ISSN: 0973-7510
E-ISSN: 2581-690X
The prevalence of multidrug-resistant Klebsiella pneumoniae is extensive, both in healthcare settings and the general population. Biofilm formation in K. pneumoniae plays a key role in infection pathogenesis and serves as important defensive strategy against antibiotics and immune evasion. This study examined the presence of efflux pumps, potential for biofilm development, and antibiotic susceptibility profiles of K. pneumoniae clinical isolates. Antibiotic susceptibility testing of K. pneumoniae isolates was performed using the disc diffusion method. All isolates were tested for efflux pump presence using the cartwheel method, and biofilm production was estimated using tissue culture plate, tube, and Congo red agar methods. PCR amplification was performed using specific primers to detect genes encoding drug resistance and biofilm formation. All 17 isolates of K. pneumoniae isolates exhibited multidrug- resistance and functional efflux pumps. Nevertheless, the capacity of these organisms to produce biofilms differed, with eight (47%) strong biofilm formers, seven (41%) moderate biofilm formers, and two (11%) weak biofilm formers. The antibiotic resistance genes, blaCTX-M , blaKPC , and blaNDM were present in 15 (88%), 11 (64%), and seven (41%) K. pneumoniae isolates, respectively. The genes, acrAB, tolC, and mdtK, encoded efflux pumps present in 12 (70%), 15 (88%), and 10 (58%) isolates, respectively. Biofilm genes, mrkD, fimH, and luxS, were present in 16 (94%) isolates. This study revealed multiple factors that lead to the notable drug resistance observed in K. pneumoniae isolates. Therefore, it is advisable to implement a holistic strategy for managing diseases caused by pathogenic bacteria.
Antibiotic Resistance, Biofilm, Klebsiella pneumoniae, Efflux Pump
Biofilms comprise a well-organized group of microorganisms bounded within a matrix structure they create themselves and are attached to either a living or non-living surface. Bacterial biofilms are widely acknowledged as important etiological factors in numerous infections, given that a substantial proportion of bacterial infections, ranging from 65% to 80%, are attributed to the development of biofilms.1 Certain physiological circumstances and genetic interactions occurring inside biofilms result in substantial augmentation of resistance to antimicrobial agents. Additionally, the acquisition of drug resistance and biofilm formation are associated.2 Klebsiella pneumoniae is a routinely isolated causative pathogen of clinically aided infections. The microorganisms in question exhibit opportunistic behavior and can produce a substantial biofilm layer, which serves as an important virulence factor. This characteristic allows the bacteria to adhere to both animate and inanimate surfaces, thereby contributing to its resistance to antimicrobial interventions.3 The severity of K. pneumoniae infections is mostly due to its virulence components, specifically type-1 and type-3 fimbriae, capsular polysaccharides, and common pilus pili. These factors facilitate biofilm formation, thereby exacerbating the pathogenesis of the bacterium.4 Multidrug-resistant (MDR) strains of K. pneumoniae are rapidly emerging and frequently present a considerable risk to patients as therapeutic options become less effective and the fatality rate increases.5 Compared with other bacteria, K. pneumoniae exhibits a greater tendency for antibiotic resistance. This is primarily due to its capability to produce resistance-mediating enzymes, such as extended-spectrum β-lactamase (ESBLs) and carbapenemase.6 In addition, active efflux-pumps are crucial for the acquisition of antibiotic resistance in microbes. Efflux pump systems are essential for removing chemical substances, such as antibiotics, from bacterial cells. This process is energy-dependent and helps the cells get rid of these compounds.7 Efflux pump activity might be related to the establishment of biofilms, resulting in an increased level of antibiotic resistance.8 The study aims to assess the functionality of efflux-pumps, biofilm production, and pattern of antibiotic-resistance trends in K. pneumoniae obtained from clinical samples.
Collection of bacterial isolates
Seventeen K. pneumoniae isolates were procured from Voluntary Health Services, Chennai, India. Since the study did not involve any human participant nor the collection of clinical specimens, informed consent was not necessary. Institutional ethical committee approval was obtained to carry out the study. All isolates were evaluated for antibiotic sensitivity, efflux pump production, and the ability to form biofilms. All experiments were performed at the Department of Microbiology, VISTAS.
Antimicrobial susceptibility testing (AST)
The AST pattern was studied using the disc diffusion method on sterile Muller–Hinton agar plates (MHA; Himedia, India) with antibiotic discs (Himedia), according to the Clinical and Laboratory Standards Institute (CLSI, 2023) guidelines.9 The isolates were tested against piperacillin (100 µg (P), amoxyclav 20/10 µg (AMC), piperacillin with tazobactam 100/10 µg (PIT), ceftazidime 30 µg (CAZ), cefepime 30 µg (CPM), cefotaxime 5 µg (CTX), imipenem 10 µg (IPM), meropenem 10 µg (MPM), ciprofloxacin 5 µg (CIP), levofloxacin 5 µg (LE), amikacin 30 µg (AK), gentamicin 10 µg (GEN), tetracycline 30 µg (TET), chloramphenicol 30 µg (C), and nitrofurantoin 300 µg (NIT). Isolates resistant to three or more groups of antibiotics were classified as MDR K. pneumoniae (MDR-KP).
Efflux-pump detection using ethidium-bromide (EtBr) cartwheel method
The detection of efflux-pumps in the isolates was carried using the fluorescent dye, EtBr, cartwheel method.10 Uniformly dispersed suspensions of 17 K. pneumoniae isolates, with a cell density of 106 cells/mL, were inoculated onto MHA plates supplemented with EtBr (1 mg/L) after incubation for 24 h at 37 °C. The plates were analyzed using a UV transilluminator to detect fluorescence.
Biofilm forming assays
Tissue culture plate method (TCP)
The potential of K. pneumoniae isolates to form biofilms was quantified using the TCP technique.11 All K. pneumoniae isolates were grown in tryptone soya casein digest broth (TSB) with and without 1% glucose (Himedia) for 24 h at 37 °C. Incubation was followed by dilution with fresh medium (1:100). Approximately 200 µL of diluted culture samples were added in 96-well microtiter plates. After a 24 h incubation, each well was stripped and washed three times with pH 7.2 phosphate-buffered saline (PBS). The plate was heat-fixed at 60 °C for 20 min. The wells were stained with 0.2 mL of 0.1% (v/v) crystal violet solution (CV) for 5 min. The wells were then cleaned four times with demineralized water. The optical density (OD) of adhered biofilms was measured at 570 nm using a Bio-Rad Microplate Reader (iMark). This experiment was repeated thrice. The average OD of the negative controls was calculated. The value of three times the standard deviation (SD) was added to the average OD and used as the OD cutoff (Odc). K. pneumoniae isolates with an OD below the threshold OD value (ODc) were considered poor biofilm-formers. Isolates with OD values >0.240, 0.120-0.240, and <0.120 were classified as strong, moderate, and weak biofilm producers, respectively.
Tube method (TM)
In the qualitative tube method, K. pneumoniae isolates were introduced into 10 mL of TSB with and without 1% glucose and incubated for 24 h at 37 °C. The contents were discarded and the cells were rinsed with PBS (pH 7.2), followed by incubation. Subsequently, the tubes were stained with a 0.5% solution of the CV dye for 5 min, washed with deionized water, and air-dried. A visible film lining the tube indicated whether biofilm developed.11
Congo red agar (CRA) method
For CRA screening, modified brain-heart infusion (BHI) medium was prepared using 37 g/L of sucrose, 50 g/L of agar, and 8 g/L of Congo red indicator dye. The Congo red suspension and BHI with sucrose medium were individually prepared and sterilized by autoclaving and mixed at 55 °C. The test organism was inoculated onto CRA media and incubated at 37 °C for 24 h. Biofilm production was characterized by the presence of black colonies with a dry and crystalline texture.12
Light microscopy analysis of biofilm detection
Several stages of biofilm development were examined using light microscopy.13 The overnight culture was diluted with fresh TSB broth with 1% glucose at a ratio of 1:100 and the resultant mixture was transferred into separate wells of a sterile 12-well plate up to the rim. A sterile glass cover slip was placed on top of the wells and incubated at 37 °C. At different time points (6 h, 12 h, 18 h and 24 h) the coverslips were removed from each well and rinsed three times using PBS (pH 7.2). They were then stained with 0.5% CV for 10 min, washed with demineralized water, and air-dried. The morphologies of different biofilm stages were observed under a light microscope.
DNA isolation and polymerase chain reaction (PCR) conditions
Chromosomal DNA was extracted from individual K. pneumoniae isolates, as described by Sambrook14 using the phenol-chloroform method. Table displays a list of specific primer sequences for the identified genes, designed and synthesized using Barcode Biosciences (Bangalore, India).
Table :
Primer sequence and annealing temperature used
| Target genes | Primer sequence from 5’ – 3’ | Annealing temperature (AT) | Reference |
|---|---|---|---|
| fimH | F-AACGGCACCTCTTACCCGTT R-TGCTCACCGGCGTCAGATAC |
58 | [1] |
| 16S rRNA | F-ATTTGAAGAGGTTGCAAACGAT R-TTCACTCTGAAGTTTTCTTGTGTTC |
55 | |
| blaKPC | F-GCTACACCTAGCTCCACCTTC R-ACAGTGGTTGGTAATCCATGC |
58 | |
| blaNDM | F-CTGCATTGATGCTGAGCGGG R-GCTGGCGGAAAACCAGATCG |
60 | |
| pgaA | F-GCAGACGCTCTCCTATGTC R-GCCGAGAGCAGGGGAATC |
58 | [15] |
| mdtk | F-GCGCTTAACTTCAGCTCA R-GATGATAAATCCACACCAGAA |
50 | [16] |
| blaCTX-M-1 | F-ACAGCGATAACGTGGCGATG R-TCGCCCAATGCTTTACCCAG |
58 | |
| tolC | F-ATCAGCAACCCCGATCTGCGT R-CCGGTGACTTGACGCAGTCCT |
60 | |
| acrAB | F-ATCAGCGGCCGGATTGGTAAA R-CGGGTTCGGGAAAATAGCGCG |
62 | |
| magA | F-GGTGCTCTTTACATCATTGC R-GCAATGGCCATTTGCGTTAG |
53 | [28] |
| rmpA | F-ACTGGGCTACCTCTGCTTCA R-CTTGCATGAGCCATCTTTCA |
57 | |
| aero | F-GCATAGGCGGATACGAACAT R-GCATAGGCGGATACGAACAT |
57 | |
| mrkD | F-CCACCAACTATTCCCTCGAA R-ATGGAACCCACATCGACATT |
56 | |
| luxS | F-GCCGTTGTTAGATAGTTTCACAG R-CAGTTCGTCGTTGCTGTTGATG |
55 | [17] |
| ecpRAB | F-CCTATGTAATTAATGGCAGGTTT R-GCTGTTCATAAAGGATGAAATATC |
55 | [18] |
The PCR was performed by combining 15 µL of the PCR Taq DNA polymerase 2× master mix red (Bio-Rad), 6.0 µL of template (DNA), 6.0 µL of sterile nuclease free water, and 1.5 µL of both primers (forward and reverse) at a concentration of 250 nM, resulting in a 30-µL reaction mixture. Thermal cycling was performed using a Nexus Gradient Eppendorf Master Cycler (Hamburg, Germany). The parameters were as follows: DNA unwinding at 94 °C (3 min), 35 cycles of denaturation at 94 °C (30 s), primer annealing at a temperature specified in Table for 30 s, extension at 72 °C (45 s), and elongation at 72 °C (7 min). A reaction mixture with all components except the DNA template was used as the negative control. The PCR results were examined using gel electrophoresis (1.5% agarose gel with EtBr). Molecular DNA markers (HiMedia) were used to confirm the product size. The gel was electrophoresed in 1× Tris-EDTA buffer at 50 V for 1.5 h. PCR amplicons were observed using a gel documentation method (Protein Simple; AlphaImager Mini Imaging).
Statistical data analysis
The association between biofilm development in various media was examined using one-way ANOVA, whereas the relationship between drug resistance and biofilm formation was analyzed using Pearson’s correlation statistics in MS Excel.
Drug resistance profile
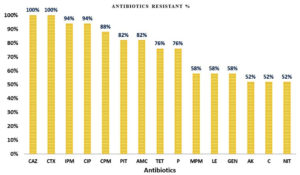
All 17 (100%) K. pneumoniae isolates were resistant to ceftazidime and cefotaxime; 16 (94%) were resistant to imipenem and ciprofloxacin; and 15 (88%) were resistant to cefepime. Additionally, 82% were resistant to piperacillin-tazobactam, and amoxicillin-clavulanate; 76% were resistant to piperacillin and tetracycline; 58% were resistant to gentamicin and meropenem; and 52% were resistant to amikacin, chloramphenicol, and nitrofurantoin (Figure 1).
Figure 1. The prevalence percentage of antibiotic resistance in K. pneumoniae isolates. CAZ: Ceftazidime; CTX: Cefotaxime; IPM: Imipenem; CIP: Ciprofloxacin; CPM: Cefepime; PIT: Piperacillin tazobactam; AMC: Amoxicillin plus clavulanic acid; TET: Tetracycline; P: Penicillin; MPM: Meropenem; LE: Levofloxacin; GEN: Gentamicin; AK: Amikacin; C: Chloramphenicol; NIT: Nitrofurantoin
Efflux pump and biofilm formation
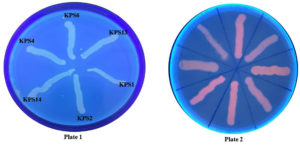
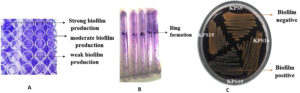
All K. pneumoniae isolates demonstrated efflux pump activity, as demonstrated by no fluorescence when exposed to UV light (Figure 2). The TCP method revealed that approximately 47% and 41% of K. pneumoniae isolates were strong and moderate biofilm producers, whereas 11% were non-biofilm producers. Additionally, 88% of the isolates tested positive for biofilm production in the tube method. Furthermore, 18% of the isolates tested positive for biofilm production in the CRA method. Figure 3 depicts biofilm formation in various experiments.
Figure 2. Efflux pump detection in K. pneumoniae clinical isolates by ethidium bromide cartwheel method (Plate 1: K. pneumoniae strains with 1 mg/L of EtBr; Plate 2: Antibiotic susceptible bacterial strains that do not possess efflux pump as control)
Figure 3. Biofilm formation in different experimental methods; A: Tissue culture method of K. pneumoniae phenotypes with strong, moderate, and weak adherence in response to biofilm formation in a microtiter plate; B: Tube method shows visible ring lining tube as positive biofilm production; C: Black crystalline colony formation in Congo red agar method indicates positive for biofilm production
Biofilm formation under a light microscope
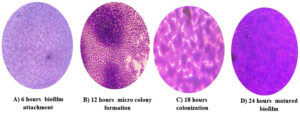
Light microscopy analysis of K. pneumoniae revealed different stages of biofilm formation, including attachment, colonization, maturation, and dispersion. Distinct biofilm morphologies were observed at each stage and different time intervals (Figure 4). Biofilms rapidly develop during each time interval from the planktonic to the mature biofilm stage. The bacteria embedded inside the biofilms were observed during the maturation stage. The multiplication and spread of organisms within the biofilm were observed during the colonization stage, resulting in the development of biofilms.
Figure 4. Light microscopic image of different stages of biofilm development in K. pneumoniae isolates using the coverslip method. The biofilm-coated coverslip stained with crystal violet dye was examined at different time points; A: Attachment of biofilm to the surface (6 hours); B: Formation of microcolony and water channels (12 hours); C: Colonization of biofilm (18 hours); D: Fully developed and stabilized biofilm (24 hours)
Prevalence of resistant, virulence, efflux pump, and biofilm-associated genes
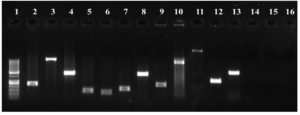
Antimicrobial resistance genes encoding ESBLs and carbapenemases were present in 15 (88%; blaCTX-M), 11 (64%; blaKPC), and 7 (41%; blaNDM) of K. pneumoniae isolates. Virulence-related genes, including aerobactin synthase, mucoviscosity-associated (magA), mucoid phenotype A (rmpA), pili formation (ecpRAB), and adhesin (pgaA) genes, were observed in 9 (52%), 8 (47%), 3 (17%), 14 (82%), and 15 (88%) of the isolates respectively. Twelve isolates (70%) had the efflux pump gene, acrAB, 15 (88%) had the tolC gene, and 10 (58%) had the mdtK gene. Sixteen isolates (94%) had biofilm-associated genes that encode type-1 (mrkD) and type-3 (fimH) adhesion fimbriae. Similarly, except for one, all the remaining 16 isolates showed the presence of the luxS gene involved in quorum sensing. Figure 5 shows a gel image of the amplified target genes in a representative KPS13 strain.
Figure 5. PCR analysis of K. pneumoniae (KPS13). Lane: 1-100 bp DNA ladder, 2-mrkD (240 bp), 3-ecpRAB (1025 bp), 4-luxS (447 bp), 5-16s (130 bp), 6-fimH (109 bp), 7-pgaA (157 bp), 8-mdtK (453 bp), 9-blaCTX-M (216 bp), 10-aero (556 bp), 11-magA (1282 bp), 12-acrAB (312 bp), 13-tolC (525 bp), [14-blaKPC (985 bp), 15-blaNDM (105 bp), 16-rmpA (535 bp) were not amplified in KPS13 strain]
Statistical analysis
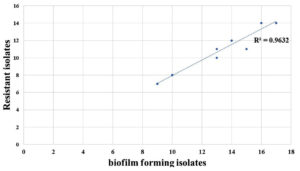
TSB supplemented with glucose significantly enhanced biofilm formation compared to that with TSB alone (p < 0.01). A correlation coefficient of R2 = 0.9632 was observed in the scatter plot, demonstrating the correlation between biofilm formation and antibiotic resistance (Figure 6).
In this study, K. pneumoniae isolated from different hospital samples showed high (100%) resistance to ceftazidime and cefotaxime, followed by imipenem and ciprofloxacin (94.4%). Earlier investigations conducted in Lagos, Nigeria, also recorded a significant and widespread resistance pattern of K. pneumoniae to β-lactam antibiotics.7
India has a high rate of antimicrobial resistance, partly because the country does not have strict laws controlling the use of antibiotic drugs. Efflux pumps facilitate the development of MDR-KP isolates. The antibiotic concentrations inside bacterial cells can be reduced via efflux pumps, which are important components for bacterial survival. Based on our findings, acrAB and tolC efflux pumps were more prevalent in the isolates than the mdtk genotype, which is consistent with other studies.19 In this study, the detection of the acrAB-tolC system in the isolates was correlated with MDR in K. pneumoniae isolates, also consistent with other reports.20
The inclusion of 1% glucose in the TSB broth significantly enhanced biofilm formation among the isolates. Therefore, the two media may have different methods of promoting biofilm formation. The TCP approach was determined to be the most appropriate among the three methods (TCP, TM, and CRA) for biofilm identification in terms of accuracy and quantification. Thus, the results are consistent with those of other reports.11,21 The TM method showed lower specificity than that with the TCP method,22 Using the CRA method, only 18% of the K. pneumoniae isolates tested positive for biofilm formation, whereas 88% of the K. pneumoniae isolates tested positive using the TCP method. Therefore, the CRA method is the least specific for biofilm detection.22
Light microscopy analysis of biofilm formation revealed different stages of biofilm development. The 6 h stage was characterized by attachment, while the 12 h stage involved the synthesis of exopolysaccharides and the formation of microcolonies. The 24 h stage marked the maturation and stabilization of the biofilms.1 This biofilm formation was consistent with previously established research on gram-negative bacteria. Furthermore, K. pneumoniae MDR isolates exhibited the highest level of biofilm formation during the biofilm maturation stage, which is consistent with previous findings on MDR Pseudomonas aeruginosa and Escherichia coli isolates.23,24
A previous study revealed that K. pneumoniae isolates derived from hospital samples exhibit a substantial presence of virulence, efflux pumps, and biofilm-related genes. The prevalence of luxS among K. pneumoniae isolates was 94%, which is similar to a study reporting a 98% prevalence.17 The genes linked to virulence were also widespread in MDR bacteria. K. pneumoniae adhesion to the mucosal lining of the host is mediated by fimbriae genes, type-1 (fimH), type-3 (mrkD), and pilus formation (ecpRAB).25 The distribution of the mrkD adhesin gene in K. pneumoniae was found to be 20% in a previous study, 18 while in our study, it was found to be 94%, confirming the high prevalence of the mrkD genotype. Additionally, this study is consistent with another study reporting an 88% prevalence of mrkD and fimH-1 genotypes.26 Furthermore, the present study reveals that the ecpA gene promotes adhesion to host tissues in 82% of the isolates, which is in accordance with previous research.18 An earlier study demonstrated that virulence adhesins are linked to strains that produce carbapenemases.27 Similar findings were observed in the present study, where 58% of the isolates harbored the carbapenem blaKPC gene. In our investigation, 15 isolates (88%) had the blaCTX-M gene, among which 53 and 33% were high and moderate biofilm producers, respectively. However, only 13% did not produce biofilms. This reveals that the blaCTX-M, aids in biofilm formation.28 This study also revealed the presence of rmpA, magA, and aerobactin virulence genes in 17%, 47%, and 52% of K. pneumoniae isolates respectively, similar to the prevalence of virulence genes in other studies.26,28 The prevalence of these genes may vary due to differences in virulence genes, which may be associated with the source of K. pneumoniae isolates in clinical samples.
Establishing broad conclusions from this study is challenging because of the limited number of isolates. Additionally, the study only focused on biofilm screening in drug-resistant pathogens. Therefore, future studies should encompass various geographical regions within the country and include larger numbers of isolates.
The association between MDR bacteria and biofilm formation is still a debatable topic. However, our data indicate that antibiotic resistance in clinical pathogens may be a decisive component in determining pathogenicity traits. The association between biofilm development and drug resistance offers valuable insights for the management of drug-resistant bacteria. The variation in the expression of various virulence factors across bacteria restricts the available treatment options and presents difficulties for microbiological laboratories in identifying them. Detecting the virulence characteristics of clinical pathogens aids in the management of medically important pathogens in hospital environments. Therefore, it is necessary to assess the rate at which virulence factors are expressed to investigate the dissemination of virulence within various bacterial populations.
ACKNOWLEDGMENTS
The authors would like to thank the Management of VISTAS for providing the Research fellowship during the study period and for their ongoing support in completing the research work.
CONFLICT OF INTEREST
The authors declare that there is no conflict of interest.
AUTHORS’ CONTRIBUTION
All authors listed have made a substantial, direct and intellectual contribution to the work, and approved it for publication.
FUNDING
This study was supported by a stipend received under Vels Research Fellowship Scheme (VRF) from the Management of VISTAS.
DATA AVAILABILITY
The datasets generated and/or analysed during the current study are available from the corresponding author on reasonable request.
ETHICS STATEMENT
This study was approved by the Institutional Ethics Committee (IEC), VISTAS, Chennai, India, vide reference number VISTAS-SPS/IEC/I/2022/04.
- Al-Bayati M, Samarasinghe S. Biofilm and Gene Expression Characteristics of the Carbapenem-Resistant Enterobacterales, Escherichia coli IMP and Klebsiella pneumoniae NDM-1 Associated with Common Bacterial Infections. Int J Environ Res Public Health. 2022;19(8):4788.
Crossref - Yaita K, Gotoh K, Nakano R, et al. Biofilm-Forming by Carbapenem Resistant Enterobacteriaceae May Contribute to the Blood Stream Infection. Int J Mol Sci. 2019;20(23):5954.
Crossref - Seifi K, Kazemian H, Heidari H, et al. Evaluation of Biofilm Formation Among Klebsiella pneumoniae Isolates and Molecular Characterization by ERIC-PCR. Jundishapur J Microbiol. 2016;9(1):e30682.
Crossref - Li B, Zhao Y, Liu C, Chen Z, Zhou D. Molecular pathogenesis of Klebsiella pneumoniae. Future Microbiol. 2014;9(9):1071-1081.
Crossref - Nirwati H, Sinanjung K, Fahrunissa F, et al. Biofilm formation and antibiotic resistance of Klebsiella pneumoniae isolated from clinical samples in a tertiary care hospital, Klaten, Indonesia. BMC Proc. 2019;13(Suppl 11):20.
Crossref - Bengoechea JA, Sa Pessoa J. Klebsiella pneumoniae infection biology: living to counteract host defence. FEMS Microbiol Rev. 2019;43(2):123-144.
Crossref - Akinpelu S, Ajayi A, Smith SI, Adeleye AI. Efflux pump activity, biofilm formation and antibiotic resistance profile of Klebsiella spp. isolated from clinical samples at Lagos University Teaching Hospital. BMC Res Notes. 2020;13(1):258.
Crossref - Reza A, Sutton JM, Rahman KM. Effectiveness of Efflux Pump Inhibitors as Biofilm Disruptors and Resistance Breakers in Gram-Negative (ESKAPEE) Bacteri. Antibiotics. 2019;8(4):229.
Crossref - CLSI, Performance Standards for Antimicrobial Susceptibility Testing, Clinical and Laboratory Standards Institute, 2023, 33rd Ed. CLSI Supplement M100.
- Martins M, McCusker MP, Viveiros M, et al. A Simple Method for Assessment of MDR Bacteria for Over-Expressed Efflux Pumps. Open Microbiol J. 2013;7:72-82.
Crossref - Christensen GD, Simpson WA, Younger JJ, et al. Adherence of coagulase-negative staphylococci to plastic tissue culture plates: a quantitative model for the adherence of staphylococci to medical devices. J Clin Microbiol. 1985;22(6):996-1006.
Crossref - Hassan A, Usman J, Kaleem F, Omair M, Khalid A, Iqbal M. Evaluation of different detection methods of biofilm formation in the clinical isolates. Braz J Infect Dis. 2011;15(4):305-311.
Crossref - Al-Shabib NA, Husain FM, Rehman MT, et al. Food color ‘Azorubine’ interferes with quorum sensing regulated functions and obliterates biofilm formed by food associated bacteria: An in vitro and in silico approach. Saudi J Biol Sci. 2020;27(4):1080-1090.
Crossref - Green MR, Sambrook J. Isolation of High-Molecular-Weight DNA Using Organic Solvents. Cold Spring Harb Protoc. 2017;2017(4):pdb.prot093450.
Crossref - Vuotto C, Longo F, Balice MP, Donelli G, Varaldo PE. Antibiotic resistance related to biofilm formation in Klebsiella pneumoniae. Pathog. 2014;5;3(3):743-58.
Crossref - Ferreira RL, da Silva BCM, Rezende GS, et al. High Prevalence of Multidrug-Resistant Klebsiella pneumoniae Harboring Several Virulence and β-Lactamase Encoding Genes in Brazilian Intensive Care Unit. Front Microbiol. 2019;9:3198.
Crossref - Shadkam S, Goli HR, Mirzaei B, Gholami M, Ahanjan M. Correlation between antimicrobial resistance and biofilm formation capability among Klebsiella pneumoniae strains isolated from hospitalized patients in Iran. Ann Clin Microbiol Antimicrob. 2021;20(1):13.
Crossref - Alcantar-Curiel MD, Blackburn D, Saldana Z, et al. Multi-functional analysis of Klebsiella pneumoniae fimbrial types in adherence and biofilm formation. Virulence. 2013;4(2):129-138.
Crossref - Maurya N, Jangra M, Tambat R, Nandanwar H. Alliance of efflux pumps with β-lactamases in multidrug-resistant Klebsiella pneumoniae isolates. Microb Drug Resist. 2019;25(8):1155-1163.
Crossref - Xu Q, Jiang J, Zhu Z, et al. Efflux pumps AcrAB and OqxAB contribute to nitrofurantoin resistance in an uropathogenic Klebsiella pneumoniae isolate. Int J Antimicrob Agents. 2019;54(2):223-227.
Crossref - Mathur T, Singhal S, Khan S, Upadhyay DJ, Fatma T, Rattan A. Detection of biofilm formation among the clinical isolates of Staphylococci: an evaluation of three different screening methods. Indian J Med Microbiol. 2006;24(1):25-29.
Crossref - Harika K, Shenoy VP, Narasimhaswamy N, Chawla K. Detection of Biofilm Production and Its Impact on Antibiotic Resistance Profile of Bacterial Isolates from Chronic Wound Infections. J Glob Infect Dis. 2020;12(3):129-134.
Crossref - Corehtash ZG, Khorshidi A, Firoozeh F, Akbari H, Aznaveh AM. Biofilm Formation and Virulence Factors among Pseudomonas aeruginosa Isolated from Burn Patients. Jundishapur J Microbiol. 2015;8(10):e22345.
Crossref - Reisner A, Krogfelt KA, Klein BM, Zechner EL, Molin S. In Vitro Biofilm Formation of Commensal and Pathogenic Escherichia coli Strains: Impact of Environmental and Genetic Factors. J Bacteriol. 2006;188:3572-3581.
Crossref - Panjaitan NSD, Horng YT, Cheng SW, Chung WT, Soo PC. EtcABC, A Putative EII complex, regulates type 3 fimbriae via CRP-cAMP signaling in Klebsiella pneumoniae. Front Microbiol. 2019;10(1):1558-1563.
Crossref - Mirzaie A, Ranjbar R. Antibiotic resistance, virulence-associated genes analysis and molecular typing of Klebsiella pneumoniae strains recovered from clinical samples. AMB Express. 2021;11(1)-122.
Crossref - de Cassia AMR, de Barros EMR, Loureiro NG, de Melo HRL, Maciel MAV, Lopes ACS. Presence of fimH, mrkD, and irp2 Virulence Genes in KPC-2-Producing Klebsiella pneumoniae Isolates in Recife-PE, Brazil. Curr Microbiol. 2014;69(6):824-831.
Crossref - Hamam SS, El Kholy RM, Zaki MES. Study of Various Virulence Genes, Biofilm Formation and Extended-Spectrum β-lactamase Resistance in Klebsiella pneumoniae Isolated from Urinary Tract Infections. The Open Microbiology Journal. 2019;13:249-255.
Crossref
© The Author(s) 2025. Open Access. This article is distributed under the terms of the Creative Commons Attribution 4.0 International License which permits unrestricted use, sharing, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.