ISSN: 0973-7510
E-ISSN: 2581-690X
It is critical to find an alternative therapeutic approach to combat Pseudomonas aeruginosa (P. aeruginosa) that can simultaneously reduce the occurrence of bacterial resistance. The tetraspanin CD9, a highly expressed membrane protein in melanocytes was chosen for this study because it is highly expressed in keratinocytes and has been implicated in the pathogenesis of bacterial infections in a previous study. The antimicrobial activity of CD9 peptides against the standard strain P. aeruginosa (ATCC 27853) and a clinical multidrug-resistant P. aeruginosa (MDR- P. aeruginosa) was studied using the disc diffusion method. The minimum inhibitory concentration (MIC) and minimum bactericidal concentration (MBC) of CD9 peptides were determined by broth microdilution assays with concentrations ranging from 1 mg/mL to 4.88×10-4 mg/mL. The antibiofilm activity of the CD9 peptides was also determined. CD9 peptides showed an 11.75 ± 2.36 mm inhibition zone against the standard P. aeruginosa strain but none against the MDR- P. aeruginosa. Both isolates had the same MIC value, 0.25 mg/mL. The MBC for the standard strain P. aeruginosa was 0.5 mg/mL, while for the MDR- P. aeruginosa strain, it was 1 mg/mL. CD9 peptides significantly inhibited up to 70% biofilm against both P. aeruginosa isolates. CD9 peptides showed a modest inhibitory effect against the standard strain P. aeruginosa but not against MDR- P. aeruginosa. Interestingly, CD9 peptides were found to be a good anti-biofilm treatment against both P. aeruginosa isolates. This study demonstrated that CD9 peptides have the potential to be an alternative antimicrobial treatment against P. aeruginosa.
Pseudomonas aeruginosa, Tetraspanin, Peptide, CD9, Antibacterial, Anti-biofilm, Antibiotic Resistance, Antibiotic Alternatives
P. aeruginosa is a motile rod-shaped Gram-negative bacterium that was first isolated by Carle Gessard in 1882 upon the discovery of blue-green pus and tissues close to infection.1 It is normally isolated from soil, or contaminated and moist surfaces such as bottles, sinks, medical equipment such as catheters, and food trays.2 P. aeruginosa is a leading cause of nosocomial infections such as skin and soft tissue infections, ventilator-associated pneumonia, and bacteraemia.2 Studies have reported that approximately 60% of infections in burn patients are due to P. aeruginosa and 15% of burned patients reported episodes of bloodstream infections2. In addition, Ng et al. recently reported that there have been increased rates of P. aeruginosa bacteraemia during the COVID-19 pandemic period due to pneumonia, which added to the complexity of COVID-19 symptoms and has led to longer hospitalisations.3
P. aeruginosa exhibits several mechanisms of antibiotic resistance, which is increasingly critical and limits the scope for effective treatment. Recently, P. aeruginosa has shown resistance to גeta-lactams, carbapenems, aminoglycosides, and fluoroquinolones as antibiotics.4 Antibiotic resistance is related to the prolonged and uncontrolled use of antibiotics. P. aeruginosa possesses b-lactamase enzymes, efflux pumps, horizontal gene transfer, and biofilm formation, which lead to drug resistance.4,5 In addition, P. aeruginosa has a quorum-sensing mechanism that is important for regulating virulence factors and biofilm formation.5 Biofilm allows P. aeruginosa to adhere to host cell surfaces and is more tolerant than planktonic cells or free-floating bacteria6. According to studies, 80% of all bacterial infections are associated with the formation of biofilms and antibiotic resistance that can evade the host immune response.7 Therefore, it is crucial to find an alternative and effective solution to this crisis to reduce the severity of P. aeruginosa pathologies and the phenomenon of multidrug resistance (MDR).
Tetraspanins are a superfamily of transmembrane proteins comprising of 33 members.8 Structurally, tetraspanins possess four transmembrane domains.8 Specific a-helices and small residues such as glycine and alanine in the transmembrane region exhibit high variability, allowing protein dynamics, helix-helix interactions, and homo- and heterodimerization to form the tetraspanin web or tetraspanin-enriched microdomains (TEMs).9 Given the broad tissue distribution and expression, tetraspanins were discovered to be involved as molecular organisers within the flexible extracellular loop region, mainly known as EC2, for bacterial attachment to host cells by interacting with bacterial cells via receptors for bacterial adhesins and promoting pathogenesis.9,10 Tetraspanins have redundant functions, which are involved in many aspects of cellular physiology including adhesion, fusion, invasion, signalling, immunological response, tumour metastasis and, key access to infectious diseases.8,10 Pathogens use tetraspanin microdomains to enter host cells and initiate infections.9
The tetraspanin CD9 was chosen for this study as it has been implicated in the pathogenesis of bacterial infections in a previous study. Tetraspanin-based treatments are emerging as a promising alternative to conventional antibiotics due to their targeted mode of action in inhibiting bacterial adhesion.10 Unlike conventional antibiotics, tetraspanin-based treatments do not kill bacteria rapidly, they only disrupt the interaction between bacterial adhesion molecules and host cell receptors, which prevents the bacteria from adhering to host cells and causing further colonisation.8,10 This targeted treatment is particularly effective against chronic infections, which are often associated with bacterial biofilms that can be resistant to conventional antibiotic treatment. Formerly, members of the human tetraspanin family have been patented and used to treat cancer, allergic diseases, and anaphylaxis.11,12 To the best of our knowledge, there has been no previous study of the direct antibacterial activity of tetraspanin peptides against P. aeruginosa.
Over the past decade, tetraspanins have shown a promising capability to counter various Gram-negative and Gram-positive bacteria as well as viruses, although they are still far from application in healthcare settings.13-15 In a pioneering study in 2011, recombinant tetraspanin and tetraspanin antibodies of CD9, CD63, and CD151 were shown to inhibit the attachment of Gram-negative bacteria (Neisseria meningitidis and Escherichia coli) and Gram-positive bacteria (Staphylococcus aureus and Streptococcus pneumoniae) to human epithelial cells.13 Another recent study by Ventress and colleagues (2016) showed that the short peptides derived directly from the sequence of the EC2 domain of tetraspanin CD9 had anti-adhesion activity against S. aureus in an epithelial cell line and primary keratinocytes and showed no adverse effects on host cells.14
Tetraspanins are eukaryotic receptors involved in direct and indirect bacterial attachment to host cells, which consequently leads to cell invasion, and short peptides derived from tetraspanins are known to have the potential to block the bacterial-host cell interaction, thus preventing host cell invasion by pathogens.8 Thus, in this study, we aimed to gain further insights into the direct effects of tetraspanin-based treatment in combination with blocking the adhesion receptor on host cells, which will confirm whether tetraspanin-based treatments exhibit dual modes of action. Therefore, we aimed to investigate for the first time the direct antibacterial effects of CD9 peptides against P. aeruginosa and to test the antibacterial and antibiofilm potential of CD9 peptides against two P. aeruginosa isolates, a standard strain (ATCC 27853) and a clinical MDR strain. We hypothesised that the CD9 peptides might have a direct inhibitory effect on bacterial isolates.
Bacterial strains
Two bacterial strains were investigated in this study: a P. aeruginosa standard strain (ATCC 27853, which served as a representative of the non-resistant strain) commonly used as a model strain for antibiotic susceptibility testing, and a multidrug-resistant P. aeruginosa (MDR- P. aeruginosa, resistant to fluoroquinolone, third-generation cephalosporins beta-lactam, and aminoglycoside antibiotics) isolated from a patient with a chest infection. The bacteria used in this study were provided by the Cluster of Pathology & Laboratory Medicine (CPDRL) UiTM Specialist Centre. The profile of drug resistance was also acquired from the databases of CPDRL. Bacteria were grown on blood agar (BA, Oxoid, UK) and maintained on the BA plate at 4÷C (short-term storage) for further analysis. The overnight bacterial suspension was prepared by inoculating bacterial colonies in Luria Bertani broth (LB, Oxoid, UK) and turbidity was adjusted to 0.5 McFarland’s (Biomerieux, USA).
CD9 Peptides
Tetraspanin CD9 was selected because of its high expression levels in keratinocytes and its involvement in the pathogenesis of bacterial infections.9 Tetraspanin CD9 peptides (MW:2195.44) were derived from the primary sequence of the large extracellular (EC2) domain of CD9, represented by a 15 amino acids sequence (EPQRETLKAIHYALN) with tetramethyl rhodamine (TMR)-tagging, as used in the pioneering work of Ventress et al. CD9 peptides were purchased from GenScript (United States) at 98% purity in lyophilised form.
Antibacterial susceptibility test
Antibacterial susceptibility testing was performed using the disc diffusion assay (Kirby-Bauer) based on Clinical and Laboratory Standards Institute (CSLI-M100) guidelines. Approximately 100 µL of the bacterial suspension (0.5 Mc Farland) was inoculated onto Mueller Hinton agar (MHA, Oxoid, Basingstoke, UK) by swabbing the entire agar plate in a back-and-forth motion. Then, 20 µL of the dissolved CD9 peptides (1 mg/mL) and 20 µL of sterile 100% (v/v) DMSO were dropped onto the sterile blank antibiotic disc and it was placed in the middle of the plate. Standard antibiotic discs (Oxoid, Basingstoke, UK) including cefotaxime (30 µg), imipenem (10 µg), and polymyxin B (300 units) were placed around it as positive controls. The plates were incubated upside down overnight at 37°C. The result of the antibacterial susceptibility test was read as the diameter of the zone of inhibition, which was measured with a ruler to the nearest millimetre (mm).
Minimum inhibitory concentration (MIC)
MIC testing was performed to determine the lowest concentration of CD9 peptides that prevents visible growth of P. aeruginosa. The MIC of CD9 peptides was measured according to a modification of the National Committee for Clinical Laboratory Standards (NCCLS) broth microdilution method, as previously described by Klubthawee et al. The MIC procedure was performed using a sterilised 96-well plate (n.a.) by aliquoting 25 µL of a standardised microbial suspension adjusted to 0.5 McFarland scale, and 25 µL of the serially diluted CD9 peptides was added to the wells at decreasing concentrations from 1 mg/mL to 4.88×10-4 mg/mL. The plate contained two control sets: a sterility control containing LB broth alone with sterile distilled water, and a growth control containing a bacterial culture with sterile LB broth media only. Plates were incubated at 37÷C for 24 hours. After incubation, absorbance values were measured at 595 nm using a microplate reader (Perkin Elmer, Victor™ X5, UK). The MIC of CD9 peptides for each P. aeruginosa isolate was recorded in the first clear well; the lowest concentration (µM) of peptides that showed no turbidity of P. aeruginosa was considered the MIC.
Minimum Bactericidal Concentration
The minimum bactericidal concentration (MBC) was determined to be the lowest concentration at which CD9 peptides were able to inhibit 99.9% growth of P. aeruginosa on the media (no detectable bacterial growth after incubation), following the method of Klubthawee et al. with some modifications. The MBC was measured by subculturing the samples from the wells used for the MIC determination on fresh agar plates. The broths used for the MBC determination were diluted @ 1:10 with sterile distilled water. One hundred microliters of the diluted broths were inoculated onto the BA by swabbing the entire agar plate in back-and-forth motions and allowing it to dry for several minutes. The plates were incubated overnight at 37°C. After incubation, the MBC value was determined as the lowest concentration at which no P. aeruginosa colony was formed on the agar plate.
Biofilm formation inhibitory test
This procedure was performed to evaluate the inhibition of mature biofilm by CD9 peptides using a crystal violet assay (CV), following the method employed by Chen and team with some modifications.31 The biofilm biomass was compared between the treated and untreated bacterial suspensions. Fifty microliters of bacterial suspension (0.5 Mc Farland) were added into the wells of a flat-bottomed 96-well microtiter plate (Jet Bio-Filtration, China). The biofilms were allowed to grow in LB media at 37°C for 48 hours. After incubation, the culture was discarded, and the wells were rinsed with sterile distilled water. An equal amount of CD9 peptides (1 mg/mL) suspended in LB broth were added to the preformed biofilm and sterile distilled water was added for the untreated wells. The plate was incubated at 37°C for another 24 hours. After incubation, the culture was discarded, and the wells were carefully washed three times with sterile distilled water. Adherent biofilms were fixed with 200 µL of 100% (v/v) methanol (Merck, USA) for 15 minutes, which was then discarded and washed again three times with sterile distilled water to remove non-adherent cells. The plate was air-dried at room temperature for 30 minutes, and then 200 µl of crystal violet (Labstain, Malaysia) was added and incubated at room temperature for 15 minutes. The excess stain was removed by washing with sterile distilled water and blotting on clean tissue papers. The adherent dye was resuspended with 200 µL of 30% (v/v) glacial acetic acid (Merck, USA) for 15 minutes at room temperature. Briefly, the contents of each well containing the dissolved dye were mixed by pipetting 125µL of the solution and transferred to a new plate. The optical density (OD) of the adherent biofilm was determined at 595 nm. The OD value of the treated wells was compared with the OD value of the untreated wells to observe the inhibitory effect of CD9 peptides. The rate of biofilm inhibition was calculated using the formula of [1- (OD treated /OD untreated)] × 100.31
Statistical analyses
All experiments were performed at least in triplicate. The Statistical Package for the Social Sciences (SPSS) version 28.0 (SPSS Inc., Chicago, US) was used for statistical analysis. Data represent the mean of three replicates ± standard deviation (SD). Data were analysed using the Mann-Whitney Test.
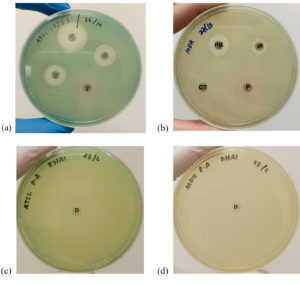
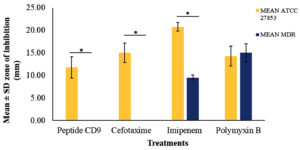
The susceptibility test showed that the CD9 peptides had an inhibitory effect only against the standard strain of P. aeruginosa, with a mean diameter of the zone of inhibition of 11.75 ± 2.36 mm, which was comparable to the effects of the other conventional antibiotics (Figure 1a). On the other hand, no zone of inhibition was observed for CD9 peptides against the MDR-P. aeruginosa isolate, which was comparable to the effect of cefotaxime (Figure 1b). The antimicrobial effect of CD9 peptides and other conventional antibiotics against both P. aeruginosa isolates is summarised in Figure 2. There was a significant difference at P ≤ 0.05 between the zones of inhibition formed by CD9 peptides against standard P. aeruginosa and MDR- P. aeruginosa. In addition, a susceptibility test for DMSO against both P. aeruginosa isolates was also performed as a control to ensure that DMSO, as a solvent for the CD9 peptides, did not interfere with the antimicrobial effect of the CD9 peptides. This study showed that DMSO did not form a zone of inhibition against either P. aeruginosa isolate (Figures 1c and d).
Figure 1. Sensitivity of CD9 peptides against P. aeruginosa standard strain (ATCC 27853) (a) and MDR- P. aeruginosa (b). No zone of inhibition by CD9 peptides against MDR isolate was detected and a similar result was obtained by cefotaxime. Susceptibility tests of 100% (v/v) DMSO against standard P. aeruginosa strain (c) and MDR- P. aeruginosa (d). DMSO caused no zone of inhibition against both isolates. Red circle: zone of inhibition by CD9 peptides, P: CD9 peptides, CT: Cefotaxime, IP: Imipenem, PB: Polymixin, D: DMSO
Figure 2. Antibacterial effects of CD9 peptides and control antibiotics against standard strain P. aeruginosa and MDR- P. aeruginosa isolate. Results are shown in mean ± SD of three replicates. * P ≤ 0.05
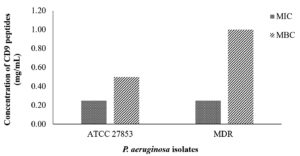
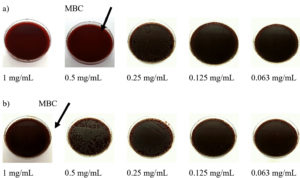
After visual inspection of growth inhibition, the MIC and MBC were examined. The MIC and MBC values of CD9 peptides for both standard strain P. aeruginosa and MDR- P. aeruginosa are shown in Figure 3. The MIC values were statistically determined based on three repeated experiments. The MIC was also determined visually by the naked eye, with no turbidity observed in the well, and the result was compared with the absorbance value. The MIC of CD9 peptides was 0.25 mg/mL for both P. aeruginosa isolates. Surprisingly, a high MBC value was observed for both isolates. For the standard strain P. aeruginosa, the MBC was the first diluted concentration, 0.5 mg/mL, while the MBC for MDR- P. aeruginosa was the initial concentration, 1 mg/mL (Figure 3 and Figure 4).
Figure 3. Minimum inhibitory concentration (MIC) and minimum bactericidal concentration (MBC) of CD9 peptides against P. aeruginosa. The results shown are of three replicates
Figure 4 (a). Representative plates of minimum bactericidal concentration of CD9 peptides against standard strain P. aeruginosa and (b) MDR- P. aeruginosa isolate
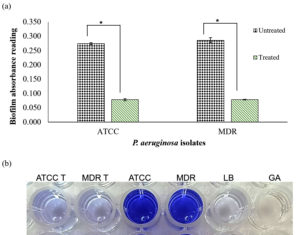
To evaluate the biofilm formation, the biofilm biomass was assessed by final crystal violet intensity. The colour intensity of crystal violet was consistent with the absorbance measurement, with a darker colour representing stronger biofilm formation and a lighter colour representing weaker biofilm formation. CD9 peptides were introduced to the overnight bacterial suspension and compared to the distilled water suspension. Biofilm formation was significantly reduced in both P. aeruginosa isolates when the cells were treated with CD9 peptides (Figure 5). The treated suspension had similar biofilm biomass to the control group. The analysis of mean absorbance values of both the treated strains of P. aeruginosa, as compared to the untreated strains, showed a statistically significant difference with a P-value ≤ 0.05. Approximately, 73.9% of the biofilm formation of the P. aeruginosa standard strain and 74.6% of the biofilm formation of MDR- P. aeruginosa was successfully inhibited by CD9 peptides treatment. However, there was no significant difference in biofilm biomass between the untreated standard strain and the MDR- P. aeruginosa isolate (P-value ≥ 0.05). Thus, the results showed that CD9 peptides have potential antibiofilm activity against P. aeruginosa.
Figure 5 (a). Biofilm inhibition assay by CD9 peptides against standard strain P. aeruginosa and MDR- P. aeruginosa isolates. (b). Representative wells photographed of biofilm inhibition by crystal violet assay against P. aeruginosa biofilm in the microtiter plate. T: treated with CD9 peptides. Controls: Luria Bertani (LB), Glacial acetic acid (GA). The data were shown as mean ± SD of absorbance reading of three replicates. * P ≤ 0.05
The development of a new antimicrobial treatment with low resistance risk is the current goal to combat microbial resistance and chronic infections. P. aeruginosa is known to utilise many resistance mechanisms including intrinsic, acquired, and adaptive resistance mechanisms, to resist antibiotics.16 P. aeruginosa usually has low outer membrane permeability, efflux pump systems, antibiotic-inactivating enzymes, and biofilm formation to confine and expel antibiotics.16 To our knowledge, this is the first study to address the potential direct antimicrobial and antibiofilm inhibitory effects of tetraspanin peptides against P. aeruginosa, as there are currently no reports highlighting the direct effect of tetraspanin peptides against this bacterium.
In this study, DMSO was used to dissolve the CD9 peptides, a water-insoluble drug, according to the manufacturer’s protocol. DMSO is known to be widely used as a solvent for water-insoluble drugs.17 It is well-established that high concentrations of DMSO attenuate the virulence factors of P. aeruginosa.17 Interestingly, however, this study showed that DMSO did not affect the growth of either of the P. aeruginosa isolates. This finding thereby ruled out DMSO influence on the efficacy of the antibacterial activity of CD9 peptides to inhibit bacteria. In addition, even though pure DMSO was used to dissolve the peptide at early stages, the final concentration was usually kept lower than the stock, as it had been diluted.
This study showed that CD9 peptides have a small inhibitory effect on the standard strain of P. aeruginosa, as seen with the small zone of inhibition formed on the agar plate. There was no green pigment formed by P. aeruginosa colonies surrounding the CD9 peptide disc on the agar plate, which is known to be released by the production of pyocyanin (blue-green) and pyoverdins1. The zone of inhibition caused by CD9 peptides was not as clear as the zone of inhibition formed by the antibiotics, which may be due to the fluorescent pink colour of the peptide. In addition, the potency of the antibiotics was stronger than that of CD9 peptides in forming a clear zone of inhibition. The efficacy of antibiotics is well-known in healthcare settings and they have been well-regulated by the FDA. Unfortunately, this study showed that a higher concentration of CD9 peptides may be needed to fully inhibit the growth of both standard and MDR- P. aeruginosa strains. The results of the current study support the theory put forward by Ventress et al. and Green et al. that tetraspanin peptides and recombinant tetraspanin peptides do not affect bacteria when added before infection to the cells. However, because the inhibition of bacterial growth does not result in bacterial death, this method cannot distinguish between the bactericidal and bacteriostatic effects of CD9 peptides.
Based on the cut-off point, the MIC and MBC results showed that the effects of CD9 peptides were considered bactericidal against the standard strain of P. aeruginosa, as the ratio of MBC to MIC was less than 4 (0.5 mg/mL: 0.25 mg/mL).18 In addition, the CD9 peptides were considered bacteriostatic against MDR- P. aeruginosa, as the ratio of MBC to MIC was equal to 4 (1 mg/mL: 0.25 mg/mL).18 This is because this treatment did not fully kill the bacteria and only slightly inhibited bacterial growth. This phenomenon can be beneficial for humans, as this treatment has been shown previously to cause no toxicity to the cells, which in turn may help in strengthening immunity and subsequently eliminating the bacteria.13,14,19,20
It was reported by Benkova et al. that the diameter of the zone of inhibition is inversely proportional to the MIC value; that is, the smaller the zone of inhibition, the higher the MIC value, and thus, higher the concentration of antimicrobial needed.21 Our study shows that a higher concentration of CD9 peptides is needed to inhibit the growth of P. aeruginosa. With these high values, a higher dose of CD9 peptides could be used as an adjuvant in any antimicrobial drug, but still be safely administered, especially if the designed peptide was less than 100 amino acids and well-regulated under the Federal Food, Drug, and Cosmetic Act (FD&C) of the FDA.22 The results from the current study also suggest that CD9 peptides may only achieve significant therapeutic effects at the tissue level by inhibiting bacterial growth by neutralising the charge on the bacterial cell membrane and/or altering the permeability of bacterial cells.20,23
Biofilm is more resistant to physical and chemical changes by different antibacterial agents up to 1000 times more than bacteria in their planktonic growth.5,6 Biofilms are a complex matrix of microbial cells composed of proteins, extracellular DNA, and exopolysaccharides, which in P. aeruginosa include alginate, Psl, and Pel, and which help the bacterial cells evade antimicrobial agents.23 Although CD9 peptides showed no visible inhibitory effect against the growth of the MDR- P. aeruginosa isolate on the agar plate, they successfully inhibited 70% of the biofilm formation of both the standard strain P. aeruginosa and the MDR- P. aeruginosa in the microtiter plate. This study showed that the untreated P. aeruginosa isolates formed a thick biofilm layer on the bottom surface of the well in the absence of any effector or inhibitor other than the broth medium and the presence of water. This condition provided an aqueous and moist surface for the bacteria to develop a biofilm attached to the bottom surface of the plate. Since DMSO did not show any inhibition in the bacterial growth in the previous section, it is suggested that it may not interfere with the efficacy of CD9 peptides as an anti-biofilm agent.
Biofilm formation is the typical characteristic of P. aeruginosa, which is often present on contaminated and moist surfaces such as bottles, sinks, and medical devices such as urine catheters.4 In addition, biofilm formation can also be favoured by the changes in the pH of the environment, which may be due to secreted bacterial metabolites.24,25 This study confirmed the inhibitory effect of CD9 peptides on P. aeruginosa biofilm formation. The effect of CD9 peptides could be due to the cross-reaction of CD9 peptides with P. aeruginosa, which may limit the ability of bacteria to form a biofilm under static conditions and may inhibit the adhesion of biofilm to host surfaces, thus reducing the potential for pathogenesis at a later stage. This inhibitory mechanism may be due to the ability of CD9 peptides to destabilise the biofilm on the surface of the wells, making the bacteria less susceptible to cell adhesion.23,26 Moreover, CD9 peptides can interfere with acidic environments during biofilm formation, as demonstrated by the reduction in the stained crystal violet.24,27 In general, biofilm formation is associated with the chronicity of bacterial infectious diseases because biofilms act as a barrier that limits the penetration of antimicrobial agents into bacteria.28 We concluded that the CD9 peptides can reduce chronicity and recurrent P. aeruginosa infections by inhibiting biofilm formation, with a lower risk of resistance development. However, crystal violet staining has the disadvantage of staining both viable and non-viable cells adhering to the surface.
This study successfully produced additional insights into the antimicrobial action of CD9 peptides against P. aeruginosa, in addition to the known anti-adherence properties. The findings in this study were consistent with our recent work which reported the antimicrobial effect of CD9 peptides against Gram-positive bacteria causing axillary malodor.29 In addition, this mode of action may be similar to D-mannose against uropathogenic Escherichia coli, which exploits strong anti-adherence properties and does not exert “antibiotic-like” activity such as destroying and killing the bacteria.30 Furthermore, this study proved that the tetraspanin CD9 peptides have an indirect inhibitory effect on bacterial attachment to host cells due to their direct inhibitory effects on P. aeruginosa and reducing the bacterial load on host cells. It is reported that the antimicrobial activity of antimicrobial peptides is influenced by a few physio-chemical properties including the length, charge, hydrophobicity, amphipathicity and hydrophobic or hydrophilic angle of the peptides.26 Positively charged amino acids of CD9 peptides such as lysine and glutamine enable the binding to the negatively charged bacterial surfaces, inducing electrostatic forces, that may cause the penetration of CD9 peptides to the cytoplasmic membrane of P. aeruginosa, without causing membrane damage and rupture.20,26
Although these results suggest that tetraspanin-based treatments cause little direct inhibition of P. aeruginosa, they prove that tetraspanins on tetraspanin enriched microdomain facilitate P. aeruginosa binding to host cells, recruit cell surface proteins, and provide bacterial adhesion interactions.9,31 Moreover, these results reflect the role of tetraspanins as indirect receptors for bacterial pathogenesis, perhaps because of their properties not only as receptors but as mediators of the adhesion platform for bacterial pathogenesis.8,10 The treatment studied here, derived from the EC2 domain of CD9, will mainly disturb the TEMs, altering the microdomain and simultaneously disrupting the interaction of tetraspanins with partner proteins on the surface of host cells.9,14 However, this study has some limitations as it was the first attempt to investigate the direct antimicrobial activity of tetraspanin CD9 against bacterial isolates. Further experiments should include other bacterial species to fully demonstrate the antimicrobial effects of tetraspanin CD9 peptides, and should further investigate the related virulence factors of P. aeruginosa, to thoroughly understand the mode of action of CD9 peptides. Due to the high cost of peptides, further research also needs to include another tetraspanins to explore the antimicrobial properties of the tetraspanin family; those shown in this study may not reflect the entire tetraspanins family.
Overall, this study demonstrated that CD9 peptides not only work as anti-adhesion inhibitors but may also have a direct effect on P. aeruginosa. This study showed a weak antibacterial effect for CD9 peptides, but they had a strong inhibitory effect on the biofilm formation of P. aeruginosa and, in turn, prevented the adhesion of bacteria to surfaces. While the direct effect was not significant compared to their anti-adhesion activity, it may still contribute to their overall efficacy as antibacterial agents. Our ongoing research will provide insights into the interactions of bacterial cells with host cells that have an impact on CD9 peptides in anti-adhesion activities. The combination of the direct bacterial inhibitory effect, the inhibition of biofilm formation, and anti-adhesion properties highlight the potential of tetraspanin-based treatments as effective agents to combat bacterial infections.
ACKNOWLEDGMENTS
The author would like to thank the Institute for Medical Molecular Biotechnology (IMMB), Universiti Teknologi MARA (UiTM) for the valuable support.
CONFLICT OF INTEREST
The authors declare that there is no conflict of interest.
AUTHORS’ CONTRIBUTION
KM, SAR and HAT conceptualized and designed the study. KM performed literature review, data acquisition and experimental studies.KM, SAR and HAT performed data and statistical analysis. KM wrote the manuscript. KM, SAR and HAT reviewed and edited the manuscript. All authors read and approved the final manuscript for publication.
FUNDING
This study was funded by Universiti Teknologi MARA, Malaysia, with reference numbers 600-RMC/GIP 5/3 (088/2021) and 600-IRMI 5/3/LESTARI (012/2019).
DATA AVAILABILITY
All datasets generated or analyzed during this study are included in the manuscript.
ETHICS STATEMENT
Not applicable.
- Gonחalves T, Vasconcelos U. Colour Me Blue: The History and the Biotechnological Potential of Pyocyanin. Molecules. 2021;26(4):927.
Crossref - Horcajada JP, Montero M, Oliver A, et al. Epidemiology and treatment of multidrug-resistant and extensively drug-resistant Pseudomonas aeruginosa infections. Clin Microbiol Rev. 2019;32(4):e00031.
Crossref - Ng QX, Ong NY, Lee DYX, et al. Trends in Pseudomonas aeruginosa (P. aeruginosa) Bacteremia during the COVID-19 Pandemic: A Systematic Review. Antibiotics. 2023;12(2):409.
Crossref - Pachori P, Gothalwal R, Gandhi P. Emergence of antibiotic resistance Pseudomonas aeruginosa in intensive care unit; a critical review. Genes Dis. 2019;6(2):109-119.
Crossref - Perez-Perez M, Jorge P, Perez Rodriguez G, Pereira MO, Lourenחo A. Quorum sensing inhibition in Pseudomonas aeruginosa biofilms: new insights through network mining. Biofouling J Bioadhesion Biofilm Res. 2017;33(2):128-142.
Crossref - Moradali MF, Ghods S, Rehm BHA. Pseudomonas aeruginosa Lifestyle: A Paradigm for Adaptation, Survival, and Persistence. Front Cell Infect Microbiol. 2017;7:39.
Crossref - Chakraborty P, Ghosh Dastidar D, Paul P, et al. Inhibition of biofilm formation of Pseudomonas aeruginosa by caffeine: a potential approach for sustainable management of biofilm. Arch Microbiol. 2020;202(3):623-635.
Crossref - Murad K, Ab Rahim S, Al-Talib H. Antimicrobial Effects of Tetraspanins: A New Turnabout in Treatment of Microorganisms. Malays J Med Sci MJMS. 2022;29(4):6-13.
Crossref - Umeda R, Satouh Y, Takemoto M, et al. Structural insights into tetraspanin CD9 function. Nat Commun. 2020;11(1):1606.
Crossref - Karam J, Mיresse S, Kremer L, Daher W. The roles of tetraspanins in bacterial infections. Cell Microbiol. 2020;22(12):1-12.
Crossref - Marni R, Chakraborty A, Malla R. Oncogenic tetraspanins: Implications for metastasis, drug resistance, cancer stem cell maintenance and diagnosis of leading cancers in females. Gene Rep. 2022;27:101548.
Crossref - Robert JMH, Amoussou NG, Mai HL, Loge C, Brouard S. Tetraspanins: useful multifunction proteins for the possible design and development of small-molecule therapeutic tools. Drug Discov Today. 2020;26(1):56-58.
Crossref - Green LR, Monk PN, Partridge LJ, Morris P, Gorringe AR, Read RC. Cooperative role for tetraspanins in adhesin-mediated attachment of bacterial species to human epithelial cells. Infect Immun. 2011;79(6):2241-2249.
Crossref - Ventress JK, Partridge LJ, Read RC, Cozens D, MacNeil S, Monk PN. Peptides from tetraspanin CD9 are potent inhibitors of Staphylococcus aureus adherence to keratinocytes. PLoS ONE. 2016;11(7):1-17.
Crossref - Fast LA, Mikulicic S, Fritzen A, et al. Inhibition of tetraspanin functions impairs human papillomavirus and cytomegalovirus infections. Int J Mol Sci. 2018;19(10):3007.
Crossref - Pang Z, Raudonis R, Glick BR, Lin TJ, Cheng Z. Antibiotic resistance in Pseudomonas aeruginosa: mechanisms and alternative therapeutic strategies. Biotechnol Adv. 2019;37(1):177-192.
Crossref - Guo Q, Wu Q, Bai D, et al. Potential Use of Dimethyl Sulfoxide in Treatment of Infections Caused by Pseudomonas aeruginosa. Antimicrob Agents Chemother. 2016;60(12):7159-7169.
Crossref - Wald-Dickler N, Holtom P, Spellberg B. Busting the Myth of “Static vs Cidal”: A Systemic Literature Review. Clin Infect Dis. 2018;66(9):1470-1474.
Crossref - Ashraf S, Chaudhry U, Raza A, Ghosh D, Zhao X. In vitro activity of ivermectin against Staphylococcus aureus clinical isolates. Antimicrob Resist Infect Control. 2018;7:27.
Crossref - Lei J, Sun L, Huang S, et al. The antimicrobial peptides and their potential clinical applications. Am J Transl Res. 2019;11(7):3919-3931.
- Benkova M, Soukup O, Marek J. Antimicrobial susceptibility testing: currently used methods and devices and the near future in clinical practice. J Appl Microbiol. 2020;129(4):806-822.
Crossref - Boutin JA, Tartar AL, van Dorsselaer A, Vaudry H. General lack of structural characterization of chemically synthesized long peptides. Protein Sci. 2019;28(5):857-867.
Crossref - Kamali E, Jamali A, Ardebili A, Ezadi F, Mohebbi A. Evaluation of antimicrobial resistance, biofilm forming potential, and the presence of biofilm-related genes among clinical isolates of Pseudomonas aeruginosa. BMC Res Notes. 2020;13(27):1-6.
Crossref - Lin Q, Pilewski JM, Di YP. Acidic Microenvironment Determines Antibiotic Susceptibility and Biofilm Formation of Pseudomonas aeruginosa. Front Microbiol. 2021;12:747834.
Crossref - Wu X, Al Farraj DA, Rajaselvam J, et al. Characterization of biofilm formed by multidrug resistant Pseudomonas aeruginosa DC-17 isolated from dental caries. Saudi J Biol Sci. 2020;27(11):2955-2960.
Crossref - Klubthawee N, Adisakwattana P, Hanpithakpong W, Somsri S, Aunpad R. A novel, rationally designed, hybrid antimicrobial peptide, inspired by cathelicidin and aurein, exhibits membrane-active mechanisms against Pseudomonas aeruginosa. Sci Rep. 2020;10:9117.
Crossref - Roy PK, Ha AJW, Mizan MdFR, et al. Effects of environmental conditions (temperature, pH, and glucose) on biofilm formation of Salmonella enterica serotype Kentucky and virulence gene expression. Poult Sci. 2021;100(7):101209.
Crossref - Brindhadevi K, LewisOscar F, Mylonakis E, Shanmugam S, Verma TN, Pugazhendhi A. Biofilm and Quorum sensing mediated pathogenicity in Pseudomonas aeruginosa. Process Biochem. 2020;96:49-57.
Crossref - Al-Talib H, Abdulwahab MH, Murad K, Amiruddin ND, Mohamed NN. Antimicrobial Effects of Tetraspanin CD9 Peptide against Microbiota Causing Armpit Malodour. Antibiotics. 2023;12(2):271.
Crossref - Scribano D, Sarshar M, Prezioso C, et al. d-Mannose Treatment neither Affects Uropathogenic Escherichia coli Properties nor Induces Stable FimH Modifications. Molecules. 2020;25(2):316.
Crossref - Chen X, Thomsen TR, Winkler H, Xu Y. Influence of biofilm growth age, media, antibiotic concentration and exposure time on Staphylococcus aureus and Pseudomonas aeruginosa biofilm removal in vitro. BMC Microbiol. 2020;20(1):264.
Crossref
© The Author(s) 2023. Open Access. This article is distributed under the terms of the Creative Commons Attribution 4.0 International License which permits unrestricted use, sharing, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.