This review provides an overview regarding the main aspects of fatal bacterial infections and antibiotics; in recent years, it has been observed that gram-negative bacteria are prevalent in infections owing to a failure to treat the infection and antibiotic resistance. This has led to the phrase “the end of the antibiotic era,” which was revealed in late 2017. This topic has gained momentum among the journalists, specialists, and broadcasters, who have developed immense interest in exploring new approaches and substitutes for antibiotics to treat infections. Several factors contribute to the increasing antibiotic resistance; these can be divided into two main categories, that is, those caused by human behaviors with respect to antibiotic use and those enacted by pathogens as they attempt to protect themselves against antibiotics. Therefore, the main purpose of this review is to discuss and summarize the most important factors and emphasize the measures to tackle drug resistance worldwide. A comprehensive studies were conducted to evaluate the reasons of antimicrobial resistance, studying different factors including bacterial strains (either positive or negative) gram bacteria, antimicrobial agent, in case of negative gram bacteria that’s mean the isolates are not inhibited by the selected antimicrobial agent or by achievable concentrations, the normal dose schedules and / or the diameters of the area in the range, that’s lead to a specific mechanisms of microbial resistance (e.g., beta-lactamase).
Drug Resistance, bacterial infections, beta-lactamase, antibiotics.
Antimicrobial resistance (AMR) or drug resistance occurs when the microbes (fungi, viruses, bacteria, and others) areexposed to antimicrobial agents (antibiotics, antifungals, antivirals, and others); some microbes that develop resistance to the majority of antimicrobial agents are termed “superbugs.” This resistance is increasing worldwide. Thus, several actions may accelerate the emergence and spread of antibiotic-resistant bacteria, such as: misusing of antibiotics, poor infection prevention and control practices, working under unsanitary conditions and mishandling food that lead to spread illness, disability, and death1. About 17 different classes of antibiotics have been produced to date
(Table 1)2.
Table (1):
Major antibiotic classes by mechanism of action.
Mechanism of action |
Antibiotic families |
|---|---|
Inhibition of cell wall synthesis |
Beta-lactams (penicillins, cephalosporins, carbapenems, monobactams); glycopeptides; cyclic lipopeptides (daptomycin) |
Inhibition of protein synthesis |
Tetracyclines; aminoglycosides; oxazolidonones (linezolid); streptogramins (quinupristin-dalfopristin); ketolides; macrolides; lincosamides |
Inhibition of DNA synthesis |
Fluoroquinolones |
Inhibition of RNA synthesis |
Rifampin |
Competitive inhibition of folic acid synthesis Inhibition |
Sulfonamides; trimethoprim |
Membrane disorganizing agents |
Polymyxins (Polymyxin-B, Colistin) |
Other mechanisms |
Metronidazole |
Modified from Levy and Marshall (2004)2
In the absence of antibiotics or antimicrobials for treatment and prevention of microbial diseases, other treatment procedures such as structure transplantation, cancer chemotherapy, diabetes therapy, and surgeries (e.g., caesarean servings or hip replacements) pose a high risk of infection.3 The total cost of healthcare may also increase depending upon drug resistant infections as well as longer stays in the hospitals and consequence more intensive care required.4
The discovery of antibiotics was a crucial moment in the history of mankind as it revolutionized scientific medicine and saved numerous lives. Unfortunately, the infectious strains are becoming resistant to antibiotics, and therefore, health professionals fear the return of the pre-antibiotic era. In this context, the bacterial genome has been examined as resistance strains, and was found that more than 20,000 possible examined bacterial genes have been successfully concluded as resistance strains5.
Antibiotic resistance against the antimicrobials was observed in the late 1950s and 1960s, among the intestinal bacteria, Salmonella sp, Shigella sp, and Escherichia coli.6 During this early period, these resistant strains caused enormous commercial losses and affected the clinical treatments worldwide; however, this phenomenon was presumed to be confined to the intestinal microbes. This misconception was later cleared in the late 1970s, after observing that Neisseria nebulae and Haemophilus influenzae were resistant to ampicillin and serotonin; moreover, few studies described their resistance to tetracycline and chloramphenicol. The increasing use of antimicrobial agents has led to several issues worldwide, specifically in developing countries, where these antibiotics are used without prescriptions. The hygiene settings prevented from transferring the confrontation and the trivial care boxes (T-box) that were manipulated for admission to new and competent antibiotics. T-boxes are structures that recognize when a cell is deficient in a specific amino acid, the building blocks of cells, and they allow bacteria to respond to this deficiency by initiating a process that generates more of that amino acid, including pathogens such as M. tuberculosis and B. anthracis, which causes the deadly anthrax disease6,7.
Klebsiella pneumoniae, is a common intestinal bacteria causing several life-threatening diseases including pneumonia, and its worldwide spread is a major cause of hospital-acquired infections, as well as bloodstream infections, infections in neonates and intensive-care unit patients; this strain is usually resistant to carbapenem antibiotics8. In some countries, due resistance to this strain, the carbapenem antibiotic had no effect in more than half of people treated for pneumonia infection8,9. Furthermore, another example of antibacterial resistance associated with urinary tract infection (UTIs), E. coli strain, a major bacterial species, and showed to be resistant to fluoroquinolone antibiotics (ciprofloxacin or norfloxacin), and Klebsiella pneumoniae is the second most important bacteria in this type of infection, the most common prescribed antibiotics to treat this infection UTIs are sulfamethoxazole, trimethoprim, fluoroquinolones, but on other countries showed that the bacterial resistance to sulfamethoxazole, trimethoprim and ciprofloxacin reached its critical7,9.
In various health facilities and societies, certain microbes are resistant to the first-line drugs that treat infections caused by Staphylococcus aureus, a common cause of severe infections in these areas. In some cases, people infected with S. aureus resistant to methylation (methyl S. aureus) have a ≥64% mortality rate even with the nonresistant forms10.
In general, frequent of uses of antibiotics has several side effects; however, it has been observed that several patients use antibiotics to treat the wrong medical conditions. It is common to use antibiotics to treat respiratory infections caused by viruses such as the cold or flu (influenza), in this case uses of antibiotics had no affect on treat this infection10. The wrong uses of antibiotics lead to a new antibiotic-resistant strains. Based on data cited for to the centers for disease control (CDC), Overuses of antibiotic are a particular problem, and showed to be higher in some regions of the world, as the Southeast. For example, carbapenems, a major class of beta lactam antibiotics, increased significantly from 2007 to 2010. carbapenems a group of broad-spectrum beta-lactam antibiotic agents are three parenteral preparations, used for treatment of severe or high-risk bacterial infections, similar to penicillins and cephalosporins11.
Factors
Human factors
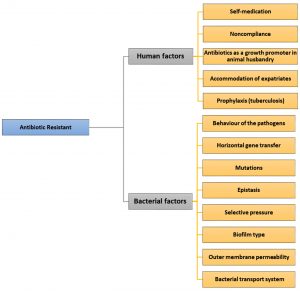
Human factors, caused by human behaviors regarding antibiotic use include: (i) self-medication, (ii) noncompliance of patients, (iii) antibiotic use for growth promotion in animal husbandry, (iv) ongoing transmission due to the lack of functional infection control programs,12 and (v) accommodation of more than ten million expatriates, mainly hailing from endemic areas, for example, tuberculosis (TB) cases from endemic places, for work purposes as well as for religious rituals carried out in Mecca and Medina, Kingdom of Saudi Arabia.
Self-medication
Access to antibiotics is a main concern in several countries worldwide, including Saudi Arabia. The accessibility of antibiotics and community approaches toward the use and misuse of antibiotics have contributed tremendously to the increasing antibiotic resistance13. In several developing countries, including Saudi Arabia, antibiotics can be obtained as a commodity without a prescription from a qualified healthcare professional14. About 90% of upper respiratory infections are caused by viruses; unfortunately, physicians experience tremendous pressure from patients to prescribe antibiotics for these viral infections, as individuals erroneously think that antibiotics can treat such infections; hence, antibiotic prescription for viral infections is observed on a daily basis, despite that the rules and regulations forbid such act15,16.
Noncompliance
Noncompliance arises due to various reasons including missing medication, early cessation of treatment as the patient starts to feel better, and inadequate access to appropriate antibiotics. Other potential reasons for noncompliance are dosage frequency, treatment duration, complexity of the treatment, and its side effects17. In addition, psychological distress can lead to noncompliance, as it has been found that some individuals do not comply because the medication changes the patient’s urine color18. Noncompliance undoubtedly promotes antibiotic resistance, as an unfinished course of antibiotics often wipes out most susceptible bacteria but allows the relatively resistant bacteria to survive and thrive19,20. The infections caused by resistant bacteria resulting from patient noncompliance are difficult to treat, as patients remain sick for a longer period of time, which can necessitate long-term hospitalization and lead to the further spread of resistant bacteria21.
Antibiotics as a growth promoter in animal husbandry
Use of antimicrobial agents as growth promoters is an efficient technique for enhancing the productivity and health of livestock. Unfortunately, the existing use of antibiotics to treat animals and encourage animal growth has led to the emergence and spread of antibiotic resistance. For instance, avoparcin is a growth promoter that is used in the agricultural systems. Avoparcin significantly contributes to the emergence and spread of vancomycin-resistant enterococcal infections in the USA22. Another example is virginiamycin, which is used as an animal feed additive in the agriculture industry. The overuse of virginiamycin can lead to the acquisition of resistance to streptogramins. As a result of streptogramin resistance, the use of virginiamycin has been banned in Denmark and throughout Europe23,24.
A study conducted in 2013 reported that Saudi Arabia has no published records on the use of antibiotics as a growth promoter, although several reports have described the isolation of various multidrug-resistant (MDR) bacteria from animal feed.25 A survey was conducted by Al-Mustafa,26 in which 23 randomly chosen poultry farms and all veterinary pharmacies in the eastern province of Saudi Arabia were identified and 29 antimicrobial agents were accessible for poultry use. Of the 29 agents, 22 antibiotics (75.9%) are vital for treating human infections. These antibiotics include ampicillin, neomycin, colistin, doxycycline, enrofloxacin, oxytetracycline, sulfamethoxazole, and erythromycin.27 The prevalence of resistance to these antibiotics is unknown due to lack of data from both human and animal subjects28.
Hospital-acquired infections (HAI)
These infections have been documented for more than a century as a serious problem disrupting excellence in healthcare29. HAI are a major source of adverse healthcare outcomes. Hospitals, primarily intensive care units, are a significant breeding ground for the emergence and spread of antibiotic-resistant bacteria, as contact between the patients and hospital staff can generate risk for cross-infection30,31. Despite various efforts considering the control of antibiotic-resistant nosocomial infections, only slight indication of better pathogen control within healthcare services has been observed in most countries, including Saudi Arabia32. To support patients with tuberculosis (TB), mainly multidrug-resistant tuberculosis (MDR-TB), patients are often hospitalized for prolonged time period33. During long-term hospitalization, the patient may be cross-infected by a different strain of TB from another patient. Genotyping data have revealed that patients have indeed been cross-infected by different drug-resistant strains during the treatment period and hospitalizations34. Additional genotyping data have revealed that Saudi Arabia is suffering from ongoing TB transmission, as indicated by high clustering rates of clades circulating in the community35. Moreover, recent sequencing data have confirmed that Saudi Arabia faces tremendous challenges due to the ongoing transmission, which is mainly caused by MDR strains36,37.
Influence of expatriates and pilgrims
Travel is recognized as a risk factor for the spread of various infectious diseases38. Saudi Arabia faces certain challenges, as it accommodates more than 10 million expatriates that primarily descend from the endemic areas, carrying diseases such as tuberculosis (TB). In addition, as Saudi Arabia hosts two holy mosques, and also welcomes an additional 10 million individuals who visit annually for religious Islamic rituals39. Those 10 million individuals stay at specific places at are in close vicinity to each other. Such conditions are ideal for the transmission and exchange of infectious diseases. Recent data have revealed that some travelers returning from the Hajj acquired New Delhi metallo-beta-lactamase-1-producing Escherichia coli and MDR Acinetobacter baumannii during the Hajj pilgrimage40. In addition, data collected from two main hospitals in Mecca revealed that ceftazidime resistance was evident in 52.7% of Pseudomonas aeruginosa, 34.4% of K. pneumoniae, and 24.6% of E. coli isolates tested. Furthermore, another report indicated that the number of septicemia incidents in Mecca increases by 16.5% during the Hajj season due to the arrival of infected individuals from various countries25,41.
Prophylaxis
Antibiotic prophylaxis can be used successfully to avoid infection in individuals with risk of bacterial infection due to other medical issues, such as recurrent cellulitis, meningococcal disease, spontaneous bacterial peritonitis associated with cirrhosis, infectious endocarditis, infections related to open fractures, prosthetic joint implantation, and wounds. Antibiotics may also be prescribed to prevent infection in individuals with destabilized immune systems, such as people receiving chemotherapy, those infected with HIV, and people who travel globally where they are likely to get an infection42,43.
In all these circumstances, an authorized physician must decide whether antibiotics are necessary. Resistance resulting from the overuse of antibiotics, along with associated increases in healthcare costs and toxicity in patients, is eminent unless antibiotic use is restricted. In addition, drugs taken to prevent infection should only be used for a short time42.
Bacterial behaviors influencing drug resistance
One of the determinants for antimicrobial activity is the bacterial status. Under antibiotic exposure, bacterial phenotypes such as susceptibility, tolerance, resistance, and persistence differ.44 These behaviors are enacted by pathogens as they attempt to protect themselves against antibiotics. Several bacteria exhibit natural (i.e., intrinsic) resistance to different types of antibiotics with various degrees45. In addition, bacteria may develop resistance by two extrinsic mechanisms: acquisition of a resistance-encoding gene from another bacterium, which occurs via horizontal gene transfer (HGT) or by genetic mutation. Both of these extrinsic mechanisms enable the bacteria to expose a novel resistance determinant. The ability of bacteria to transfer resistance genes among themselves significantly contributes to the spread of antibiotic resistance46. Bacterial factors include (i) horizontal gene transfer, (ii) physical or chemical mutations, (iii) epistasis, (iv) selective pressures, (v) biofilms, and (vi) outer membrane permeability by bacterial transport systems (Fig. 1).
Horizontal Gene Transfer (HGT)
HGT is known as the transfer of genetic material from one species to another species47. Genetic transfer occurs by plasmids, bacteriophages, and transposons that can move from the chromosomal- and plasmid-associated resistance genes to disparate bacterial hosts. In addition, DNA released from dead bacteria can be obtained and recombined into the bacterial genome, thus emerging in new strains48. Mobile genetic elements, such as plasmids, bacteriophages, genomic islands, and transposons, play a significant role in the progression of various bacteria and enable the spread of genes associated with drug resistance, pathogenicity, and fitness, causing the emergence of “hospital superbugs.”49, 50 Mechanisms of HGT vary widely among the bacteria, and it acts as a key mediator of antibiotic resistance and spread51.
Mutations
The mutation would cause a change in the genomic structure of the bacteria and produce new species and traits, including bacterial resistance to antibiotics. Mutations divided into three types spontaneous (natural), physical or chemical, and genetic manipulation by transferring genes from bacteria to other bacteria using different cloning methods that acquired the modified bacteria (mutant strain) new characteristic over than the wild type such as resistance to antibiotics, salt tolerance, and others. Some mutations allow bacteria to synthesize enzymes that inhibit antibiotics, whereas others may remove the antibiotic targets from the cell, thus inhibiting the antibiotic activity or restricting antibiotic entry into the cell52. When bacterial populations are treated with these specific antibiotics, resistant bacteria will be able to reproduce and can increase the number of bacteria, and the end result is the a group of different bacteria resistant mainly to the specific antibiotic. Mutations caused by antibiotic resistance can have a clinical significant impact on the effectiveness of specific antibiotic groups or specific bacterial pathogens. Mutations can also modify conditions in which the resistance genes are spread. In the long run, mutations may be crucial in the development and diversification of acquired resistance elements52,53.
Epistasis is defined as the interaction between different genes, which is essential in both molecular and quantitative genetics54. In recent years, Epistasis has become a hot topic in complex disease genetics, for complex traits such as diabetes, asthma, hypertension and multiple sclerosis. This is probably due to complicating factors such as an increased number of contributing loci and susceptibility alleles, incomplete penetrance, and contributing environmental effects55. Epistasis is notable phenotypic differences among individuals with the same genotype at one locus depend on their genotypes at another locus56. The progress of epistasis is mainly missing from classical population genetics research57. In the past decade, however, this topic has received immense attention as the data related to the monogenetic systems have become accessible58,functional59, or physiological60.
Epistasis is the effect of one gene (locus), which is reliant on the presence of one or extra “modifier genes,” that is, the genetic background61. On the other hand, this term indicates that the phenotypic effect of one gene is masked by a disparate gene (locus). Thus, epistatic mutations possess disparate effects in combination than individually. It was primarily a concept from genetics but is nowadays utilized in biochemistry, computational biology, and evolutionary biology, due to the gene interaction, managing the nonlinear effects. Epistasis has a colossal impact on the form of evolutionary landscapes, that leads to profound consequences for progress and phenotypic trait evaluability62. Epistasis can considerably affect antimicrobial resistance. Moreover, the combinations of resistance mutations may crucially affect the development of multidrug resistance63.
At the genotypic level, epistasis occurs by the one locus gene masks, or prevents the phenotype of another locus gene for ascertaining fitness while not noticeable at phenotype level, as described by Lunzer64. In other theoretical studies, on the progress of recombination, the form of the fitness used as a function of number of deleterious mutations shown to be critical65. In contrast, negative epistasis, under certain conditions of the recombination (or sex), is favorable, with increasing mutation number and linear decrease in the log fitness66. Contradictory to the positive epistasis, an approximate numerical treatment by Azevedo67, revealed that the deleterious mutations can be recombined and should be targeted toward negative (synergistic) epistasis, and whether recombination can influence epistasis68.
Selective pressure
The use of antibiotics in abundance is associated with the significant rise of multidrug resistance and the way antibiotics are used. Bacteria can also gain more resistance to antibiotics in response to environmental stresses; however, these antibiotics provide selective pressure that can lead to the emergence of several bacteria exposed to mutated antibiotics or that obtain slices of DNA to develop antibiotic resistance. DNA slices may encode these multidrug pumps and enable the flow of various antibacterial agents from the cell. The selection pressures resulting from the use and misuse of antibiotics significantly support the phenomenon of antibiotic resistance69,71.
Biofilm formation
Biofilm is defined as a self-produced polymer matrix comprising polysaccharides, proteins, and the genetic material (DNA), which is the common mode of growth for most microorganisms in natural and medical systems and leads to protective bacterial growth that promotes the survival of bacteria in hostile environments72. Nevertheless, in biofilms, the prevalence of antibiotics is assumed to be extremely weak, due to nutrient constraints, slow growth, adaptive stress responses, and the formation of a series of coherent cells that form a multilayered defense system73. Therefore, according to drug resistance, each gene and gene product can be used as a target for developing new chemotherapeutic agents; however, the presence of biofilm formation, that facilitate chronic infections and antibiotic resistance.74 Thus, bacterial biofilm-specific resistance is considerable higher than any antibiotic resistance in planktonic bacteria. Consequently, biofilm-related infections are more problematic to treat and more prone to recurrence. The relation between biofilm and antibiotic resistance is essential for biomedical researchers75.
The bacterial cells cannot be affected by the antibiotics used without genetic modifications; these types of cells are recognized as persister cells. In contrast, resistant cells have the ability to grow in the presence of antibiotics, and do not produce inhibitory cells in the presence of these antibiotics76,77.
Outer membrane permeability and bacterial transport systems
The outer membrane of microorganisms crucially provides an additional layer of cellular defense to the organism without interfering with the exchange of essential substances necessary for cell survival. In addition, most of the antibiotics used have intracellular targets and cross the bacterial cell envelope to have an effective impact. Thus, the outer membrane of microorganisms is a difficult barrier that various antibiotics must cross in order to inhibit their growth. The lipid-mediated route is the main pathway that antibiotics can use to cross the bacterial outer membrane. The lipid and protein contents of the outer membranes have a strong potent effect on the bacterial sensitivity to various antibiotics, and resistance to these drugs relate to changes in these large molecules78.
Bacterial transport systems comprising structured protein channels may also influence antibiotic resistance in microbes by assisting the bacteria in both the acquisition of essential nutrients and the expulsion of toxic molecules, such as host bactericidal molecules and antibiotics. Consequently, bacterial transport systems play a significant role in both intrinsic bacterial drug resistance and the failure of antibiotic treatment78-80.
As described in this study, numerous factors contribute to the rise and spread of drug-resistant pathogens. Some factors are caused by human behaviors, whereas others are caused by bacterial behaviors. As professionals, we can only manage the factors caused by human behaviors, as we cannot stop the bacterial pathogens from finding ways to survive. Therefore, the only way to stop the spread of resistant pathogens is to break the cycle of ongoing transmission.
ACKNOWLEDGMENTS
I would like to offer my special thanks to Dr. Amir Elsayed Tantawy. Advice given by him has been a great help in writing this review.
FUNDING
This research was funded by the Deanship of Scientific Research at Princess Nourah bint Abdulrahman University through the Fast-track Research Funding Program.
ETHICS STATEMENT
This article does not contain any studies with human or animal subjects performed by the author.
AVAILABILITY OF DATA
The data used to support the findings of this study is included with in the article.
- Aslam B, Wang W, Arshad MI, Khurshid M, Muzammil S, Rasool MH, Nisar MA, Alvi RF, Aslam MA, Qamar MU, Salamat MKF, Baloch Z. Antibiotic resistance: a rundown of a global crisis. Infect Drug Resist, 2018; 11: 1645–1658.
Crossref - Levy SB, Marshall B. Antibacterial resistance worldwide: causes, challenges and responses. Nat Med, 2004; 10: 122-129.
Crossref - Roca I, Akova M, Baquero F, Carlet J, Cavaleri M, Coenen S, Cohen J, Findlay D, Gyssens I, Heuer OE, Kahlmeter G, et al. The global threat of antimicrobial resistance: science for intervention. New Microbes New Infect, 2015; 6: 22–29.
Crossref - Nathan C, Cars O. Antibiotic resistance-problems, progress, and prospects. N Engl J Med, 2014; 371: 1761–1763.
Crossref - Davies J, Davies D. Origins and evolution of antibiotic resistance. Microbiol Mol Biol Rev, 2010; 74: 417–433.
Crossref - Musgrove MT, Jones DR, Northcutt JK, Cox NA, Harrison MA, Fedorka-Cary PJ, Ladely SR. Antimicrobial resistance in Salmonella and Escherichia coli isolated from commercial shell eggs. Poult Science, 2006; 85: 1665-1669.
Crossref - Hoffman SJ, Caleo GM, Daulaire N, Elbe S, Matsoso P, Mossialos E, Rizvi Z, Rottingen JA. Strategies for achieving global collective action on antimicrobial resistance. Bull World Health Organ, 2015; 93(12): 867–876.
Crossref - Paczosa KM, Mecsas J. Klebsiella pneumoniae: Going on the Offense with a Strong Defense. Microbiol Mol Biol Rev, 2016; 80: 629–661.
Crossref - Codjoe FS, Donkor ES. Carbapenem Resistance: A Review. Med Sci (Basel). 2018; 6(1): 1-28.
Crossref - Kateete DP, Bwanga F, Seni J, Mayanja R, Kigozi E, Mujuni B, Ashaba FK, Baluku H, Christine F. Kallander NK, Rutebemberwa E, Asiimwe BB, Joloba ML. CA-MRSA and HA-MRSA coexist in community and hospital settings in Uganda Antimicrobial. Resistance and Infection Control, 2019; 8:94-103.
Crossref - Zhang D, Cui K, Lu W, Bai H, Zhai Y, Hu S, Li H, Dong H, Feng W, Dong W. Evaluation of carbapenem use in a tertiary hospital: antimicrobial stewardship urgently needed. Antimicrob Resist Infect Control, 2019; 8: 5- 12.
Crossref - Allcock S, Young EH, Holmes M, Gurdasani D, Dougan G, Sandhu MS, Solomon L, Torok ME. Antimicrobial resistance in human populations: challenges and opportunities. Glob health epidemiol genom 2017; 2(e4)1-7.
Crossref - Piddock LJ. The crisis of no new antibiotics-what is the way forward. Lancet Infect Dis, 2012; 12(3):249-253.
Crossref - Nafisah SB, Nafesa SB, Alamery AH, Alhumaid MA, AlMuhaidib HM, Al-Eidan FA. Over-the-counter antibiotics in Saudi Arabia, an urgent call for policy makers. J Infect Public Health, 2017; 10(5): 522-526.
Crossref - Pechere JC. Patients interviews and misuses of antibiotics. Clinical Infectious Diseases, 2001; 3: 170-173.
Crossref - Mudur G. Developing countries must balance access to antibiotics with action to curb resistance. BMJ, 2011; 343: 1-12.
Crossref - Leung E, Weil DE, Raviglione M, Nakatani H. World health organization. World health day antimicrobial resistance technical working group. The WHO policy package to combat antimicrobial resistance. Bull World Health Organ, 2011; 89(5): 390-392.
Crossref - Patel PH, Rathod S, Chuahan B, Rathod H, Pethani J, Shah P. Changing trend of antibiotic susceptibility pattern of common gram-negative Bacilli isolated from medical intensive care unit of tertiary care hospital ahmedabad, Gujarat, India. Journal of Drug Discovery and Therapeutics, 2013; 1(4): 16-20.
- Kardas P. Patient compliance with antibiotic treatment for respiratory tract infections. J Antimicrob Chemother, 2002; 49(6): 897-903.
Crossref - Cals JW, Hopstaken RM, Le Doux PH, Driessen GA, Nelemans PJ, Dinant GJ. Dose timing and patient compliance with two antibiotic treatment regimens for lower respiratory tract infections in primary care. Int J Antimicrob Agents, 2008; 31(6): 531-536.
Crossref - Li B and Webster TJ. Bacteria antibiotic resistance: New challenges and opportunities for implant-associated orthopaedic infections. J Orthop Res, 2018; 36(1): 22–32.
Crossref - Stobberingh E, van den Bogaard A, London N, Driessen C, Top J, Willems R. Enterococci with glycopeptide resistance in turkeys, turkey farmers, turkey slaughterers, and (sub)urban residents in the south of The Netherlands: evidence for transmission of vancomycin resistance from animals to humans. Antimicrob Agents Chemother, 1999; 43(9): 2215-21.
Crossref - Bager F, Aarestrup FM, Madsen M, Wegener HC. Glycopeptide resistance in Enterococcus faecium from broilers and pigs following discontinued use of avoparcin. Microbial Drug Resistance, 1999; 5(1): 53-66.
Crossref - Aarestrup FM. Characterization of glycopeptide-resistant enterococcus faecium (GRE) from broilers and pigs in Denmark: genetic evidence that persistence of GRE in pig herds is associated with co-selection by resistance to macrolides. Journal of Clinical Microbiology, 2000; 38(7): 2774-2777.
Crossref - Economou V, Gousia P. Agriculture and food animals as a source of antimicrobial-resistant bacteria. Infect Drug Resist, 2015; 8: 49–61.
Crossref - Zowawi HM, Balkhy HH, Walsh TR, Paterson DL. beta-Lactamase production in key gram-negative pathogen isolates from the Arabian Peninsula. Clin Microbiol Rev, 2013; 26(3): 361-380.
Crossref - Al-Mustafa ZH, Al-Ghamdi. MS. Use of Antibiotics in the poultry industry in Saudi Arabia: Implications for public health. Annals of Saudi Medicine, 2002; 22: 4-7.
Crossref - Al-Obeid QS, CherkaouiJH. Francois SP. First Detection of an Invasive Staphylococcus Aureus Strain (D958) with reduced susceptibility to glycopeptides in Saudi Arabia. Journal of Clinical Microbiology, 2010; 48: 2190-2204.
Crossref - Singh S, Shukla S, Tandia N, Kumar N, Paliwal R. Antibiotic residues: A global challenge. Pharma Science Monitors, 2014; 5(3):184-197.
- Avci M, Ozgenc O, Coskuner SA, Olut A. Hospital acquired infections (HAI) in the elderly: comparison with the younger patients. Arch Gerontol Geriatr, 2012; 54(1): 247-50.
Crossref - Flaherty JP, Weinstein RA. Nosocomial infection caused by antibiotic-resistant organisms in the intensive-care unit. Infect Control Hosp Epidemiol, 1996; 17(4): 236-248.
Crossref - Gold HS, Moellering RC, JR. Antimicrobial-drug resistance. N Engl J Med, 1996; 335(19): 1445-1453.
Crossref - Cornejo-Juarez P, Vilar-Compte D, Perez-Jimenez C, Namendys-Silva SA, Sandoval-Hernandez S, Volkow-Fernandez P. The impact of hospital-acquired infections with multidrug-resistant bacteria in an oncology intensive care unit. Int J Infect Dis, 2015; 31: 31-34.
Crossref - Muto CA. Why are antibiotic-resistant nosocomial infections spiraling out of control?. Infect Control Hosp Epidemiol, 2005; 26(1): 10-12.
Crossref - Bieh KL, Weigel R, Smith H. Hospitalized care for MDR-TB in Port Harcourt, Nigeria: a qualitative study. BMC Infect Dis, 2017; 17: 50-59.
Crossref - Sinanovic E, Ramma L, Vassall A, Azevedo V, Wilkinson L, Ndjeka N. Impact of reduced hospitalisation on the cost of treatment for drug-resistant tuberculosis in South Africa. Int J Tuberc Lung Dis, 2015; 19(2): 172–178.
Crossref - Al-Hajoj SA, Zozio T, Al-Rabiah F, Mohammad V, Al-Nasser M, Sola C. First insight into the population structure of Mycobacterium tuberculosis in Saudi Arabia. J Clin Microbiol, 2007; 45(8): 2467-2473.
Crossref - Al-Hajoj SA. Tuberculosis in Saudi Arabia: Can we change the way we deal with the disease? Journal of Infection and Public Health, 2010; 3: 17-24.
Crossref - Schepisi MS, Sotgiu G, Contini S, Puro V, Ippolito G, Girardi E. Tuberculosis transmission from healthcare workers to patients and co-workers: a systematic literature review and meta-analysis. PLoS One, 2015; 10(4): 1-15.
Crossref - Chen YP, Shaffer M. The influence of expatriate spouses’ coping strategies on expatriate and spouse adjustment: An interdependence perspective. Journal of Global Mobility, 2018; 6(1): 20-39.
Crossref - Khan MA, Faiz A. Antimicrobial resistance patterns of Pseudomonas aeruginosa in tertiary care hospitals of Makkah and Jeddah. Ann Saudi Med, 2016; 36(1): 23–28.
Crossref - Leangapichart T, Gautret P, Griffiths K, Belhouchat K, Memish Z, Raoult D, et al. Acquisition of a high diversity of bacteria during the hajj pilgrimage, including Acinetobacter baumannii with blaOXA-72 and Escherichia coli with blaNDM-5 Carbapenemase Genes. Antimicrob Agents Chemother, 2016; 60(10): 5942-5948.
Crossref - Alzeer AH. Respiratory tract infection during Hajj. Ann Thorac Med, 2009; 4(2): 50-53.
Crossref - Osmon DR. Antimicrobial prophylaxis in adults. Mayo Clin Proc, 2000; 75(1): 98-109.
Crossref - Enzler MJ, Berbari E, Osmon DR. Antimicrobial prophylaxis in adults. Mayo Clin Proc, 2011; 86(7): 686-701.
Crossref - Brauner A, Fridman O, Gefen O, Balaban NQ. Distinguishing between resistance, tolerance and persistence to antibiotic treatment. Nat Rev Microbiol, 2016; 14: 320–330.
Crossref - Prestinaci F, Pezzotti P, Pantosti A. Antimicrobial resistance: a global multifaceted phenomenon. Pathog Glob Health. October 2015; 109(7): 309–318.
Crossref - Dutta C, Pan A. Horizontal gene transfer and bacterial diversity. J Biosci, 2002; 27(1): 27-33.
Crossref - Martinez JL. Bottlenecks in the transferability of antibiotic resistance from natural ecosystems to human bacterial pathogens. Front Microbiol, 2011; 2: 265-304.
Crossref - Frost LS, Leplae R, Summers AO, Toussaint A. Mobile genetic elements: the agents of open source evolution. Nat Rev Microbiol, 2005; 3(9): 722-732.
Crossref - Dobrindt U, Hochhut B, Hentschel U, Hacker J. Genomic islands in pathogenic and environmental microorganisms. Nat Rev Microbiol, 2004; 2(5): 414-424.
Crossref - Millan A. Evolution of Plasmid-Mediated Antibiotic Resistance in the Clinical Context. Trends in Microbiology, 2018; 26: 978-985.
Crossref - Gillespie SH. Evolution of drug resistance in Mycobacterium tuberculosis: clinical and molecular perspective. Antimicrob Agents Chemother, 2002; 46(2): 267-274.
Crossref - Wintersdorff CJV, Penders J, van Niekerk JM, Mills ND, Majumder S, van Alphen LB, et al. Dissemination of antimicrobial resistance in microbial ecosystems through horizontal gene transfer. Front Microbiol, 2016; 7(173): 1-10.
Crossref - Woodford N, Ellington MJ. The emergence of antibiotic resistance by mutation. Clin Microbiol Infect, 2007; 13(1): 5-18.
Crossref - Ehrenreich IM. Epistasis: Searching for Interacting Genetic Variants Using Crosses. Genetics, 2017; 206: 531–535.
Crossref - Weinreich DM, Watson RA, Chao L. Sign epistasis and genetic constraint on evolutionary trajectories. Evolution, 2005; 59: 1165–1174.
Crossref - Phillips PC. Epistasis—the essential role of gene interactions in the structure and evolution of genetic systems. Nat Rev Genet, 2008; 11: 855-867.
Crossref - Shapiro B, Rambaut A, Pybus OG, Holmes EC. A phylogenetic method for detecting positive epistasis in gene sequences and its application to RNA virus evolution. Mol Biol Evol, 2006; 23: 1724–1730.
Crossref - Hansen TF, Wagner GP. Modeling genetic architecture: a multilinear model of gene interaction. Theor Pop Biol, 2001; 59: 61–86.
Crossref - Cheverud JM, Routman EJ. Epistasis and its contribution to genetic variance components. Genetics, 1995; 139: 1455–1461.
- Gros PA, Le Nagard H, Tenaillon O. The evolution of epistasis and its links with genetic robustness, complexity and drift in a phenotypic model of adaptation. Genetics, 2009; 182(1): 277–293.
Crossref - Ayati M, Koyuturk M. Prioritization of genomic locus pairs for testing epistasis. Proceedings of the 5th ACM conference on bioinformatics, computational biology, and health informatics. BCB 14. New York, NY, USA. 2014; 240–248.
Crossref - Wong A. Epistasis and the Evolution of Antimicrobial Resistance. Front Microbiol, 2017; 8(246): 1-12.
Crossref - Lunzer M, Miller SP, Felsheim R, Dean AM. The biochemical architecture of an ancient adaptive landscape. Science, 2005; 310: 499–501.
Crossref - Keightley PD, Otto SP. Interference among deleterious mutations favors sex and recombination in finite populations. Nature, 2006; 443: 89–92.
Crossref - Kondrashov AS. Deleterious mutations and the evolution of sexual reproduction. Nature, 1988; 336: 435–440.
Crossref - Azevedo RBR, Lohaus R, Srinivasan S, Dang KK, Burch CL. Sexual reproduction selects for robustness and negative epistasis in artificial gene networks. Nature, 2006; 440: 87–90.
Crossref - Michalakis Y, Roze D. Epistasis in RNA viruses. Science, 2004; 306: 1492–1493.
Crossref - Morgan DJ, Okeke IN, Laxminarayan R, Perencevich NE, Weisenberg. S. Non-prescription antimicrobial use worldwide: a systematic review. The Lancet Infectious Diseases, 2011; 11(9): 692–701.
Crossref - Zowawi HM, Harris PN, Roberts MJ, Tambyah PA, Schembri MA, Pezzani MD, et al. The emerging threat of multidrug-resistant Gram-negative bacteria in urology. Nat Rev Urol, 2015; 12(10): 570-584.
Crossref - Zowawi HM. Antimicrobial resistance in Saudi Arabia. An urgent call for an immediate action. Saudi Med J, 2016; 37(9): 935-940.
- McDougald D, Rice SA, Barraud N, Steinberg PD. Should we stay or should we go: mechanisms and ecological consequences for biofilm dispersal. Nat Rev Microbiol, 2012; 10: 39-50.
Crossref - Stewart PS. Mechanisms of antibiotic resistance in bacterial biofilms. Int J Med Microbiol, 2002; 292(2): 107-113.
Crossref - Hoiby N, Bjarnsholt T, Givskov M, Molin S, Ciofu O. Antibiotic resistance of bacterial biofilms. Int J Antimicrob Agents, 2010; 35(4): 322-332.
Crossref - Kaplan JB. Antibiotic-induced biofilm formation. Int J Artif Organs, 2011; 34(9): 737-51.
Crossref - Wood TK, Knabel JS, Kwan BW. Bacterial Persister Cell Formation and Dormancy. Applied and Environmental Microbiology, 2013; 79: 7116-7121.
Crossref - Zhange Y. Persisters, persistent infections and the Yin–Yang model. Emerg Microbes Infect, 2014; 3(1): e3.
Crossref - Delcour AH. Outer membrane permeability and antibiotic resistance. Biochim Biophys Acta, 2009; 1794: 808-816.
Crossref - Ben-Kahla I, Al-Hajoj S. Drug-resistant tuberculosis viewed from bacterial and host genomes. Int J Antimicrob Agents, 2016; 48(4): 353-360.
Crossref
© The Author(s) 2020. Open Access. This article is distributed under the terms of the Creative Commons Attribution 4.0 International License which permits unrestricted use, sharing, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.