate, heptahydrate: 0.5 g/l; sodium pyruvate: 0.030 g/l; agar: 15 g/l). 100 µl aliquot of serially diluted samples (10-1 to 10-7), were spread plated on the medium and incubated at 44 ± 1 °C for samples from Jakrem hot spring and at 37 ± 1 °C for samples from Garampani hot spring for 72-96 h. Following incubation, distinct colonies that appeared on the agar media was counted, and morphological characteristics of the colonies, including shape, size, elevation, pigmentation, and color, were recorded. Further purification of the colonies was employed by repeated sub-culturing and, finally stored on Nutrient Agar slants at 4 °C and 20% glycerol stocks at -20 °C. Furthermore, purified bacterial isolates were subjected to microscopic examination and biochemical tests such as gram staining, citrate utilization, oxidase, catalase for the genus level identification.4,17
Test phytopathogenic fungi and maintenance
In the present study, three different phytopathogenic fungi-Sclerotinia sclerotiorum (ITCC 4042), Fusarium oxysporum f. sp. pisi (ITCC 4814), and Corynespora cassiicola (ITCC 6748) were obtained from the Institute of Advanced Study in Science and Technology (DST-IASST), Boragaon, Assam, India. Additionally, one phytopathogenic fungus, Colletotrichum capsici (NCBI GenBank Accession ID: KY124644), was obtained from the Microbial Ecology Laboratory, Department of Botany, Gauhati University, Assam, India. All fungal isolates were cultured on Potato Dextrose Agar (PDA; HiMedia) slants and incubated at 26 ± 1 °C for 48-72 h. The cultures were then stored at 4 °C until further use.4
In vitro assessment of antagonistic activity against selected phytopathogens
Bacterial isolates were tested for potential antagonism against the test phytopathogens in vitro. The activity was conducted through a preliminary screening using dual-culture analysis. For this activity, agar plates (Potato Dextrose Agar (PDA), HiMedia) were inoculated with a fungal plug (5 mm2) at the center. Further, the isolates were spot inoculated opposite of the inoculated fungal plug on the agar plate. A plate containing only a fungal plug (without bacterial inoculation) was also inoculated simultaneously and marked as control for calculation of percentage (%) inhibition by the isolates. The inoculated plates were further incubated at 26 ± 1 °C for 120 h. Each activity was performed in triplicates and percentage (%) inhibition was calculated.1
% inhibition = (1 – FG/CG) × 100
Where,
FG = Fungal Growth on the bacteria treated plate (mm)
CG = Fungal Growth on control plate (mm)
Microscopic observations
Scanning electron microscope (SEM) analysis was conducted for observation of morphological changes in the phytopathogenic mycelium produced due to inhibitory effect of the potent bacterial isolates. Briefly, the mycelium was taken from the bacteria-fungi interaction zone and fixed in 3% glutaraldehyde for one hour. Following fixation, the sample was dehydrated by passing it through increasing concentrations of ethanol that ranged from 10%-100%. After dehydration, the sample was gold-coated and imaged under different magnifications through scanning electron microscope (ZEISS, Sigma 300 VP).4
Metabolite extraction and characterization
Extraction of metabolite from the antagonistic isolates was performed by mass multiplication in broth medium (Nutrient Broth (NB), HiMedia). Briefly, 100 µl inoculum of potent isolate (10° cells/ml) was inoculated in 150 ml of sterilized broth medium and incubated under continuous shaking (120 rpm) for 120 h at 30 ± 1 °C. After incubation, at 10,000 rpm, centrifugation of the cultured medium was carried out for 15 min and cell free supernatant was collected. The supernatant was mixed thoroughly with equal proportion of ethyl acetate (1:1) solvent using a separating funnel. From the mixture, fraction consisting of solvent (ethyl acetate) was collected and was again mixed with equal proportion of the same solvent (ethyl acetate). The process was repeated again and the final fraction was evaporated by a rotary evaporator at <40 °C to dryness and, the remained metabolites was dissolved in methanol.18 For identification of biologically active compounds in the crude extract, gas chromatography-mass spectrometry (GC-MS) analysis was further carried out. The analysis used helium (He) as the carrier gas in the GC-MS instrument (Clarus 600C MS; liquid auto-sampler; model: Clarus 680 GC), Perkin Elmer (USA). The obtained peaks in the chromatogram were analyzed using NIST-2014 software.19
Determination of biocontrol efficacy on Brassica juncea (mustard green)
Determination of biocontrol efficacy by potent bacterial isolates against S. sclerotiorum was evaluated using Brassica juncea (mustard green) in vitro. For the present study, seeds of B. juncea (cultivar number IC 597866) were collected from the National Bureau of Plant Genetic Resources (NBPGR), Umiam, Meghalaya. Briefly, surface sterilization of the seeds was carried out by treating the mustard seeds with a solution of sodium hypochlorite (2.5%) for a duration of 10 min followed by several washed steps with sterile water (double distilled) for removal of any trace’s sodium hypochlorite. After sterilization, seeds were dried under laminar air flow (LAF) chamber on sterile filter paper. For bacterization, 100 µl overnight grown cultures of potent isolates (106 cells/ml) were inoculated in Nutrient Broth (NB) medium (150 ml) and incubated under shaking conditions (120 rpm) for 24 h at 30 ± 1 °C. Following incubation, centrifugation of the inoculated NB medium was performed at 10000 rpm for 10 min for harvesting bacterial cells. Distilled water (sterile) was further used to suspend the harvested cells and using haemocytometer count, the final density was adjusted to 106 cells/ml.1 Further, 0.1% carboxymethyl cellulose (CMC) was mixed with the bacterial cells as binder, and coated on the surface-sterilized seeds followed by further drying for additional 2 h inside the laminar air flow cabinet. After proper drying, seeds were transferred aseptically onto water agar plates. For the seed assay, the phytopathogen; S. sclerotiorum was cultured in PDB medium, and incubated for 120 h at 28 ± 1 °C. After incubation, the broth medium was passed through 4 layers of cheesecloth for filtering out the mycelium, and the collected supernatant was centrifuged at 10000 rpm for 10 min. Following centrifugation, the supernatant was discarded and the harvested spores was suspended in sterilized distilled water. The concentration of spore suspension was further determined using a hemocytometer count and, the final density was adjusted to 106 spores/ml by using sterilized distilled water.4 For the assay, each bacterized seed was inoculated with 10 µl of spore suspension (106 spores/ml) and incubated at 26 ± 1 °C for 24-120 h. Seeds without bacterization, and inoculated with phytopathogen spore suspension was marked as negative control, while seeds without bacterization, and inoculated only distilled water (sterile) was marked as positive control. Each replication for the experiment contained 15 seeds and conducted in triplicates. The biocontrol potential of the isolates was later evaluated based on the following scoring system1,20:
Score description
- No germination
- Germinated seed showing both seed coat and plumule infection
- Either seed coat or plumule infection
- Healthy seeds with no infection
Score = n × 0
Where,
n corresponds to seeds number in each score category, and
0 = score assigned (for each treatment)
Seed bacterization study
Seed bacterization study was conducted as per methodology described by Rahman et al.21 with some modifications. Briefly, surface sterilization of the seeds was carried as per methodology described above. Further, the seeds were dipped into cell suspension of potent bacterial isolates (106 cells/ml) mixed with 0.1% carboxymethyl cellulose (CMC) as a binder for 30 min. Further, 5 bacterized seeds were placed in a straight-line at the center together with S. sclerotiorum mycelial plug (5 mm) on the opposite sides. S. sclerotiorum inoculated seeds without bacterization was marked as control. Plates were incubated at 26 ± 1 °C for 24-120 h and conducted in triplicates (each replication contain 5 seeds).21
In vitro determination of biocontrol activity on detached leaf assay
The biocontrol efficiency of isolates on mustard leaves was assessed following the methodology described by Shimizu et al.22 with some modifications. Briefly, surface sterilization of the seeds was carried as per methodology described above and germinated in plastic pots which contained sterilized soil (200 g soil autoclaved twice for 15 min at 121 °C at a 24-hour interval) for 3 weeks in an open field. After the emergence of 5-6 fully expanded true leaves, surface sterilization of the leaves was performed using solution of sodium hypochlorite (5%) for 10 min followed by several washed steps with sterile water (double distilled) to remove any traces sodium hypochlorite. For bacterization, cell suspension of potent bacterial isolates (10° cells/ml) mixed with 0.1% carboxymethyl cellulose (CMC) as a binder was coated on the surface of sterilized leaves. Bacterized leaves were then further air dried inside Laminar Air Flow (LAF) chamber and transferred finally onto petri plates containing filter paper (sterile) moistened with distilled water (sterile) under aseptic conditions. 10 µl spore suspension (106 spores/ml) of S. sclerotiorum was then inoculated on each bacterized leaf at the center. Mustard leaves without bacterization and inoculated only with the S. sclerotiorum spore suspension was marked as control. Plates were further incubated at 26 ± 1 °C for 144 h and each treatment was conducted in triplicates and biocontrol efficacy was calculated.1
Biocontrol efficacy = dc – dt/dc × 100
Where,
dc = control leaf lesion diameter
dt = treated leaf lesion diameter
Attenuated total reflection (ATR) analysis
Detection and identification of functional groups in the crude metabolites extracted from three potent bacterial isolates antagonistic to Sclerotinia sclerotiorum were performed using Attenuated Total Reflection (ATR) spectroscopy. For metabolite extraction, the selected bacterial isolates were mass-cultured in Nutrient Broth (NB; HiMedia). Briefly, 100 µl of bacterial inoculum (106 cells/ml) was inoculated into 150 ml of sterilized NB and incubated at 30 ± 1 °C for 120 h under continuous shaking at 120 rpm. After incubation, the culture broth was centrifuged at 10,000 rpm for 15 min to obtain the cell-free supernatant. The supernatant was extracted with ethyl acetate in a 1:1 (v/v) ratio using a separating funnel. The organic (ethyl acetate) phase was collected and subjected to two additional extractions with fresh ethyl acetate to ensure maximum metabolite recovery. The combined ethyl acetate fractions were evaporated to dryness under reduced pressure at temperatures below 40 °C using a rotary evaporator. The resulting dried crude extract was finely ground using a micro mortar and pestle. ATR spectra of the crude extract were recorded in the range of 500-4000 cm-1, with four scans performed over the entire range using an integrated spectrometer and built-in plotter.4
Analysis for antimicrobial peptides (AMP) biosynthesis gene cluster
The analysis was conducted using three sets of primers viz. fengycin (FENDF: GGCCCGTTCTCTAAATCCAT & FENDR: GTCATGCTGACGAGAGCAAA), surfactin (SRFAF: TCGGGACAGGAAGACATCAT & SRFAR: CCACTCAAACGGATAATCCTGA) and iturin (ITUCF: GGCTGCTGCAGATGCTTTAT & ITUCR: TCGCAGATAATCGCAGTGAG) by PCR based method for detection of antimicrobial peptides (AMP) biosynthesis genes involved in antagonism from 3 potent isolates. The reaction mixture (15 µl) contained 1X PCR buffer, 0.2 mM dNTPs, 1 µM forward primer, 1 µM reverse primer, MgCl₂‚ (2 mM), Taq DNA polymerase (1.25 U) and genomic DNA (approximately 60 ng). PCR was performed with the following conditions- 94 °C (2 min), 35 cycles of 94 °C (30 s), annealing-58 °C (1 min), extension-72 °C (1.5 min) and extension step (final) at 72 °C (7 min). Further, the amplified products were analyzed in agarose gel (1.2%) and images were captured using a gel documentation system.4,23
Molecular identification of antagonist bacterial isolates
Genomic DNA from all the antagonistic isolates was extracted according to the manufacturer’s instructions provided in HiPurA® Bacterial Genomic DNA Purification Kit (HiMedia, MB505). Universal primers viz. 27F (5′-AGAGTTTGATCCTGGCTCAG-3′) and 1492R (5′-TACGGYTACCTTGTTACGACTT-3′) were used for amplification of the extracted DNA through PCR in a thermal cycler. The reaction mixture contained dATP (0.2 mM), dCTP (0.2 mM), dGTP (0.2 mM), dTTP (0.2 mM), 27F (1 µM), 1492R (1 µM), MgCl₂‚ (2 mM), Taq DNA polymerase (1.25 U), and genomic DNA (1 µl) with the reaction conditions: 95 °C (2 min), continued by 35 cycles of 95 °C (30 s), 53 °C (30 s), and 72 °C (1.5 min), and final extension of 72 °C (7 min). The finally obtained amplified products were analyzed in agarose gel (1.2%) and images were captured using a gel documentation system. Further purification and gene sequencing of the amplified products were carried out at Unigenome (Unipath Specialty Laboratory). After gene sequencing, obtained sequences were subjected to BLAST based search on NCBI database for determination of expect value (e-value). Phylogenetic tree was later constructed using the neighbor-joining method in MEGA7, and the obtained sequences were finally submitted to the NCBI, GenBank database.4,23
Isolation of bacteria
The present study recorded a total of 96 distinct bacterial isolates from both the hot springs, of which 43 isolates from Garampani and, 53 isolates from Jakrem hot spring, respectively. The colony-forming unit (CFU) was recorded as 2.07 x 102 CFU/ml for Garampani hot spring and 1.36 x 102 CFU/ml for Jakrem hot spring, respectively. In the present study, recorded mean temperature of water samples was 36.16 ± 0.25 °C with a pH of 8.43 ± 0.18 for Garampani hot spring. In contrast, water samples from Jakrem hot spring recorded higher mean temperature of 44.96 ± 0.2 °C, with a pH of 8.54 ± 0.02 respectively.
In vitro assessment of antagonistic activity against selected phytopathogens
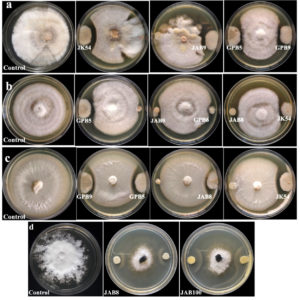
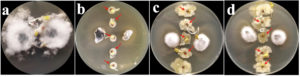
8 bacterial isolates (5 isolates from Jakrem and 3 isolates from Garampani hot spring) were recorded with positive activity against the test phytopathogens through dual-culture analysis. Microscopic examination revealed that all the 8 potent bacterial isolates were gram-positive, rod-shaped bacteria. Moreover, all the 8 bacterial isolates also showed positive for catalase and oxidase activities. However, only 4 bacterial isolates i.e. JK54, JAB8 and JAB100 isolated from Jakrem hot spring and, GPB6 isolated from Garampani hot spring was recorded positive for citrate utilization activity (Table 1). The dual culture assay revealed 6 bacterial isolates (3 isolates from Jakrem and 3 isolates from Garampani hot spring) were antagonistic to Fusarium oxysporum with highest activity shown by isolate GPB9 with an inhibition of 48.52 ± 2.46% and lowest activity by isolate GPB6 with an inhibition of 17.05 ± 2.17% (Figure 1a). Similarly, 6 bacterial isolates (3 isolates from Jakrem and 3 isolates from Garampani hot spring) showed antagonistic activity to Corynespora cassiicola with highest activity recorded by isolate GPB9 with an inhibition of 43.55 ± 3.009% and lowest activity was recorded by isolate JAB9 with an inhibition of 6.06 ± 2.94% (Figure 1b). Again, 4 bacterial isolates (2 isolates from Jakrem and 2 isolates from Garampani hot spring) were antagonistic to Colletotrichum capsici with maximum activity recorded by isolate GPB5 with 50.92 ± 1.33% inhibition of followed by isolate JK54 with 43.14 ± 2.46% inhibition, isolate GPB9 with 40.85 ± 1.69% inhibition and isolate JAB8 recorded with lowest activity with an inhibition of 24.01 ± 1.79% respectively (Figure 1c). However, only 3 bacterial isolates isolated from Jakrem hot spring was antagonistic to Sclerotinia sclerotiorum with maximum activity recorded by bacterial isolate JAB100 with an inhibition % of 25.2 ± 2.29 followed by bacterial isolate JAB8 with an inhibition % 19.54 ± 1.18 and bacterial isolate JAB1 with lowest activity against Sclerotinia sclerotiorum with an inhibition % of 9.24 ± 1.32 respectively (Figure 1d). The present study recorded, only 1 bacterial isolate, JAB8 out of the total, isolated from Jakrem hot spring displayed antagonistic activity to all the test phytopathogens tested in the present study (Table 2).
Table (1):
Morphological and biochemical characteristics of antagonistic bacterial isolates
Bacterial ID |
Cell morphology |
Gram staining |
Catalase |
Oxidase |
Citrate utilization |
|---|---|---|---|---|---|
JK54 |
Rod |
+ |
+ |
+ |
+ |
JAB1 |
Rod |
+ |
+ |
+ |
– |
JAB8 |
Rod |
+ |
+ |
+ |
+ |
JAB9 |
Rod |
+ |
+ |
+ |
– |
JAB100 |
Rod |
+ |
+ |
+ |
+ |
GPB5 |
Rod |
+ |
+ |
+ |
– |
GPB6 |
Rod |
+ |
+ |
+ |
+ |
GPB9 |
Rod |
+ |
+ |
+ |
– |
(+ denotes positive activity; – denotes no activity)
Table (2):
Antagonistic activity of potent bacterial isolates against fungal phytopathogens
Bacterial ID |
Fusarium oxysporum |
Corynespora cassiicola |
Colletotrichum capsici |
Sclerotinia sclerotiorum |
|---|---|---|---|---|
JK54 |
31.17 ± 2.53 |
38.27 ± 0.53 |
43.14 ± 2.46 |
– |
JAB1 |
– |
– |
– |
9.24 ± 1.32 |
JAB8 |
18.62 ± 2.34 |
16.28 ± 1.26 |
24.01 ± 1.79 |
19.54 ± 1.18 |
JAB9 |
13.27 ± 2.55 |
6.06 ± 2.94 |
– |
– |
JAB100 |
– |
– |
– |
25.2 ± 2.29 |
GPB5 |
34.75 ± 3.93 |
39.27 ± 2.28 |
50.92 ± 1.33 |
– |
GPB6 |
17.05 ± 2.17 |
7.72 ± 1.22 |
– |
– |
GPB9 |
48.52 ± 2.46 |
43.55 ± 3.009 |
40.85 ± 1.69 |
– |
(Values are mean of 3 replications ± standard deviation; – denotes no activity)
Figure 1. Growth suppression and inhibition of phytopathogens by selected bacterial isolates under dual-culture treatment (a: Fusarium oxysporum f.sp. pisi; b: Corynespora cassiicola; c: Colletotrichum capsici and d: Sclerotinia sclerotiorum; JK54, JAB8, JAB9, JAB100: bacterial isolates isolated from Jakrem hot spring; GPB5, GPB6, GPB9: bacterial isolates isolated from Garampani hot spring)
Microscopic observations
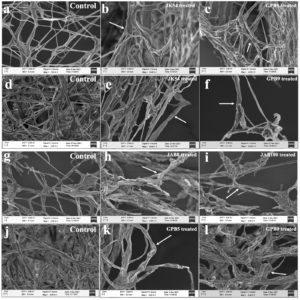
Morphological changes due to interaction with the antagonist bacterial isolates viz. JK54, JAB1, JAB8, JAB9, JAB100 from Jakrem hot spring and, GPB5, GPB6, GPB9 from Garampani hot spring in dual culture assay recorded deformities in the mycelium of test phytopathogens under scanning electron microscope (SEM) analysis. SEM analysis recorded alterations in the mycelium of phytopathogens to varying degrees due to bacterial interactions observed in the dual culture assay. Morphological changes in the mycelium of treated sets included hyphal distortion, shrinkage, and irregularities. The hyphal distortion included flaccid depressed regions along the mycelium of phytopathogenic fungi Corynespora cassiicola and Colletotrichum capsici. Other irregularities such as curling, deformed mycelium, plasmolyzed, shrunken and exposed ruptured sites due to leakage of cellular contents were observed along the mycelium of phytopathogenic fungi Fusarium oxysporum and Sclerotinia sclerotiorum after bacterial interaction. However, in contrast, the hyphal segments of the control sets exhibited no such deformities and maintained consistent homogeneity throughout the observation period (Figure 2).
Figure 2. Morphological deformities in mycelia indicated by arrows observed under scanning electron microscope (SEM) after bacterial isolates interactions (a: Corynespora cassiicola (control); b: C. cassiicola co-inoculated with JK54; c: C. cassiicola co-inoculated with GPB5; d: Only Fusarium oxysporum f.sp. pisi (control); e: F. oxysporum f.sp. pisi co-inoculated with JK54; f: Fusarium oxysporum f.sp. pisi co-inoculated with GPB9; g: Only Sclerotinia sclerotiorum (control), h: S. sclerotiorum co-inoculated with JAB8, i: S. sclerotiorum co-inoculated with JAB100; j: Only Colletotrichum capsici (control); k: C. capsici co-inoculated with GPB5; l: C. capsici co-inoculated with GPB9)
Characterization of extracted metabolites
GC-MS analysis of crude extract
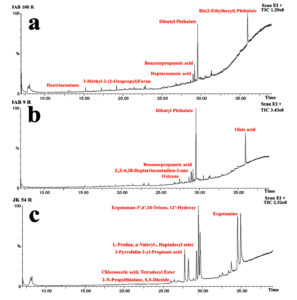
A total of 44 antimicrobial and bioactive compounds was recorded from the crude metabolite of antagonistic bacterial isolates through GC-MS analysis (Table 3). Bioactive compounds such as; Benzenepropanoic acid, Nonanoic acid, 1,2-Benzenedicarboxylic acid, Dibutyl Phthalate, 3-Methyl-2-(2-Oxypropyl) Furan; 3-N-Hexylthiolane, N-Decanoic acid, Phthalic acid, Oleic acid; Oxirane, Decyl; 2-Nonadecanone 2,4-dinitrophenylhydrazine; Heptacosanoic acid, Octadecanoic acid, Z,Z-6,28-Heptatriactontadien-2-one were some of the common bioactive compounds detected in the crude metabolite of majority of potent bacterial isolates through peak analysis. However, production of bioactive compounds such as Ergotamine; Erucic acid; Ergotaman-3’,6′,18-Trione 12’-Hydroxy, Hexahydropyrrolizin-3-one; (3-Aminopropyl) Dibutylborane; 3-Methyldec-3-ene; L-Proline, L-Leucine identified through peak analysis were recorded only for bacterial isolate JK54. Other compounds such as Pentanoic acid, Octanoic acid, N-Decanoic acid were detected only in the crude metabolite produced by bacterial isolate JAB1. Similarly, Cyclohexanebutanoic acid; Citronellol bioactive compounds were detected only in the crude metabolite produced by bacterial isolate JAB8 and 9-Hexadecenoic acid, Sulforous acid, compounds were only detected in the crude metabolite produced by bacterial isolate JAB100 through peak analysis (Figure 3).
Table (3):
List of bioactive compounds identified in the crude metabolite of antagonist bacterial isolates
No. |
Compound Name |
Activity |
Bacterial ID |
Ref. |
|---|---|---|---|---|
1. |
3-Methyl-2-(2-Oxypropyl) Furan |
Antimicrobial |
JAB1, JAB8, JAB9, JAB100, GPB5, GPB6, GPB9 |
24,25 |
2. |
Z,Z-6, 28-Heptatriactontadien- 2-one |
Vasodilator |
JAB100, JAB8, JAB9, GPB5, GPB6, GPB9 |
26 |
3. |
Oxirane, Decyl |
Antimicrobial, adhesive |
JAB8, JAB100, JAB9 |
26 |
4. |
BIS (2-Ethylhexyl) Phthalate |
Antibacterial |
JAB1, JAB8, GPB5, GPB9 |
27 |
5. |
3-N-Hexylthiolane, S,S-dioxide |
Antimicrobial, Antiviral |
JAB1, JAB8, JAB9, GPB5, GPB6, GPB9, JK54 |
26 |
6. |
Dibutyl Phthalate |
Antimicrobial |
JAB1, JAB100, JAB9, GPB5 |
28 |
7. |
Phthalic acid, butyl hexyl ester |
Antimicrobial |
JAB1, JAB100, JAB9, GPB5, GPB6 |
29 |
8. |
1,2-Benzenedicarboxylic acid |
Antimicrobial |
JAB1, JAB100 |
30,31 |
9. |
Pentanoic acid, 4-Methyl- |
Antimicrobial |
JAB1, GPB5 |
32,33 |
10. |
Pentanoic acid, 3-Methyl- |
Antioxidant, Anti-inflammatory |
JAB1 |
34 |
11. |
Beta-L- Arabinopyranoside Methyl |
Catechol-O- methyltransferase inhibitor, Antioxidant |
JAB1, GPB5 |
26 |
12. |
Benzenepropanoic acid 3,5-bis (1 1- dimethylethyl)-4-Hydroxy- Methyl ester |
Antifungal, Antioxidant |
JAB1, JAB100, GPB6 |
35 |
13. |
Nonanoic acid |
Antimicrobial |
JAB1, GPB5 |
26 |
14. |
2,5,6-trimethyl 1,3 Oxathiane |
Antidiabetic, Antioxidant |
JAB1, GPB5 |
36 |
15. |
N-Decanoic acid |
Antibacterial, Antifungal |
JAB1, GPB5 |
26 |
16. |
Octanoic acid |
Antibacterial |
JAB1 |
26 |
17. |
N-Decanoic acid |
Antibacterial, Antifungal |
JAB1 |
26 |
18. |
Oleic acid |
Anticancer, Antimicrobial, Antifungal |
JAB8, JAB9, GPB5, GPB6, GPB9 |
26,37 |
19. |
11,14-Eicosadienoic acid, Methyl ester |
Anti-inflammatory, Antioxidant |
JAB8, JAB9 |
26 |
20. |
Oleyl alcohol, Trifluoroacetate |
Used as a nonionic surfactant, emulsifier, emollient |
JAB8, JAB9, GPB5, GPB6, GPB9 |
26 |
21. |
Palmitoleic acid |
Anti-inflammatory |
JAB8, GPB9 |
26 |
22. |
Cyclohexanebutanoic acid |
Effect on CNS, Antioxidant |
JAB8 |
26 |
23. |
2-Nonadecanone 2,4- dinitrophenylhydrazine |
Antimicrobial |
JAB8, JAB9 |
26 |
24. |
Citronellol |
Antimicrobial |
JAB8 |
38,39 |
25. |
1,2-Benzenedicaroboxylic acid, Dionyl Ester |
Antimicrobial |
JAB1, JAB8, JAB9, GPB5, GPB9 |
30,31 |
26. |
1,2-benzenedicarboxylic acid, butyl 2-Ethylhexyl ester |
Antifungal |
GPB5, JAB1, JAB100 |
40 |
27. |
Octadecanoic acid |
Antioxidant |
JAB100, JK54 |
30 |
28. |
9-Octadecanoic acid |
Antihypertensive |
JAB100, JK54 |
30 |
29. |
9-Hexadecenoic acid, Methyl ester, (Z) |
Antimicrobial |
JAB100 |
41 |
30. |
Sulforous acid, Butyl Dodecyl ester |
Antibacterial, Antioxidant, Antifungal |
JAB100 |
42,43 |
31. |
Heptacosanoic acid, 25-Methyl-, Methyl ester |
Antimicrobial |
JAB100 |
26 |
32. |
Hentriacontane |
Antioxidant, Anti-inflammatory, Antifungal |
JAB100 |
43,44 |
33. |
Chloroacetic acid, Tetradecyl Ester |
Antifungal |
JK54 |
45 |
34. |
Ergotamine |
Antibacterial, Antifungal |
JK54 |
46,47 |
35. |
2-N-Propylthiolane, S,S-Dioxide |
Antioxidant |
JK54 |
48 |
36. |
3-Pyrrolidin-2-yl-Propionic acid |
Antibacterial |
JK54 |
49 |
37. |
Ergotaman-3′,6′,18-Trione, 12′- Hydroxy |
Antimicrobial |
JK54 |
49 |
38. |
Erucic acid |
Antifungal |
JK54 |
50 |
39. |
Hexahydropyrrolizin-3-one |
Antimicrobial, Anti-inflammatory |
JK54 |
51 |
40. |
Pyrrolo[1,2-a]Pyrazine-1,4-dione, Hexahydro-3-(2-methylpropyl)- |
Antimicrobial, Algicidal, Antifungal |
JK54 |
51 |
41. |
(3-Aminopropyl) Dibutylborane |
Antimicrobial |
JK54 |
52 |
42. |
3-Methyldec-3-ene |
Antibacterial |
JK54 |
53 |
43. |
L-Proline, n-valeryl-, Heptadecyl ester |
Antimicrobial |
JK54 |
54 |
44. |
L-Leucine, n-cyclopropylcarbonyl-, Pentadecyl ester |
Antibacterial |
JK54 |
55 |
Figure 3. Total Ion Chromatogram (TIC) obtained from Gas Chromatography-Mass Spectrometry (GC-MS) analysis of the crude extracts of JAB100 (a), JAB9 (b), and JK54 (c), showing the profile of bioactive compounds present in each extract
Determination of biocontrol efficacy on Brassica juncea (mustard green)
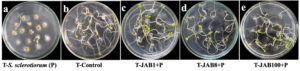
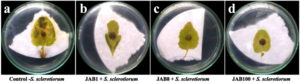
In vitro inoculation of mustard seeds with the phytopathogen alone (negative control-P) or in combination with antagonistic bacterial isolates showed differences in the germination of the plumule and radicle compared to the control (water only). Out of the 45 seeds treated with the fungal pathogen S. sclerotiorum (negative control), 16 seeds failed to germinate, 23 seeds exhibited both plumule and seed coat infection, and 5 seeds showed either seed coat or plumule infection and only 1 seed was recorded with healthy germination, resulting in a total score of 36 for that treatment set. In contrast, seeds inoculated only with water (positive control) recorded 1 seed with both plumule and seed coat infection, 1 seed with either seed coat or plumule infection and 43 seeds with healthy growth and no infection symptoms, leading to a total score of 129 for the treatment set. The results also indicated increased germination rates in mustard seeds with both plumule and radicle length and no visible symptoms of infection when bacterized with cell suspensions of the antagonistic bacterial isolates together with S. sclerotiorum (Figure 4). In presence of S. sclerotiorum (P), co-inoculation of mustard seeds bacterized with cell suspension of bacterial isolate JAB1 recorded a total score of 134 with 44 seeds showed healthy germination of both plumule and radicle and only 1 seed was recorded with seed coat infection under in vitro analysis. Moreover, co-inoculation of mustard seeds bacterized with cell suspension of bacterial isolate JAB100 and bacterial isolate JAB8 together with S. sclerotiorum recorded no visible symptoms of either plumule or seed coat infection and all the 45 mustard seeds recorded healthy germination of both plumule and radicle with a total score of 135 (Table 4).
Table (4):
In vitro screening for biocontrol efficiency of antagonist bacterial isolates against S. sclerotiorum on mustard seeds
Treatment ID |
No germination (nx0) |
Germinated seed with both seed coat and plumule infection (nx1) |
Either seed coat or plumule infection (nx2) |
Seeds with no infection (nx3) |
Score |
|---|---|---|---|---|---|
-Control (P) |
16 |
23 |
5 |
1 |
36 |
+Control |
0 |
1 |
1 |
43 |
129 |
JAB1+P |
0 |
0 |
1 |
44 |
134 |
JAB8+P |
0 |
0 |
0 |
45 |
135 |
JAB100+P |
0 |
0 |
0 |
45 |
135 |
-Control (P): Only S. sclerotiorum inoculated seeds; +C: Only sterile distilled water seeds inoculated seeds; JAB1, JAB8 and JAB100: bacterial isolate ID; Score = n×0, where n is the number of seeds in each score category, and 0 = score assigned for each treatment
Figure 4. Seed germination assay on water agar plates inoculated with only S. sclerotiorum (a) – negative control), inoculated with only sterile distilled water (b) – positive control), JAB1 bacterized seeds co-inoculated with S. sclerotiorum (c), JAB8 bacterized seeds co-inoculated with S. sclerotiorum (d) and JAB100 bacterized seeds co-inoculated with S. sclerotiorum (e)
Seeds bacterization study
The result of seed bacterization study recorded reduced incidence of S. sclerotiorum infection with higher percentage of seedling germination in bacterized sets of seeds when compared with control. In control sets (seeds with only S. sclerotiorum), out of 15 seeds only 3 seeds showed germination while other failed to germinate and were subsequently colonised by S. sclerotiorum mycelia. In contrast, bacterization of mustard seed recorded inhibition on the mycelial development of S. sclerotiorum along with increased seed germination with maximum activity by bacterial isolate JAB100 followed by bacterial isolate JAB8 and bacterial isolate JAB1 bacterized seeds (Figure 5).
Figure 5. In vitro biocontrol activity of bacterized seeds against S. sclerotiorum (a: mustard seeds with only S. sclerotiorum; b: JAB1 bacterized seeds; c: JAB8 bacterized seeds; d: JAB100 bacterized seeds showing protection from S. sclerotiorum infection; arrows indicate seed germination)
In vitro determination of biocontrol activity on detached leaf assay
Bacterization of surface sterilized mustard leaves recorded reduction in lesion diameter with higher biocontrol efficacy against S. sclerotiorum when compared with control (without bacterization). In control sets (inoculated with S. sclerotiorum only), the average diameter of lesions i.e. 12.33 ± 1.30 mm was recorded. However, in treated sets, bacterization of leaves with antagonist isolates and further inoculation of the bacterized leaves with S. sclerotiorum, the lesion diameter reduced to 4.21 ± 0.31 mm for bacterial isolate JAB100 followed by 4.45 ± 0.38 mm for bacterial isolate JAB1, and 4.99 ± 0.08 mm for bacterial isolate JAB8 respectively. Biocontrol efficacy of the potent bacterial isolates was recorded in the order of 65.39 ± 6.48% for bacterial isolate JAB100, 62.24 ± 3.67% for bacterial isolate JAB1 and 59.14 ± 4.64% for bacterial isolate JAB8 when compared with the control sets (Figure 6).
Figure 6. In vitro biocontrol efficiency of potent bacterial isolates on detached mustard leaf against S. sclerotiorum. (a) negative control showing large infected lesions (12.33 ± 1.30 mm) (b, c, d) bacterized mustard leaf showing reduced lesion diameter after 7 days of inoculation (JAB1: 4.45 ± 0.38 mm, JAB8: 4.99 ± 0.08 mm and JAB100: 4.21 ± 0.31 mm)
Attenuated total reflection (ATR) analysis
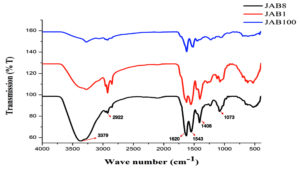
Extracted metabolite from the 3 potent bacterial isolates antagonistic to Sclerotinia sclerotiorum was subjected to attenuated total reflection analysis for identification of functional groups. The analysis recorded the occurrence of similar functional groups in the crude metabolite extracted from all the bacterial isolates which included O-H stretch for alcohols (3379 cm-1), C-H stretch for alkanes (2922 cm-1), C=C stretch for conjugated alkenes (1620 cm-1), N-O stretch for nitro compounds (1543 cm-1), O-H bending for carboxylic acids (1408 cm-1), C-N stretch for amines (1073 cm-1) (Figure 7).
Figure 7. Attenuated Total Reflection (ATR) analysis for identification of functional groups in the extracted crude metabolite by 3 potent bacterial isolates antagonistic to Sclerotinia sclerotiorum (JAB1, JAB8, JAB100: bacterial isolates ID isolated from Jakrem hot spring)
Analysis for antimicrobial peptides (AMP) biosynthesis gene cluster
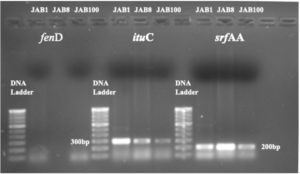
PCR analysis detected the occurrence of 2 antimicrobial peptides (AMP) biosynthesis genes in isolates antagonistic to the phytopathogen viz. S. Sclerotiorum. The analysis recorded srfAA and ituC AMP genes were recorded from all the bacterial isolates after subjecting the extracted genomic DNA to PCR analysis using specific primers. PCR product of 200 bp (srfAA) and 300 bp (ituC) was detected and documented on Gel-Documentation system. Moreover, genomic DNA from all the bacterial isolates were recorded negative for fenD antimicrobial peptide (AMP) genes (Figure 8).
Figure 8. Detection of ituC (iturin) and srfAA (surfactin) antimicrobial peptide biosynthesis gene through agarose gel electrophoresis. (fenD AMP gene was recorded negative for all the 3 bacterial isolates; JAB1, JAB8, JAB100: bacterial isolates ID isolated from Jakrem hot spring)
Molecular identification of antagonist bacterial isolates
Based on 16S rRNA sequences, molecular identity of antagonist bacterial isolates were identified as Bacillus velezensis (Accession No.-OR754585.1 for bacterial isolate JAB1), Bacillus proteolyticus (Accession No.: OR755781.1 for bacterial isolate JAB8), Bacillus cereus (Accession No.: PQ357480 for bacterial isolate JAB9), Bacillus sp. (Accession No.: OR755825.1 for bacterial isolate JAB100), Bacillus tequilensis (Accession No.: OR826593.1 for bacterial isolate JK54), Bacillus subtilis (Accession No.: PQ357330 for bacterial isolate GPB5), Bacillus tequilensis (Accession No.: PQ357343 for bacterial isolate GPB6) and Bacillus cereus (Accession No.: PQ357479 for bacterial isolate GPB9) (Figure 9).
The present study recorded isolation of 96 distinct bacterial isolates from Garampani and Jakrem hot spring water samples through serial dilution technique. The hot springs of Garampani and Jakrem represent unique ecological niches, often overlooked in microbial bioprospecting studies. Isolation of 96 distinct isolates from such extreme environments not only highlights their rich microbial diversity but also underlines the potential for discovering novel biocontrol agents. Preliminary screening through dual culture assays identified 8 bacterial isolates as potential antagonists against various test phytopathogens. The antagonistic activities by bacterial isolates are well-documented in the literatures, associated with diverse array of secondary metabolites, lytic enzymes, volatile organic compound productions, and the induction of host defense responses.21,56-58
The antagonist isolates in this study showed synthesis of antifungal compounds, effectively inhibiting the germination of phytopathogens by altering hyphal morphology, as observed through microscopic observations. SEM analysis revealed variations and irregularities in hyphal morphology due to bacterial interactions, while no such deformities were observed in the control sets. These structural aberrations in the phytopathogenic hyphae can be inferred due to antibiosis and lytic enzyme production.57 Phytopathogens inhibition by antagonistic bacterial isolates through the production of wide array of antifungal compounds, as demonstrated by SEM analysis and GC-MS, is particularly significant. These findings underscore the robust inhibitory mechanisms employed by Bacillus species, ranging from cell wall degradation to membrane disruption, for controlling phytopathogen effectively.
Numerous Bacillus spp. and its related genera have been reported with the production of secondary metabolites, hydrolytic enzymes, and induction of host defenses, providing biological control against phytopathogens.21,56-58 The genus Bacillus possesses unique survival traits, such as endospore formation and the production of antibiotics and metabolites that can antagonize a variety of phytopathogens, including Sclerotinia, Colletotrichum, Rhizoctonia, and Fusarium.4,21 The commercial viability of these Bacillus isolates is further enhanced by their resilience, including endospore formation and environmental adaptability and thereby, making them potential candidate for large-scale field applications. This aligns with the global trend toward harnessing microbial agents for integration of pest management strategies in agriculture.
For example, reduction in Fusarium wilt incidence in watermelon was achieved through Bacillus sp. WB application.59 Additionally, in vitro inhibition of rice pathogens such as Fusarium semitectum, Curvularia lunata and Helminthosporium oryzae by Bacillus amyloliquefaciens strain BAS23 was documented by Saechow et al.60 Biocontrol activity of B. subtilis isolated from rhizosphere of cotton against F. oxysporum had also been reported.61 Moreover, Jangir et al.31 isolated strains of Bacillus antagonistic to F. oxysporum f. sp. lycopersici from tomato rhizosphere. Reduction in the incidence of root rot by B. velezensis YW 17 against F. oxysporum in ginseng had also been reported.62 Similarly, antagonistic activity of Bacillus velezensis (Y6 and F7) against F. oxysporum and Ralstonia solanacearum had also been reported.63 The wide-range antagonistic activity of these isolates, effective against a number of phytopathogenic fungi suggests their potential application across multiple crops. This broad utility could provide benefits of economic significance, especially for crops like mustard, where diseases caused by Sclerotinia have been historically difficult to manage.
The current study identified several antimicrobial compounds, including 1,2-Benzenedicarboxylic acid, nonanoic acid, dibutyl phthalate, and oleic acid, through Gas chromatography-mass spectrometry (GC-MS) analysis. Notably, 1,2-Benzenedicarboxylic acid derivatives were produced by most bacterial isolates. Shabanamol et al.40 reported the antifungal activity of these compounds, specifically 1,2-Benzenedicarboxylic acid, butyl 2-Ethylhexyl ester from Lysinibacillus sphaericus KJ872548 against Rhizoctonia solani. Fatty acid derivatives are commonly extracted from Bacillus and Paenibacillus members.40,64 Chen et al.65 highlighted the antifungal activity of nonanoic acid produced by Burkholderia cenocepacia ETR-B22. Antifungal potential of nonanoic acid against various pathogens had been documented in various studies.66,67 Antifungal activity of Dibutyl phthalate produced by Streptomyces strain BITDG-11 against F. oxysporum f. sp. cubense had been reported.68 Bacillus spp. were also reported to produce variety of cyclic lipopeptides viz. surfactin, iturin, fengycins involved in antagonism against the phytopathogen.4,69
In the present study, Attenuated Total Reflection (ATR) analysis detected various peaks corresponding to alcohol, alkanes, alkenes, nitro compounds, carboxylic acids and amines. The presence of alcohol, alkane was earlier reported from the metabolite extracted from B. safensis STJP.70 Phenolic compounds and alcohols were earlier reported to be effective in controlling leaf spot disease caused by Alternaria alternata.71 Moreover, the presence of C-H, C-N stretch and, O-H bending from the siderophore extract of Bacillus albus had also been previously reported.72 The production of aromatic and aliphatic compounds by Bacillus spp. had been reported with antagonistic activity against pathogenic fungi.73 FTIR spectrum of crude lipopeptides extracted from B. velezensis NKMV 3 also recorded the presence of -OH, C-H stretching.74 In the present study, S. sclerotiorum antagonising isolates were also recorded positive for iturin, surfactin and negative for fengycin biosynthesis gene when examined by PCR analysis. These antimicrobial peptides disrupt membrane permeability of the phytopathogens along with the induction of disease resistance in the host plant and are synthesized non-ribosomally as non-ribosomal peptide synthetases (NRPSs) or polyketide synthases (PKSs).75-77 Hazarika et al.78 reported the presence of 5 biosynthetic gene cluster in B. subtilis that mediated antagonistic activity against sugarcane phytopathogen. Antagonistic activity of Bacillus strains against anthracnose causing Colletotrichum acutatum in Andean lupin seeds was mediated by lipopeptide genes.79 Confirmation of molecular identity was conducted through gene sequencing of 16S rRNA, for all antagonistic isolates, categorizing them within the genus Bacillus. Bacillus amyloliquefaciens mediated inhibition of white mold disease caused by S. sclerotiorum in mustard had been reported.21 Similarly, Hu et al.80 reported the inhibition of S. sclerotiorum in oilseed rape through the application of B. subtilis BY-2 under field conditions. The biocontrol activity of five different strains of rhizospheric Bacillus against S. sclerotiorum was documented by Vinodkumar et al.81 Additionally, Hu et al.58 reported biocontrol activity through the production of volatile metabolites by B. cereus CF4-51, which degraded mycelial cell walls and interfered with membrane permeability. Additionally, Hu et al.58 reported that Bacillus cereus CF4-51 exhibited biocontrol activity through the production of volatile metabolites, which degraded fungal mycelial cell walls and disrupted membrane permeability. Owing to their robust secretion of extracellular enzymes and high growth rates, members of the genus Bacillus are often regarded as industrial workhorses, with most species generally categorized as safe.82 Bacillus amyloliquefaciens subsp. plantarum and Bacillus tequilensis, both isolated from the rhizosphere, have demonstrated antagonistic activity against Corynespora cassiicola in pot trials.83 Beyond direct biocontrol, many Bacillus species are known to induce systemic resistance (ISR) in plants through the emission of volatile organic compounds, which are effective against a broad spectrum of phytopathogens.84 For instance, the induction of defense-related compounds by Bacillus subtilis EXB-123 was reported to inhibit Colletotrichum capsici in chilli.85 Similarly, B. subtilis was shown to enhance chilli seed germination and suppress Colletotrichum gloeosporioides OGC1 under in vitro conditions.86 In vivo application of Bacillus sp. strain M10 also resulted in effective protection of chilli pepper and tomato fruits against C. capsici.87 Moreover, Brevibacillus laterosporus BPM3, isolated from Garampani hot spring, was reported to exhibit antagonistic activity against multiple phytopathogens, including Fusarium oxysporum f. sp. ciceri, F. semitectum, Magnaporthe grisea, and Rhizoctonia oryzae.88
Further studies focusing on the field efficacy of these isolates under different agro-climatic conditions, combined with genome sequencing, could provide insights into the genetics that is mediating the antagonistic properties. This would aid in developing genetically enhanced isolates with superior biocontrol potential. Therefore, the use of Bacillus spp. for the management of cosmopolitan phytopathogens, presents an ideal approach due to their self-perpetuating nature, selective and sustainable control of target pathogens, low cost, and environmentally friendly characteristics in agriculture.4
This study successfully isolated 96 bacterial isolates from the Garampani and Jakrem hot water springs, identifying eight with significant antagonistic activity against various phytopathogens. The observed antagonism is linked to the production of secondary metabolites, lytic enzymes, and host defense responses, as evidenced by alterations in hyphae when observed through scanning electron microscopy. Bacillus species exhibited remarkable potential as biological control agents, with metabolites like 1,2-Benzenedicarboxylic acid and nonanoic acid contributing to their efficacy. Moreover, presence of surfactin (srfAA) and iturin (ituC) AMP genes was also recorded from the genomic DNA of potent isolates. Notably, 3 of these isolates demonstrated effectiveness against Sclerotinia sclerotiorum in mustard plants, highlighting their promise for sustainable agricultural practices. Molecular identification confirmed their classification within the genus Bacillus, emphasizing their role in managing phytopathogens. Further exploration of these isolates could enhance crop health and productivity while reducing reliance on chemical inputs in agriculture.
ACKNOWLEDGMENTS
The authors would like to express their deepest gratitude to (L) Prof. Dhruva Kumar Jha, Former Professor, Department of Botany, Gauhati University for his immense guidance and tireless support. The authors are also thankful to Department of Botany (UGC-DRS I and DST-FIST), Gauhati University for providing necessary facilities, National Bureau of Plant Genetic Resources (NBPGR), Umiam, Meghalaya for providing mustard seeds, Guwahati Biotech Park, Amingaon for carrying out the GC-MS analysis, and Institute of Advanced Study in Science and Technology (IASST), Boragaon for providing the phytopathogens used in the present study. Finally, the author (AK) would like to thank Council of Scientific and Industrial Research (CSIR) for the financial support received under CSIR-UGC NET JRF scheme (F.No. 16-9 (June 2019)/2019(NET/CSIR); UGC-Ref. No.:598/ (CSIR-UGC NET JUNE 2019).
CONFLICT OF INTEREST
The authors declare that there is no conflict of interest.
AUTHORS’ CONTRIBUTION
AK, JR and KT conceptualized the study. AK collected the samples and performed the experiment. AK, JR and KT performed data analysis and interpretation. KT performed supervision and data validation. AK and JR wrote the original draft. KT wrote and reviewed the manuscript. JR and KT edited the manuscript. All authors read and approved the final manuscript for publication.
FUNDING
This work was supported by funding received under the CSIR-UGC NET JRF scheme, Government of India.
DATA AVAILABILITY
The datasets generated and/or analysed during the current study are available from the corresponding author on reasonable request.
ETHICS STATEMENT
Not applicable.
- Goswami M, Deka S. Isolation of a novel rhizobacteria having multiple plant growth promoting traits and antifungal activity against certain phytopathogens. Microbiol Res. 2020;240:1-17.
Crossref - Glick BR. Plant growth promoting bacteria:mechanisms and applications. Scientifica. 2012;2012(1):963401.
Crossref - Correa OS, Soria MA. Potential of Bacilli for Biocontrol and Its Exploitation in Sustainable Agriculture. In: Maheshwari, D. (eds) Plant Growth and Health Promoting Bacteria. Microbiology Monographs, vol 18. Springer, Berlin, Heidelberg. 2011:197-209.
Crossref - Kumar A, Rabha J, Jha DK. Antagonistic activity of lipopeptide-biosurfactant producing Bacillus subtilis AKP, against Colletotrichum capsici, the causal organism of anthracnose disease of chilli. Biocatal Agric Biotechnol. 2021;36:102133.
Crossref - Szalewski DA, Hinrichs VS, Zinniel DK, Barletta RG. The pathogenicity of Aspergillus fumigatus, drug resistance, and nanoparticle delivery. Can J Microbiol. 2018;64(7):439-453.
Crossref - Strange RN, Scott PR. Plant disease: A threat to global food security. Annu Rev Phytopathol. 2005;43(1):83-116.
Crossref - Evans N, Baierl A, Semenov MA, Gladders P, Fitt BD. Range and severity of a plant disease increased by global warming. J R Soc Interface. 2008;5(22):525-531.
Crossref - Chakraborty S. Potential impact of climate change on plant-pathogen interactions. Australas Plant Pathol. 2005;34(4):443-448.
Crossref - Jayaprakashvel M, Mathivanan N. Management of plant diseases by microbial metabolites. In: Maheshwari, D. (eds) Bacteria in Agrobiology: Plant Nutrient Management. Springer, Berlin, Heidelberg. 2011:237-265.
Crossref - Sahay H, Yadav AN, Singh AK, Singh S, Kaushik R, Saxena AK. Hot springs of Indian Himalayas: Potential sources of microbial diversity and thermostable hydrolytic enzymes. 3 Biotech. 2017;7(2):118.
Crossref - Badhai J, Ghosh TS, Das SK. Taxonomic and functional characteristics of microbial communities and their correlation with physicochemical properties of four geothermal springs in Odisha, India. Front Microbiol. 2015;6:1166.
Crossref - Verma JP, Jaiswal DK, Krishna R, Prakash S, Yadav J, Singh V. Characterization and screening of thermophilic Bacillus strains for developing plant growth promoting consortium from hot spring of Leh and Ladakh region of India. Front Microbiol. 2018;9:1293.
Crossref - Debnath S, Rawat D, Mukherjee AK, Adhikary S, Kundu R. Applications and constraints of plant beneficial microorganisms in agriculture. Biostimulants in Plant Science. 2019:1-26.
Crossref - Tabbene O, Ben Slimene I, Bouabdallah F, Mangoni ML, Urdaci MC, Limam F. Production of anti-methicillin-resistant Staphylococcus activity from Bacillus subtilis sp. strain B38 newly isolated from soil. Appl Biochem Biotechnol. 2009;157(3):407-419.
Crossref - Radhakrishnan R, Hashem A, Abd_Allah EF. Bacillus: A biological tool for crop improvement through bio-molecular changes in adverse environments. Front Physiol. 2017;8:667.
Crossref - Kambura AK, Mwirichia RK, Kasili RW, Karanja EN, Makonde HM, Boga HI. Bacteria and Archaea diversity within the hot springs of Lake Magadi and Little Magadi in Kenya. BMC Microbiol. 2016;16(1):136.
Crossref - Fasina KA, Adesetan TO, Oseghale F, Egberongbe HO, Aghughu OO, Akpobome FA. Bacteriological and phytochemical assessment of Ficus asperifolia Linn. infusion. Biomed Res Int. 2020;2020(1):9762639.
Crossref - Lertcanawanichakul M, Pondet K, Kwantep J. In vitro antimicrobial and antioxidant activities of bioactive compounds (secondary metabolites) extracted from Streptomyces lydicus A2. J Appl Pharm Sci. 2015;5(2):017-21.
Crossref - Sarkar RD, Kalita MC. Green synthesized Se nanoparticle-mediated alleviation of salt stress in field mustard, TS-36 variety. J Biotech. 2022;359:95-107.
Crossref - Borah SN, Goswami D, Lahkar J, Sarma HK, Khan MR, Deka S. Rhamnolipid produced by Pseudomonas aeruginosa SS14 causes complete suppression of wilt by Fusarium oxysporum f. sp. pisi in Pisum sativum. Bio Control. 2015;60(3):375-385.
Crossref - Rahman MME, Hossain DM, Suzuki K, et al. Suppressive effects of Bacillus spp. on mycelia, apothecia and sclerotia formation of Sclerotinia sclerotiorum and potential as biological control of white mold on mustard. Australas Plant Pathol. 2016;45(1):103-117.
Crossref - Shimizu M, Yazawa S, Ushijima Y. A promising strain of endophytic Streptomyces sp. for biological control of cucumber anthracnose. J Gen Plant Pathol. 2009;75(1):27-36.
Crossref - Dowarah B, Agarwal H, Krishnatreya DB, Sharma PL, Kalita N, Agarwala N. Evaluation of seed associated endophytic bacteria from tolerant chilli cv. Firingi Jolokia for their biocontrol potential against bacterial wilt disease. Microbiol Res. 2021;248:126751.
Crossref - Al-Wathnani H, Ara I, Tahmaz RR, Bakir MA. Antibacterial activities of the extracts of cyanobacteria and green algae isolated from desert soil in Riyadh, Kingdom of Saudi Arabia. Afr JBiotechnol. 2012;11(38):9223-9229.
Crossref - Karthik R, Saravanan R, Ebenezar KK, Sivamalai T. Isolation, purification, and characterization of avian antimicrobial glycopeptide from the posterior salivary gland of Sepia pharaonis. Appl Biochem Biotechnol. 2015;175(3):1507-1518.
Crossref - Ralte L, Khiangte L, Thangjam NM, Kumar A, Singh YT. GC-MS and molecular docking analyses of phytochemicals from the underutilized plant, Parkia timoriana revealed candidate anti-cancerous and anti-inflammatory agents. Sci Rep. 2022;12(1):3395.
Crossref - Lotfy WA, Mostafa SW, Adel AA, Ghanem KM. Production of di-(2-ethylhexyl) phthalate by Bacillus subtilis AD35: Isolation, purification, characterization and biological activities. Microb Pathog. 2018;124:89-100.
Crossref - Roy RN, Laskar S, Sen SK. Dibutyl phthalate, the bioactive compound produced by Streptomyces albidoflavus 321.2. Microbiol Res. 2006;161(2):121-126.
Crossref - Makhwitine JP, Kumalo HM, Ndlovu SI, Mkhwanazi NP. Epigenetic induction of secondary metabolites production in endophytic fungi Penicillium chrysogenum and GC-MS analysis of crude metabolites with anti-hiv-1 activity. Microorganisms. 2023;11(6):1404.
Crossref - Balasundari T, Boominathan M. Screening of bioactive compounds by GC-MS, antimicrobial activity and in silico studies in Cynodon dactylon L. Pers leaves. World J Sci Res. 2018;3(1):7-15.
- Jangir M, Pathak R, Sharma S, Sharma S. Biocontrol mechanisms of Bacillus sp., isolated from tomato rhizosphere, against Fusarium oxysporum f. sp. lycopersici. Biol Control. 2018;123:60-70.
Crossref - Mohamad OAA, Li L, Ma J-B, et al. Evaluation of the antimicrobial activity of endophytic bacterial populations from Chinese traditional medicinal plant licorice and characterization of the bioactive secondary metabolites produced by Bacillus atrophaeus against Verticillium dahliae. Front Microbiol.2018;9:924.
Crossref - Sharma D, Pramanik A, Agrawal PK. Evaluation of bioactive secondary metabolites from endophytic fungus Pestalotiopsis neglecta BAB-5510 isolated from leaves of Cupressus torulosa D. Don. 3 Biotech. 2016;6(2):210.
Crossref - Solesi OA, Adesina FC, Adebayo-Tayo BC, Abiodun AS. Gas chromatography/mass spectrometry (GC-MS) analysis of Jatropha curcas latex and its antimicrobial activity on clinical isolates. World J Adv Res Rev. 2020;8(1):012-018.
Crossref - Elamin MM, Abdelrahim NA, Elhag DEA, Joseph MR, Hamid ME. Bioactive pyrrole-pyrazine derivative from a novel Bacillus species and review of the literature. Afr J Pharm Pharmacol. 2021;15(8):138-151.
Crossref - Dhanisha SS, Drishya S, Guruvayoorappan C. Traditional knowledge to clinical trials:A review on nutritional and therapeutic potential of Pithecellobium dulce. J Basic Clin Physiol. 2022;33(2):133-142.
Crossref - Jiang L, Wang W, He Q, et al. Oleic acid induces apoptosis and autophagy in the treatment of Tongue Squamous cell carcinomas. Sci Rep. 2017;7(1):11277.
Crossref - Gochev V, Wlcek K, Buchbauer G, et al. Comparative evaluation of antimicrobial activity and composition of rose oils from various geographic origins, in particular Bulgarian rose oil. Nat Prod Commun. 2008;3(7):1934578X0800300706.
Crossref - Dangol S, Poudel DK, Ojha PK, et al. Essential oil composition analysis of Cymbopogon species from eastern Nepal by GC-MS and chiral GC-MS, and antimicrobial activity of some major compounds. Molecules. 2023;28(2):543.
Crossref - Shabanamol S, Thampi M, Sajana P, Varghese S, Karthika S, George TK, Jisha MS. Characterization of the major antifungal extrolite from rice endophyte Lysinibacillus sphaericus against Rhizoctonia solani. Arch Microbiol. 2021;203(5):2605-2613.
Crossref - Rahman MM, Ahmad SH, Mohamed MTM, Ab Rahman MZ. Antimicrobial compounds from leaf extracts of Jatropha curcas, Psidium guajava, and Andrographis paniculata. Sci World J. 2014;2014(1):635240.
Crossref - Arulkumar A, Rosemary T, Paramasivam S, Rajendran RB. Phytochemical composition, in vitro antioxidant, antibacterial potential and GC-MS analysis of red seaweeds (Gracilaria corticata and Gracilaria edulis) from Palk Bay, India. Biocatal Agric Biotechnol. 2018;15:63-71.
Crossref - Seerangaraj V, Suruli K, Vijayakumar U, et al. Isolation and characterization of bioactive compounds for Bacillus cereus and Bacillus subtilis from Oreochromis mossambicus and Labeorohita. Int J Pharm Sci Rev Res. 2017;43(2):71-77.
- Naqvi SF, Javaid A, Khan IH. Fungicidal activity of stem extract of Chenopodium murale L. against the pathogen of Fusarium wilt of tomato. Allelopathy Journal. 2023;59(1):69-80.
Crossref - Rajaofera MJN, Wang Y, Dahar GY, et al. Volatile organic compounds of Bacillus atrophaeus HAB-5 inhibit the growth of Colletotrichum gloeosporioides. Pestic Biochem Physiol. 2019;156:170-176.
Crossref - Nchabeleng MM, Fonkui TY, Ezekiel G. The Indiscriminate Chemical Makeup of Secondary Metabolites Derived from Endophytes Harvested from Aloe barbadensis Miller in South Africa’s Limpopo Region. Molecules. 2024;29(6):1297.
Crossref - Ajijah N, Fiodor A, Kazimierczuk K, et al. Pseudomonas protegens ML15 and Trichoderma koningiopsis Tr21 Co-Culture:A potent strategy for Suppressing Fusarium cerealisinfections in wheat through Augmented antifungal Metabolite production. Biol Control. 2024;198:105621.
Crossref - Khan MS, Yusufzai SK, Kaun LP, Shah MD, Idris R. Chemical composition and antioxidant activity of essential oil of leaves and flowers of Alternanthera sessilis red from Sabah. J Appl Pharm Sci. 2016;6(12):157-161.
Crossref - VeilumuthuP, Christopher JG. Characterization of Secondary Metabolites Derived from Tomato Endophyte-Streptomyces sp. Shanivit. Curr Trends Biotechnol Pharm. 2022;16(Suppl 1):141-152.
Crossref - Maharani SV, Shinde NS, Wani PV, Patil AS. Antifungal appraisal of Burkholderia gladioli Strain VIMP03 (JQ867372) against Ceratocystis paradoxa. Asian J Pharm Clin Res. 2017;10(3):221-226.
Crossref - Goh JXH, Tan LTH, Law JWF, et al. Streptomyces sp. MUM 195J: A Promising Probiotic for Controlling Vibrio parahaemolyticus Infection in Aquaculture. Prog Microbes Mol Biol. 2024;7(1):a0000443.
Crossref - Alrumman SA, Mostafa YS, Al-Qahtani STS, Sahlabji T, Taha TH. Antimicrobial activity and GC-MS analysis of bioactive constituents of Thermophilic bacteria isolated from Saudi hot springs. Arab JSci Eng. 2019;44(1):75-85.
Crossref - Dheepa R, Vinodkumar S, Renukadevi P, Nakkeeran S. Phenotypic and molecular characterization of chrysanthemum white rust pathogen Puccinia horiana (Henn) and the effect of liquid based formulation of Bacillus spp. for the management of chrysanthemum white rust under protected cultivation. Biol Control. 2016;103:172-186.
Crossref - Sharma N, Koul M, Joshi NC, Dufossé L, Mishra A. Fungal-bacterial combinations in plant health under stress: physiological and biochemical characteristics of the filamentous fungus Serendipita indica and the Actinobacterium Zhihengliuella sp. ISTPL4 under in vitro arsenic stress. Microorganisms. 2024;12(2):405.
Crossref - Rammali S, Hilali L, Dari K, et al. Antimicrobial and antioxidant activities of Streptomyces species from soils of three different cold sites in the Fez-Meknes region Morocco. Sci Rep. 2022;12(1):17233.
Crossref - Sun G, Yao T, Feng C, Chen L, Li J, Wang L. Identification and biocontrol potential of antagonistic bacteria strains against Sclerotinia sclerotiorum and their growth-promoting effects on Brassica napus. Biol Control. 2017;104:35-43.
Crossref - Ansary WR, Prince FRK, Haque E, et al. Endophytic Bacillus spp. from medicinal plants inhibit mycelial growth of Sclerotinia sclerotiorum and promote plant growth. Z Naturforsch C. 2018;73(5-6):247-256.
Crossref - Hu J, Dong B, Wang D, Meng H, Li X, Zhou H. Genomic and metabolic features of Bacillus cereus, inhibiting the growth of Sclerotinia sclerotiorum by synthesizing secondary metabolites. Arch Microbiol. 2023;205(1):8.
Crossref - Xu W, Wang K, Wang H, et al. Evaluation of the biocontrol potential of Bacillus sp. WB against Fusarium oxysporum f. sp. niveum. Biol Control. 2020;147:104288.
Crossref - Saechow S, Thammasittirong A, Kittakoop P, Prachya S, Thammasittirong SNR. Antagonistic activity against dirty panicle rice fungal pathogens and plant growth- promoting activity of Bacillus amyloliquefaciens BAS23. J Microbiol Biotechnol. 2018;28(9):1527-1535.
Crossref - Gajbhiye A, Rai AR, Meshram SU, Dongre AB. Isolation, evaluation and characterization of Bacillus subtilis from cotton rhizospheric soil with biocontrol activity against Fusarium oxysporum. World J Microbiol Biotechnol. 2010;26(7):1187-1194.
Crossref - Wei J, Zhao J, Suo M, Wu H, Zhao M, Yang H. Biocontrol mechanisms of Bacillus velezensis against Fusarium oxysporum from Panax ginseng. Biol Control. 2023;182:105222.
Crossref - Cao Y, Pi H, Chandrangsu P, et al. Antagonism of two plant-growth promoting Bacillus velezensis isolates against Ralstonia solanacearum and Fusarium oxysporum. Sci Rep. 2018;8(1):4360.
Crossref - Sreenivasulu B, Paramageetham C, Sreenivasulu D, Suman B, Umamahesh K, Babu GP. Analysis of chemical signatures of alkaliphiles using fatty acid methyl ester analysis. J Pharm Bioallied Sci. 2017;9(2):106-114.
Crossref - Chen JH, Xiang W, Cao KX, et al. Characterization of volatile organic compounds emitted from endophytic Burkholderia cenocepacia ETR-B22 by SPME-GC-MS and their inhibitory activity against various plant fungal pathogens. Molecules. 2020;25(17):3765.
Crossref - Li W, Long Y, Mo F, et al. Antifungal activity and biocontrol mechanism of Fusicolla violacea J-1 against soft rot in kiwifruit caused by Alternaria alternata. J Fungi. 2021;7(11):937.
Crossref - Jang YW, Jung JY, Lee IK, Kang SY, Yun BS. Nonanoic acid, an antifungal compound from Hibiscus syriacus Ggoma. Mycobiology. 2012;40(2):145-146.
Crossref - Zhang L, Zhang H, Huang Y, Peng J, Xie J, Wang W. Isolation and evaluation of rhizosphere actinomycetes with potential application for biocontrolling Fusarium wilt of banana caused by Fusarium oxysporum f. sp. cubense tropical race 4. Front Microbiol. 2021;12:763038.
Crossref - Agarwal H, Dowarah B, Baruah PM, Bordoloi KS, Krishnatreya DB, Agarwala N. Endophytes from Gnetum gnemon L. can protect seedlings against the infection of phytopathogenic bacterium Ralstonia solanacearum as well as promote plant growth in tomato. Microbiol Res. 2020;238:126503.
Crossref - Prakash J, Arora NK. Novel metabolites from Bacillus safensis and their antifungal property against Alternaria alternata. Antonie Van Leeuwenhoek. 2021;114(8):1245-1258.
Crossref - Wang H, He W, Dong XA, Wang H, Dong F. In situ FT-IR investigation on the reaction mechanism of visible light photocatalytic NO oxidation with defective g-C3N4. Sci Bull. 2018;63(2):117-125.
Crossref - Chandwani S, Dewala S, Chavan SM, Paul D, Kumar K, Amaresan N. Genomic, LC-MS, and FTIR analysis of plant probiotic potential of Bacillus albus for managing Xanthomonas oryzae via different modes of application in rice (Oryza sativa L.). Probiotics and Antimicrobial Proteins. 2024;16(5):1541-1552.
Crossref - Puskarova A, Buekova M, Krakova L, Pangallo D, Kozics K. The antibacterial and antifungal activity of six essential oils and their cyto/genotoxicity to human HEL 12469 cells. Sci Rep. 2017;7(1):8211.
Crossref - Vignesh M, Shankar SRM, MubarakAli D, Hari BNV. A novel rhizospheric bacterium: Bacillus velezensis NKMV-3 as a biocontrol agent against Alternaria leaf blight in tomato. Appl Biochem Biotechnol. 2022;194:1-17.
Crossref - Ongena M, Jacques P. Bacillus lipopeptides: versatile weapons for plant disease biocontrol. Trends Microbiol. 2008;16(3):115-125.
Crossref - Raaijmakers JM, De Bruijn I, Nybroe O, Ongena M. Natural functions of lipopeptides from Bacillus and Pseudomonas: more than surfactants and antibiotics. FEMS Microbiol Rev. 2010;34(6):1037-1062.
Crossref - Mihalache G, Balaes T, Gostin I, Stefan M, Coutte F, Krier F. Lipopeptides produced by Bacillus subtilis as new biocontrol products against fusariosis in ornamental plants. Environ Sci Pollut Res. 2018;25:29784-29793.
Crossref - Hazarika DJ, Goswami G, Gautom T, et al. Lipopeptide mediated biocontrol activity of endophytic Bacillus subtilis against fungal phytopathogens. BMC Microbiol. 2019;19(1):1-13.
Crossref - Yanez-Mendizabal V, Falconi CE. Efficacy of Bacillus spp. to biocontrol of anthracnose and enhance plant growth on Andean lupin seeds by lipopeptide production. Biol Control. 2018;122:67-75.
Crossref - Hu X, Roberts DP, Xie L, et al. Formulations of Bacillus subtilis BY-2 suppress Sclerotinia sclerotiorum on oilseed rape in the field. Biol Control. 2014;70:54-64.
Crossref - Vinodkumar S, Nakkeeran S, Renukadevi P, Malathi VG. Biocontrol potentials of antimicrobial peptide producing Bacillus species: multifaceted antagonists for the management of stem rot of carnation caused by Sclerotinia sclerotiorum. Front Microbiol. 2017;8:446.
Crossref - Schallmey M, Singh A, Ward OP. Developments in the use of Bacillus species for industrial production. Can J Microbiol. 2004;50(1):1-7.
Crossref - Riddech N, Sritongon K, Phibunwatthanawong T. Production of plant growth promoting antagonistic Rhizobacteria to promote cucumber growth and control leaf spot disease (Corynespora cassiicola). Chiang Mai J Sci. 2017;44(1):72-82.
- Compant S, Duffy B, Nowak J, Clement C, Barka EA. Use of plant growth-promoting bacteria for biocontrol of plant diseases:Principles, mechanisms of action, and future prospects. Appl Environ Microbiol. 2005;71(9):4951-4959.
Crossref - Ramanujam B, Honnur Basha HB, Vinaya Hemannavar VH, Chowdappa P, Rangeshwaran R. Induction of defense related enzymes and phenols in chilli plants by Bacillus subtilis against anthracnose pathogen, Colletotrichum capsici. Indian Phytopathol. 2012; 65(4): 382-385.
- Ashwini N, Srividya S. Potentiality of Bacillus subtilis as biocontrol agent for management of anthracnose disease of chilli caused by Colletotrichum gloeosporioides OGC1. 3 Biotech. 2014; 4:127-136.
Crossref - Srikhong P, Lertmongkonthum K, Sowanpreecha R, Rerngsamran P. Bacillus sp. strain M10 as a potential biocontrol agent protecting chili pepper and tomato fruits from anthracnose disease caused by Colletotrichum capsici. BioControl. 2018; 63(6): 833–842.
Crossref - Saikia R, Gogoi DK, Mazumder S, Yadav A, Sarma RK, Bora TC, Gogoi BK. Brevibacillus laterosporus strain BPM3, a potential biocontrol agent isolated from a natural hot water spring of Assam, India. Microbiol. Res. 2011; 166(3): 216-225.
Crossref
© The Author(s) 2025. Open Access. This article is distributed under the terms of the Creative Commons Attribution 4.0 International License which permits unrestricted use, sharing, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.