ISSN: 0973-7510
E-ISSN: 2581-690X
Scrub typhus (ST) is a re-emerging zoonotic disease caused by Orientia tsutsugamushi and is transmitted by chiggers. Serological tests targeting IgM and/or IgG antibodies play a major role in the detection of ST cases. Orientia 56kDa genome is common target for the molecular diagosis of ST to identify the prevalence of specific serotypes of O. tsutsugamushi in and around Puducherry by targeting 56kDa gene with the application of phylogenetic analysis. This prospective laboratory-based study was conducted in a tertiary care teaching hospital, from November 2105 to March 2018. Blood samples were collected from out-patients/in-patients, and those tested positive for ST IgM ELISA (n=140) and an equal number of negative samples were archived and anonymized. Genomic DNA was extracted and analyzed by using Nested PCR and LAMP assay. The positive products were purified and sequenced. Phylogenetic tree was constructed based on the sequences. Among 280 samples, 45 (16.1%) N-PCR and 102 LAMP (36.43%) positivity was observed for 56kDa gene. Forty-one N-PCR positive products were sequenced and accession numbers were obtained (MG601875 to MG601917) for the isolates. Phylogenetic analysis was done by Maximum Likelihood methods and this study has showed that 32.3% are similar to the Karp isolates. Molecular diagnosis of Scrub typhus has become essential in case of doubtful serology and early acute phase of illness. Gene sequencing result indicates that most of them were different from the existing ones, which may belong to the newer strains. The identification of newer strains will be helpful in future for development of scrub typhus vaccine.
Scrub Typhusin Puducherry, Orientia tsutsugamushi, LAMP Assay, Eschar, Nested PCR
Scrub typhus (ST) is re-emerging infectious zoonotic disease caused by an obligate intracellular gram-negative bacterium Orientia tsutsugamushi, which occur globally.1-4 The infection is mostly caused by the chiggers’ larvae breed on human’s blood in contrast to the adults on plants. Children may get affected accidentally by walking with bare foot and other activities like sitting or lying on the grass vegetation where the larval mite breeds. Patient’s symptoms start from 6-21 days after mite bite.1 Other non-specific findings like fever, headache, rash, lymphadenopathy etc. can be observed in different febrile conditions such as Dengue, Malaria, Leptospirosis, and other hemorrhagic fevers. Rapid Immunochromatographic Test (RICT), ELISA can be used for routine diagnosis and IFA needs to be imported and more technical expertise and cost affordable for the patients.1-2 Molecular methods play a vital role in the early diagnosis of ST and other rickettsial infections to confirm the disease.4-6 Orientia 56kDa is the most common target for serology as well as molecular diagnosis.7-10 More number of ST cases are missed due to the late appearance of antibodies in the serological tests.1 This can be overcome by new diagnostic assays viz., PCR, Nested PCR (N-PCR), Real Time PCR (qPCR) and Loop-Mediated Isothermal Amplification (LAMP) which helps in the early detection of O. tsutsugamushi DNA in blood samples.3-18 LAMP is one of the additional molecular methods for the rickettsial diagnosis and easy to perform, with four to six primers in isothermal temperature and it does not require any costly instruments. To the best of our knowledge, only a few reports are available in world database for the diagnosis of ST using LAMP.3-5 Hence, ST N- PCR and LAMP were employed in this study by targeting 56kDa, to identify the prevalent genotypes circulating in and around Puducherry by using Phylogenetic analysis.
Study Participants
The Out-patients/In-patients attending Mahatma Gandhi Medical College and Research Institute(MGMCRI), Puducherry during the time period from November 2015 to March 2018 were included in the study. The study was conducted with the approval from Institutional Human Ethical Committee (IHEC), (IHEC/24/2020) MGMCRI, Puducherry. After getting consent from the patients, the blood specimens were collected from suspected febrile patients from General Medicine and Pediatrics were analyzed for ST IgM ELISA at Department of Microbiology and the molecular work at Centre for Interdisciplinary Research Facility (CIDRF), MGMCRI Campus, Puducherry.
Molecular diagnosis of Scrub Typhus
Buffy coat preparation
About 2 ml of whole blood was collected in a tube containing Ethylenediaminetetraacetic acid (K2EDTA) anti-coagulant. This was transferred into 15ml falcon tubes and 4ml of phosphate buffered saline (PBS) and inverted 2 to 3 times. Blood sample was diluted by adding 1ml of HiSep LSM to the new falcon tube and centrifuged at 1500 rpm for 30 minutes at 15-25°C. Transfer the upper layer of plasma into a separate Eppendorf tube. Collected the white layer (Buffy coat) in a separate falcon tube and added 3 more volumes of PBS. Centrifuged at 260g for 10 mins and without any brake. Remove the supernatant and finally add 500µl of PBS immediately transferred to 1.5ml Micro centrifuge tubes and stored at -80°C for the extraction of genomic DNA. 19 About 200µl buffy coat samples were used for the genomic DNA extractions as per the procedure of DNA Blood Mini Kit (Qiagen, Germany). The purity of the extracted DNA was determined by absorbance (A) using Eppendorf Spectrophotometer. A260/A280 ratios were in the range of 1.7-1.8 for the 280 samples. The samples were aliquoted and stored in 0.2ml PCR tubes at 80°C till the completion of the molecular tests.
Nested PCR
N-PCR outer and inner primer sets targeting 56kDa gene and cycling conditions were followed as described by Furuya et al., 20 in25.0µlreaction volume. PCR was carried out in Applied Biosystem, VeritiDx 96-wells Thermal cycler (Thermo Fisher Scientific, Singapore) and post-amplification, the amplicon with specific product size of 483-bp was visualized by 1.5 % agarose gel electrophoresis and documented by Gel Documentation system (Bio-Rad, USA).
Loop-Mediated Isothermal Amplification (LAMP)
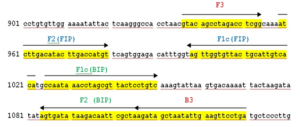
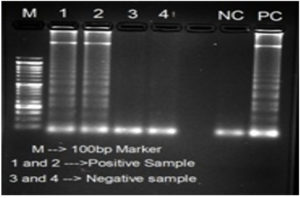
The LAMP primers (forward inner primer (FIP), backward inner primer (BIP), forward outer primer (F3) and backward outer primer (B3)) targeting 56kDa gene were designed from a highly conserved region of O. tsutsugamushi Hualien – 5 strain (Accession no. AY243357.1) using the Primer Explorer version 4.0 http://loopamp.eiken.co.jp/e/:,is using for screening specimens. The location and the sequences of the primers were shown in Figure 1 and Table represents the custom synthesized LAMP primers by Sigma Aldrich, Bangalore. LAMP Reaction was conducted in 25µl of reaction volume comprising of 7.5µlNuclease free water, 3.5µl 1.4mM DNTPs 1.0µl of each 40µM of Forward and Reverse Innerprimersand1.0µl of each 5µM of Forward and Reverse Outer Primers,3.5µl of 5M Betaine, 2.5µl10X Isothermal Amplification Buffer II, 1.5µl of 8mM MgSo4, 1.0µlBst 3.0 DNA Polymerase (8000 Units/ml), 0.5µl 3mM HydroxyNaphthol blue and 1.0µl genomic DNA. After amplification, primarily visual observation of the reaction was done and interpretation was made based on the color change from Sky blue to purple for positive and negative reactions, respectively (Figure 2). Further confirmation was made by loading the LAMP amplicon in 1.5% agarose gel electrophoresis stained with Ethidium bromide solution for the presence of ladder like pattern in positive samples (Figure 3).
Table:
Details of LAMP primers used for 56kDa gene targets
Primer |
5′ Sequence 3′ |
Positions |
|---|---|---|
F3 |
GTACAGCCTAGACCTCGG |
937-954 |
B3 |
TCAGGAACTTCAATATTAGCTATC |
1107-1130 |
FIP |
TGTGACAATGCAGTAACACCAACTTTTTATCTTGACATACTTGACCATGT |
959-980 /999-1022 |
BIP |
CCAATAAACCTAGCGTTACTCCTGTTTTTTAGCGAATTGTCTTATATCACT |
1084-1106/1025-1048 |
Note: F3 & B3 – Forward and Backward primers; FIP & BIP – Forward and Backward Inner Primers
Figure 1. Location and sequences of the LAMP primers for 56kDa gene
Note: Location of outer (B3 +F3) and inner primers (FIP+BIP) for Orientia tsutsugamushi using Hualien – 5 strain (GenBank Accession no. AY243357.1). Forward inner primer and backward inner primers contain two separate sequences (F1c + F2 and B1c + B2, respectively)
Figure 2. Image for Visual Detection of LAMP products: Upper row: 1 & 2 – DNA Samples positive for 56kDa gene target for O. tsutsugamushi, 3 and 4 – Sample negative for O. tsutsugamushi DNA; Lower row: NC – Negative control, PC – Positive control
Figure 3. Analysis of LAMP products in agarose gel electrophoresis: M – 100bp DNA ladder; Samples 1 & 2 – Positive and Samples 3 & 4 – Negative; NC – Negative Control and PC – Positive control
Phylogenetic analysis
Both N-PCR and LAMP positive products were purified by Qiagen DNA Purification kit and the products were sequenced by Applied Biosystems ABI3730XL (Eurofins Genomics India Private Limited, Whitefield, Bangalore) for specificity of the amplicon. The sequences obtained from the study was submitted and accession numbers were procured from the GenBank database (National Centre for Biotechnology Information –NCBI). The sequences were aligned using Clustal W and the Evolutionary analysis was constructed using of MEGA version 11.0.8 The 56-kDa TSA gene of O. tsutsugamushi isolates were aligned with reference sequences with a total of 68 nucleotide sequences. One thousand bootstrap value replicates were analyzed to estimate the reliability of the node. The evolutionary tree was inferred by using the Maximum Likelihood method and Kimura 2-parameter model. Initial tree(s) for the heuristic search were obtained automatically by applying Neighbor-Join and BioNJ algorithms to a matrix of pairwise distances estimated using the Maximum Composite Likelihood (MCL) approach, and then selecting the topology with superior log likelihood value. Bootstrap values more than 75% were considered to be significant.
Statistical analysis
Percentages were calculated for categorical variables. Mean and standard deviations (SD) were calculated with 95% confidence interval for numerical variables. Sensitivity, specificity, positive and negative predictive value was calculated using Med Calc’s online software. Chi-square test was applied for numerical variables and Fisher’s exact test for small number samples. Statistical tests were analyzed by Quick Calcs, GraphPad Prism. P values ≤ 0.05 were considered statistically significant.
Out of 280 participants screened, 170 (60.71%) were male who were between the age of 9 to 82 years and 110 (39.29%)were females aged from one year to 80 years. Mean ± SD age for the patients with 95% confidence interval has been recorded as 32.94 ± 21.29 (30.04 – 35.82) and voluntary blood donors as 29.78 ± 6.47 (28.25 – 31.31). The patients’ duration of illness was recorded as 3–15 days (Mean duration ±SD,8.8 ± 2.7 days [8.4 – 9.2]). Totally, 280 samples screened by N-PCR and LAMP, 45 (16.1%) and 102 (36.43%) were positive for 56kDa gene. About 10 representative LAMP positive samples were confirmed for specificity by sequencing the amplified products. Diagnostic evaluation of LAMP was calculated using N-PCR is kept as reference standard. Sensitivity, specificity, PPV & NPV with confidence interval are 74.0% (58.0-85.6%), 70.6% (64.3-76.3%), 32.3% (26.8-38.4%) and 93.2% (89.4-95.7%) respectively. Forty-one N-PCR and two LAMP positive products of 56kDa, of O. tsutsugamushi isolates were sequenced and submitted to GenBank (NCBI) for accession numbers. Nine strains viz., MG601895.1, MG601913.1, MG601885.1, MG601911.1, MG601880.1, MG601899.1, MG601905.1, MG601881.1, MG601892.1 were closely related with 85% homology of Taiwan and Haryana strains. One strain MG601906.1 had similar sequences with Odisha, AIIMS and Japan.
Ten isolates from the present study, MG601891.1, MG601896.1, MG601893.1 and MG601901.1, MG601917.1, MG601915.1, MG601914.1, MG601916.1, MG601876.1 and MG601878.1were highly 92-97% homology with Karp and Boryong strains from CMC Vellore, Gorakhpur, Mizoram, Korean and Japan, Taiwan, New Guinea, Vietnam, AIIMS New Delhi, Puducherry and Karnataka.
Three study sequences MG601890.1, MG601898.1, MG601886.1 are 99% homology with the Kawasaki reference sequences from Odisha, PGIMER, Japan and China. In the present study, four sequences MG601912.1, MG601889.1, MG601879.1, MG601883.1 were 98% homology with Taiwan and Chennai strains. Another seven study sequences MG601897.1, MG601903.1, MG601909.1, MG601910.1, MG601904.1, MG601882.1, MG601887.1 are closely clustered with four Gilliam reference strains from Japan and China. Two study sequences MG601875.1 and MG601877.1 were 99% homology with four reference strains from CMC Vellore, IGMC Shimla and Shillong.
Remaining three reference isolates from Japan and CMC Vellore were showed 94-98% homology with the six study sequences MG601917.1, MG601915.1, MG601876.1, MG601914.1, MG601878.1, MG601916.1. Phylogenetic tree was constructed by Maximum Likelihood method based on 56kDa sequences of O.tsutsugamushi and presented in the Figure 4. In this study, only 12 sequences had significant bootstrap value more than 90% and remaining 29 sequences are less than 75% which are non-significant. Geographical locations of participants reporting to hospital at Puducherry and flow of study procedures were shown in Figure 5.
Figure 4. Geographical locations of participants reporting to MGMCRI, Puducherry and flow of study procedures. The image depicts the geographical locations of patients reporting to MGMCRI, Puducherry. The map includes number of cases reported from each location
Molecular diagnosis of Scrub typhus has become essential in case of doubtful serology and early acute phase of illness. Presence of O. tsutsugamushi DNA could be demonstrated either in the Buffy coat or eschar biopsy samples. Conventional PCR, Nested PCR and Real Time PCR are being regularly used for this accurate detection.3 Recently, LAMP assay has been introduced as an additional molecular test.4 The LAMP primers were newly designed in this study and further used for screening the specimens.
According to Kala et al.,21 56 kDatsa gene is mostly used for the characterization of the O. tsutsugamushi strains.
Paris et al., reported 83% sensitivity and 88% specificity for ST LAMP which is quite higher when compared to the present study. Usually, LAMP yields more sensitivity due to its specific primer design and incorporation of six primers including loop primers in a single reaction tube.22
In the present study, the sequences from the accession numbers MG601895.1, MG601913.1, MG601885.1 were similar with Taiwan strain MW495789.1, MG601911.1, MG601880.1, MG601899.1, MG601905.1, MG601881.1, MG601892.1 were closely related with 85% homology of two more strains (ON208816.1 & ON208820.1) from Artemis Hospitals, Gurugram, Haryana. One strain MG601906.1 had similar sequences with Odisha (MK650206.1), AIIMS (MT452573.1) and Japan (AB751258.1)
Four isolates from the present study, MG601891.1, MG601896.1, MG601893.1 and MG601901.1 were highly similar with six Karp strains from Korean and Japan. Two more Boryong strains from Japan were closely clustered with above study sequences. One strain from CMC Vellore (OP068209.1) formed sub lineages with two strains from Gorakhpur (MF197390.1 & MF197397.1) and Mizoram were isolated from the mite population. Seven Karp strains AY222639.1, AY956315.1, OQ055155.1, GU128876.1, KY319228.1, KT970968.1, MH570187.1 from Taiwan, New Guinea Mizoram, Vietnam, AIIMS New Delhi, Puducherry and Karnataka have separate clusters which are in ranges from 92%-97% closely related with following six study sequences viz., MG601917.1, MG601915.1, MG601914.1, MG601916.1, MG601876.1 and MG601878.1.
Three study sequences MG601890.1, MG601898.1, MG601886.1 are 99% homology with the Kawasaki reference sequences from Odisha (MK650206.1), PGIMER (OM775457.1 &OM775459.1), Japan (AF173039.1 & M63383.1) and China (KC456660.1). Four study sequences MG601912.1, MG601889.1, MG601879.1, MG601883.1 were 98% homology with Taiwan (AY222631.1) and Chennai (MH003840.1) strains. Another seven study sequences MG601897.1, MG601903.1, MG601909.1, MG601910.1, MG601904.1, MG601882.1, MG601887.1 are closely clustered with four Gilliam reference strains from Japan (AF173035.1 & AF173033.1) and China (U19903.1 & (AF050669.1). Two study sequences MG601875.1 and MG601877.1 were 99% homology with four reference strains from CMC Vellore (KC153085.1), IGMC Shimla (KF777305 & KF777299) and Shillong (KF777329.1).
Remaining three reference isolates from Japan (AF173041.1 & (M63382.1) and CMC Vellore (KC153082.1) showed 94-98% homology with the study sequences MG601917.1, MG601915.1, MG601876.1, MG601914.1, MG601878.1, MG601916.1. Only one reference sequence from Himachal Pradesh (DQ530440.1) was showed unique when compared to the study references.
The study sequences have 98-100% of maximum identity with the other reference strains available in the NCBI database. In the present study, most of our 56kDa isolates are closely related to Karp and Gilliam, followed by Boryong and some strains are closely related to Kawasaki strains. In a total of 42 patients, 55% of sequences were related to the Karp serotype followed by 17%, were Gilliam type in Japan variant respectively. One of the studies from Pondicherry also proved that most of the sequences were matched with the Karp group strains.11 A study was conducted by Patricia et al., 11 in Puducherry comprised that 15 of their sequences were correlated with the Karp group and another 6 from Vellore strains, respectively. They also indicated that 12 of their sequences form a separate group and did not match with the other genotypes that were included as a reference strain. Phylogenetic analysis of 16 sequences from Sri Lanka was related with Karp, Kato, JGV and JGA (Kawasaki group), Gilliam, Kuroki- Boryong genotypes. Karp is one of the major dominant genotypes of O. tsutsugamushi varying from 40-64.4% and it was reported in many countries. Regarding the genotypes of O. tsutsugamushi circulating in India, Patricia et al.,11 from Puducherry reported positivity of 42.9% for Karp and 17.1% for Kato. Usha et al.,10 from Andhra Pradesh also had a higher prevalence of Karp strains (70.4%) followed by Kawasaki (29.6%). Varghese et al., 13 from Tamil Nadu and other parts of India observed that they have Kato as the majority strain with 61.5% followed by Karp (27.1%), Neimang (8.7%) and Ikeda (2.3%). Recently, Thakur et al., was recorded that majority of the circulating strains of O. tsutsugamushi in New Delhi were close to Gilliam and Karp.20 According to Biswal et al.,14 from Chandigarh, Boryong occupied the highest prevalence with 63.4% followed by Karp (23.6%), Gilliam (11.8%) and Kawasaki (1.2%). A similar observation of highest positivity of Boryong genotype was recently reported by Ramaiah et al.,16 from Manipal, Karnataka. The present study had recorded 32.29% moderate positivity for Karp like strains and 17.7% are grouped with a Kawasaki strains. Most of our isolates (50.01%) have highest homology with that of the strains reported from India, Korea, Japan and China. Extension of the research work to cover a wider geographical area in regions surrounding Puducherry might help in identifying additional genotypes of O. tsutsugamushi. Development of a Multiplex PCR targeting all three genes would help in the rapid confirmation for the diagnosis of ST. The present study concluded that the study sequences have 98-100% maximum identity with the other reference strains available in the GenBank database. Gene sequencing result indicates that most of the study sequences were related to those who submitted from India as well as overseas. Few of our study sequences were different from the existing ones, which may belong to the newer strain. Hence, this newer sequence may further be identified and it will be helpful in future for development of scrub typhus vaccine.
ACKNOWLEDGMENTS
The authors would like to thank Chancellor, Vice-Chancellor, Dean Research, Vice President (Research, Innovation and Development) Sri Balaji Vidyapeeth (Deemed to be University) and Research scholars, Mahatma Gandhi Medical Advanced Research Institute (MGMARI) for providing the facilities for the work.
CONFLICT OF INTEREST
The authors declare that there is no conflict of interest.
AUTHORS’ CONTRIBUTION
AR, JP and SS conceptualized the study. AR and JP carried out the molecular work and collected the dataset for the study. AS constructed Phylogenetic tree analysis for the study sequences. AR, JP and SS drafted the manuscript. SS and PP approved the manuscript for publication.
FUNDING
None.
DATA AVAILABILITY
All datasets generated or analyzed during this study are included in the manuscript.
ETHICS STATEMENT
This study was approved by the Institutional Human Ethics Committee (IHEC), Mahatma Gandhi Medical College & Research Institute, Pillaiyarkuppam, Pondicherry, India (IHEC, Project Number 2015/10/01-dated 30th November 2015).
INFORMED CONSENT
Written informed consent was obtained from the participants before enrolling in the study.
- Rahi M, Gupte MD, Bhargava A, Varghese GM, Arora R. DHR-ICMR guidelines for diagnosis & management of Rickettsial diseases in India. Indian J Med Res. 2015;141(4):417-22.
Crossref - Anitharaj V, Stephen S, Pradeep J, et al. Serological Diagnosis of Acute Scrub Typhus in Southern India: Evaluation of InBios Scrub Typhus Detect IgM Rapid Test and Comparison with other Serological Tests. J ClinDiagn Res. 2016;10(11):DC07-DC10.
Crossref - Kim DM, Park G, Kim HS, et al. Comparison of Conventional, Nested, and Real-Time Quantitative PCR for Diagnosis of Scrub typhus. J Clin Microbiol. 2011;49(2):607-12.
Crossref - Huber E, Ji D, Howell L, et al. Loop-mediated isothermal amplification assay targeting the 47-kDa gene of Orientia tsutsugamushi: A rapid and sensitive alternative to Real-Time PCR. J Med Microb Diagn. 2012;1(4):1-4.
Crossref - Notomi T, Okayama H, Masubuchi H, Yonekawa T, Watanabe K, Amino N, Hase T. Loop-mediated isothermal amplification of DNA. Nucleic Acids Res. 2000;28(12):E63.
- Paris DH, Blacksell SD, Nawtaisong P, et al. Diagnostic Accuracy of a Loop-Mediated Isothermal PCR Assay for Detection of Orientia tsutsugamushi during Acute Scrub Typhus Infection. PLoS Negl Trop Dis. 2011;5(9):e1307.
Crossref - Thapa S, Hamal P, Chaudhary NK, Sapkota LB, Singh JP. Burden of scrub typhus among patients with acute febrile illness attending tertiary care hospital in Chitwan, Nepal. BMJ Open. 2020;10(9):e034727.
Crossref - Kumar S, Stecher G, Li M, Knyaz C, Tamura K. MEGA X: Molecular Evolutionary Genetics Analysis across computing platforms. Mol Biol Evol. 2018;35(6):1547-1549.
Crossref - Nguyen HLK, Pham HTT, Nguyen TV, et al. The genotypes of Orientia tsutsugamushi, identified in scrub typhus patients in northern Vietnam. Trans R Soc Trop Med Hyg. 2017;111(3):137-139.
Crossref - Usha K, Kumar E, Kalawat U, Kumar BS, Chaudhury A, Gopal DVRS. Molecular characterization of Orientia tsutsugamushi serotypes causing scrub typhus outbreak in southern region of Andhra Pradesh, India. Indian J Med Res. 2016;144(4):597-603.
- Patricia KA, Hoti SL, Kanungo R, Jambulingam P, Nazeer Y Shashikala N, Naik A, Mookappan S. Occurrence of Orientia tsutsugamushi Genotypes in Areas of UnionTerritory of Puducherry and Tamil Nadu State, India. J Infect Dis Pathol. 2017;2:1000124.
- Kumar GV, Beena PM. Molecular Characterization of Orientia tsutsugamushi in Domestic Rodents in Kolar Region, Karnataka. Asian J Epidemiol. 2017;10(3):123-127.
Crossref - Varghese GM, Janardhanan J, Mahajan SK, et al. Molecular epidemiology and genetic diversity of Orientia tsutsugamushi from patients with Scrub typhus in 3 regions of India. Emerg Infect Dis. 2015;21(1):64-69.
Crossref - Biswal M, Kumar A, Sharma N, Bhalla A, Singh S, Sethi S. Genetic diversity of Orientia tsutsugamushi strains from patients in North India. Int J Infect Dis. 2016;45(Suppl 1):166-167.
Crossref - Koraluru M, Bairy I, Singh R, Varma M, Stenos J. Molecular confirmation of scrub typhus infection and characterization of Orientia tsutsugamushi genotype from Karnataka, India. J Vector Borne Dis. 2016;53(2):185-187.
- Ramaiah A, Koraluru MC, Dasch GA. Complexity of type-specific 56 kDa antigen CD4 T-cell epitopes of Orientia tsutsugamushi strains causing scrub typhus in India. PLoS ONE. 2018;13(4):e0196240.
Crossref - Anitharaj V, Stephen S, Pradeep J, Pooja P, Preethi S. Validation of Geno-Sen’s scrub typhus real time polymerase chain reaction kit by its comparison with a serological ELISA test. J Global Infect Dis. 2017;9(3):108-112.
Crossref - Anitharaj V, Stephen S, Pratheesh P. Scrub typhus in Puducherry, India: Application of nested PCR targeting three different genes – 56 kDa, 47 kDa and groEL of Orientia tsutsugamushi and comparison with ST IgM ELISA. J Vector Borne Dis. 2020;57(2):147-152.
Crossref - Sontyana B, Govatati S, Tamanam RR. A Quick And Easy Method For The Isolation Of Peripheral Blood Mononuclear Cells From Whole Blood By Density Gradient Centrifugation Using Hisep Lsm Medium. Int. J. Of Adv. Res. 2016;4:1774-1778.
- Furuya Y, Yoshida Y, Katayama T, et al. Specific Amplification of Rickettsia tsutsugamushi DNA from Clinical Specimens by Polymerase Chain Reaction. J Clin Microbiol. 1991;24(11):2628-2630.
Crossref - Kala D, Gupta S, Nagraik R, Verma V, Thakur A, Kaushal A. Diagnosis of scrub typhus: recent advancements and challenges. 3 Biotech. 2020;10(9):396.
Crossref - Thakur CK, Chaudhry R, Gupta N, et al. Scrub typhus in patients with acute febrile illness: a 5-year study from India. QJM: An Intl J Medicine. 2020;113(6):404-410.
Crossref
© The Author(s) 2023. Open Access. This article is distributed under the terms of the Creative Commons Attribution 4.0 International License which permits unrestricted use, sharing, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.