ISSN: 0973-7510
E-ISSN: 2581-690X
The increase of pathogens in milk is threatening for the human beings. This is an investigation on overall microbiological analysis of raw and pasteurized milk as well as the milk-based beverages and also determining the antibiotic resistance pattern of isolated Escherichia coli and Staphylococcus aureus. A total of 100 samples (raw milk, pasteurized milk, mattha, lassi and laban) were taken from various locations of the capital city of Bangladesh, Dhaka. Total Viable Count, Total Coliform Count and Yeast and Moulds Count were performed as the microbiological inspection of selected samples. E.coli and Staphylococcus aureus were identified by conducting morphological analysis, gram-staining and biochemical tests. Antibiotic resistance pattern of isolated Escherichia Coli and Staphylococcus aureus were also detected with 11 commonly used antibiotics by conducting disc-diffusion method, following the CLSI guideline. The TVC range was the highest in raw milk samples (3.8×104 – 4.1×108 cfu/ml), and the lowest in pasteurized milk samples (1.2×102 – 5.4×103 cfu/ml), while 70% raw milk and 10% pasteurized milk samples strains were above the acceptable limit of Food and Drug Administration (FDA). Thirty-six Escherichia coli and thirty-two Staphylococcus aureus were isolated from all the 100 milk and milk-based beverage samples. The isolated Escherichia coli strains were most resistant to Penicillin G (81.58%), Erythromycin (78.94%) and Ampicillin (73.68%), and isolated Staphylococcus aureus strains were most resistant to Penicillin G (90.62%), Ampicillin (81.25%) and Methicillin (71.87%). respectively. Public awareness is needed to reduce the redundant use of antibiotics.
Raw milk, Pasteurized milk, Disc-diffusion method, Antibiotic resistance pattern, Beverages
Milk is considered as vital food in terms of nutrition, especially for children and adults, and it is pondered as significant to compensate the protein deficiency in human and other mammals1. Milk can be contaminated by various sources during the milking procedure in dairy farms to the milk industry- consequently, microbiological quality assessment along with the entire production chain is crucial to take obligatory measure to indemnify human health2. Because of the short shelf life and higher moisture content milk is considered as an immense growing medium for pathogen such as, Campylobacter jejuni, Staphylococcus aureus, Escherichia coli, Yersinia enterocolitica, Listeria monocytogenes, which can cause different food-borne diseases3. Pasteurization is the method of heating milk for a certain time at a predetermined temperature to destroy pathogenic microbes and this process can uplift the shelf life of a food product but, it doesn’t perish the slow growing and spore producing microorganisms4. Inappropriate handling after pasteurization can re-contaminate the pasteurized milk5. Pasteurization of milk and milk products is an emerging requirement of consumers found in developed countries. It is compulsory for all milk samples to be pasteurized in Canada, and no one can sell and distribute unpasteurized milk illegally under the regulation of Food and Drug6. FAO, FDA, EEC are the leading regulatory authorities for milk and milk products all over the world7. The acceptable limits for raw and pasteurized milk given by FDA are, TVC, <1×105 (cfu/ml) and <1×104 (cfu/ml) and coliform <1×103 and <10 (cfu/ml) respectively but no E.coli is acceptable in 1ml of raw milk and pasteurized milk8. Enterohemorrhagic Escherichia coli O157:H7 is the most dominant food borne pathogen causing different diseases in human9. Staphylococcus aureus is commonly known as pathogenic bacteria, which is found in hospitals and in the community. There are several foodborne intoxications caused by Staphylococcus aureus which are connected to the contaminated milk consumption10-12. Fungi are conventional contaminants of milk and dairy products and they can generate damages in milk, such as bad odor and flavor, decolonization, disorganization of structure and as well as economic losses13-16. Moulds that causes spoilage like Aspergillus and Penicillium, produce fungal toxins in milk and dairy products17-20. The prevalence of antibiotic resistance in Escherichia coli and Staphylococcus aureus, with high virulence potential is alarming in south Asia. Scarcity of appropriate data on milk-borne diseases and pathogens from countries with high infection record, such as Bangladesh, obstructs the controlling of disease and infections11. In order to have milk and milk products free from contamination of bacteria and other microbes, Hazard Analysis Critical Control Point (HACCP) must be obeyed, from milk collection through milk processing to milk storage22.
In Bangladesh, although a lot of work has been conducted based on milk as well as milk based products for assuring quality and welfare, but these are not sufficient at all. In this regard, this study was planned after considering the public health aspect of Bangladesh to ensure the best quality of milk and dairy based products for consumption of mankind by inspection and investigation of the pathogenic microorganisms as well as find out the effective antibiotics against food-borne pathogens.
Collection of Samples
An integrate set of 100 samples (n=100) were taken from various locations of the capital city of Bangladesh, Dhaka including, raw cow milk (n=20), pasteurized milk (n=20), mattha (n=20), lassi (n=20) along with laban (n=20). Among them, dairy farms were the collection source of raw cow milk samples whereas branded pasteurized milk samples were collected from grocery shops, also the mattha, lassi, labang (traditional milk based beverages of Dhaka city) samples were collected from local street vendors. Before sample collection the collecting tubes were cleaned aseptically23. The samples were analyzed immediately in the laboratory.
Sample Processing
Tenfold serial dilution of each sample was prepared in autoclaved saline water. Initially one milliliter of raw sample was mixed with nine milliliter of saline in test tube in order to prepare 101 dilutions and this process was repeated until 107 dilutions for every samples24-25.
Total Viable Count (TVC)
0.1 ml samples from 101–107 dilution were poured with fresh Plate Count Agar (PCA) plate. Incubation was done for 24 hours at 37°C. After that, colonies on plates were observed. Screening was done in order to identify isolated colonies and the real number of bacteria was counted in colony forming unit in (cfu/ml)26-28.
Total Coliform Count (TCC)
Total Coliform Count was done by applying spread plate method on Mac Conkey agar medium. 0.1 ml of each samples from, 101–105 dilution were spread on Mac Conkey agar medium. Pink colonies were considered for the counting of Total Coliform Count (cfu/ml) after the period of incubation26,27,29.
Yeast & Moulds Count (YMC)
For counting yeast and mold, 0.1 milliliter of samples from 101–105 dilution were spread properly on sterile Potato Dextrose Agar (PDA) plate. The PDA was mixed with 10% of lactic acid before it confers to the petri dish. The procedure was done by following ideal spread plate method. After incubation, the number of viable yeast & mould from plates had been counted in (cfu/ml)30-32.
Isolation and identification of Escherichia coli and Staphylococcus aureus
Spread plate technique on EMB and BPA agar media had done for isolation of desired E.coli and S. aureus colonies respectively. E.coli and S. aureus were identified by conducting morphological, gram-staining and biochemical tests (Catalase, Oxidase, Mobility, Indole, MR and VP)23.
Antibiogram of isolated Escherichia coli and Staphylococcus aureus
Kirby-Bauer procedure was applied to examine antimicrobial drug resistance pattern of pathogens33. The antibiotics tested for both E.coli and for S. aureus were – Methicillin (MET) 5μg, Penicillin G (P) 10μg, Ampicillin (AMP) 25μg, Amoxycillin (AML) 25μg, Gentamicin (GEN) 10μg, Streptomycin (S) 10μg, Azithromycin (AZM) 15μg, Erythromycin (E) 35μg, Ciprofloxacin (CIP) 5μg, Tetracycline (TIC) 30μg and Imipenem (IMP) 10μg.The Mueller-Hinton agar plates (containing pure culture and antimicrobial discs) were examined after proper incubation and zone diameters of each antibiotic discs of complete inhibition were measured through the millimeter scale. The results were translated according to the interpretation table of the CLSI and EUCAST34, European Committee on Antimicrobial Susceptibility Testing (2018). European Committee on Antimicrobial Susceptibility Testing (2019)35,23,36,37. The zone diameter results were compared with both CLSI and EUCAST standard zone diameter break points because, all the antibiotic resistance break points for each bacteria were not available in only one guideline. Escherichia coli ATCC@ 25922 and Staphylococcus aureus ATCC@ 29213 were used as control strains.
Microbiological assessment of raw milk and pasteurized milk
The TVC range of raw cow milk samples was the highest (3.8×104 – 4.1×108 cfu/ml) whereas the lowest TVC range was observed in case of pasteurized milk samples (1.2×102 – 5.4×105 cfu/ml). In terms of TVC, only 30% of raw cow milk had been found safe for customer consumption considering the limits given by FDA while, 90% of the pasteurized branded milk was of good quality (Fig. 1). 30% of raw milk was contaminated with coliform and six E.coli strain was detected (Table 1). The highest yeast and moulds were observed in raw milk (6.1×102 cfu/ml), while 40% found beyond the acceptable limit of FDA (Table 1).
Table (1):
Microbial count of raw cow milk and branded pasteurized milk in different microbiological parameters.
Microbiological testing parameters |
Microorganisms in raw milk (cfu/ml) |
FDA standard for raw milk (cfu/ml) |
Number and percentage of raw milk beyond the acceptable limit |
Microorganisms in pasteurized milk (cfu/ml) |
FDA standard for pasteurized milk (cfu/ml) |
Number and percentage of pasteurized milk beyond the acceptable limit |
|---|---|---|---|---|---|---|
TVC |
3.8×104 – 4.1×108 |
<1×105 |
14 (70%) |
1.2×102 – 5.4×105 |
<1×104 |
2 (10%) |
TCC |
7.0×101 – 2.2×103 |
<1×103 |
6 (30%) |
0 – 7.1×101 |
<10 |
2 (10%) |
Yeast and Mould Count |
1.8×101– 6.1×102 |
<1×102 |
10 (50%) |
0 – 9.5×101 |
<1×102 |
8 (40%) |
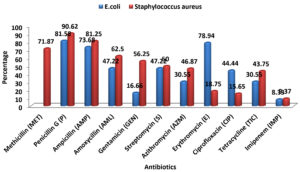
Fig. 1. Antimicrobial resistance patterns of isolated E. coli and S.aureus against commercial antibiotics
Microbiological assessment of milk based beverages (mattha, lassi, laban)
Different non-branded milk based beverages were collected to analyse the quality based on different microbiological parameters. In terms of TVC different ranges of results were observed, such as- mattha (7.8×103– 1.6×106 cfu/ml), lassi (5.8×103– 3.1×107 cfu/ml) and laban (1.9×104– 2.3×106 cfu/ml). In case of total coliform count, highest count was in mattha (2.2×103) and the lowest count was in lassi (3.1×101). Yeast and moulds were highest in mattha (7.3×102) and lowest in lassi (7.2×101). (Table 2).
Table (2):
Microbial count of mattha, lassi, laban (milk based beverages) in different microbiological parameters.
Microbiological testing parameters |
Mattha (cfu/ml) |
Lassi (cfu/ml) |
Labang (cfu/ml) |
|---|---|---|---|
TVC |
7.8×103– 1.6×106 |
5.8×103– 3.1×107 |
1.9×104– 2.3×106 |
TCC |
4.6×101-2.2×103 |
3.1×101– 8.4×102 |
7.0×101– 3.3×102 |
Yeast and Mould Count |
1.1×102-7.3×102 |
7.2×101– 6.3×102 |
8.5×101– 3.6×102 |
There has no national or international standard limitation for Mattha, Lassi and Labang.
Gram staining & Biochemical Identification
Gram-staining and specific biochemical tests were performed for the identification of Escherichia coli and Staphylococcus aureus. The suspected colonies from EMB and BPA medium which were observed as gram negative, rod and gram positive, cocci under the light microscope respectively, were chosen for the biochemical test. Catalase test and VP test were negative and other tests were positive for E.coli. On the other hand, catalase, MR and VP test were positive and other test results were negative for the isolated Staphylococcus aureus. (Table 3).
Table (3):
Morphological, gram staining and biochemical tests of Escherichia coli and Staphylococcus aureus.
Source and Colony morphology |
Gram Staining |
Catalase |
Oxidase |
Mobility |
Indole |
MR |
VP |
Suspected microorganism |
|---|---|---|---|---|---|---|---|---|
EMB agar (round colony with green metallic sheen) |
– |
+ |
– |
+ |
+ |
+ |
– |
E.coli |
BPA (black colony with halo zone) |
+ |
+ |
– |
– |
– |
+ |
+ |
Staphylococcus aureus |
Number of pathogens according to their source
The highest number of E. coli was identified from mattha (10 out of 20) and laban
(10 out of 20) sample and the lowest were identified from pasteurized milk (2 out of 20) samples. On the other hand, the highest (11 out of 20) and the lowest (3 out of 20) Staphylococcus aureus was present in raw and pasteurized milk respectively (Table 4).
Table (4):
The number of E.coli and S.aureus in contaminated samples.
Microorganisms |
Raw milk |
Pasteurized milk |
Mattha |
Lassi |
Labang |
Total |
|---|---|---|---|---|---|---|
E.coli |
6 |
2 |
10 |
8 |
10 |
36 |
Staphylococcus aureus |
11 |
3 |
8 |
5 |
5 |
32 |
Antibiogram
Disc-Diffusion Method was performed on Mueller-Hinton agar plate with 11 antibiotics commonly used in medical world for the treatment of disease associated with E.coli and S.aureus. Zone of inhibition were measured and compared with the zone diameter interpretive standard of CLSI & EUCAST. (Table 5).
Table (5):
Antibiotics used for determination of antibiotic resistance pattern.
Names of antibiotics |
Antibiotic conc. |
|---|---|
Methicillin (MET) |
5μg |
Penicillin G (P) |
10μg |
Ampicillin (AMP) |
25μg |
Amoxycillin (AML) |
25μg |
Gentamicin (GEN) |
10μg |
Streptomycin (S) |
10μg |
Azithromycin (AZM) |
15μg |
Erythromycin (E) |
35μg |
Ciprofloxacin (CIP) |
5μg |
Tetracycline (TIC) |
30μg |
Imipenem (IMP) |
10μg |
Antibiotic resistance pattern of isolated E.coli and Staphylococcus aureus
There were 36 E.coli strains and 32 S.aureus strains isolated from all the 100 milk and milk based beverages samples. Antibiotic resistance patterns in those isolated pathogens were tested with 11 commercially available antibiotics. After performing the disc diffusion method, isolated Escherichia coli were found most resistant to Penicillin G (81.58%), Erythromycin (78.94%) and Ampicillin (73.68%), (Fig. 1). The Staphylococcus aureus, were most resistant to Penicillin G (90.62%), Ampicillin (81.25%) and Methicillin (71.87%) (Fig. 1). Penicillin G and Ampicillin both were tested against the Enterobacteriaceae (Escherichia coli) considering the antibiotics using frequency of the people of Bangladesh.
This study was performed for investigating the harmful microorganisms in raw milk, pasteurized milk and milk based beverages available in Dhaka city and also for the determination of the antimicrobial resistance pattern of the strains- Escherichia Coli and Staphylococcus aureus taken from those samples with some commercial antibiotics. In this study, Total Viable Count for raw milk samples ranged from (3.8×104 – 4.1×108 cfu/ml). Unfortunately, 70% of raw cow milk and 10% of pasteurized branded milk samples were found beyond the acceptable limit of FDA. A study was done by Jubaida et al38 on pasteurized, UHT and flavored milk and they found the TVC range in pasteurized milk between 2.3×102 (cfu/ml) to 4.69×103 (cfu/ml). In their study, only one pasteurized branded milk was found contaminated with coliform, but the number was under acceptable limit (<10/ml coliform in pasteurized milk according to BSTI). In our study we found almost similar result considering the study of Jubaida et al38. We found 2 pasteurized milk samples contaminated with coliform group among the 20 samples. Some evidence showed that dairy products such as lassi, mattha, and yogurt in Dhaka city yielded a total viable count of 102–104 cfu/ml. In our study we found slightly different results with mattha (7.8×103 – 1.6×106 cfu/ml), lassi (5.8×103– 3.1×107 cfu/ml) and laban (1.9×104 – 2.3×106 cfu/ml). Among the 100 samples, 36% were contaminated with Escherichia coli and 32% with Staphylococcus aureus39, conducted their study to examine the microbial contamination within common milk products of Dhaka, Bangladesh. Their samples included mattha, sweetened yogurt, lassi and many other dairy products. All samples were found contaminated with bacteria and fungus within 102-104 (cfu/ml) and 102-103 (cfu/ml). In our study we found, the highest population of yeast and moulds were in mattha (7.3×102 cfu/ml) and the lowest in lassi (7.2×101 cfu/ml). A study was performed by Nushrat et al40, on microbial quality of dairy beverages commonly found in Dhaka. They had found (3.6×107 cfu/ml) and (1.7×103 cfu/ml) TVC in laban and strawberry milk shakes. They also found multi-drug resistant bacteria in some of their samples and all strains showed resistance against both Ampicillin and Colistin. Our study also identified multiple drug resistance pathogens in raw, pasteurized milk and milk based beverages of Dhaka city. In the present study, through disc-diffusion method, isolated Escherichia coli strains were most resistant to Penicillin G (81.58%), Erythromycin (78.94%) and Ampicillin (73.68%), and isolated Staphylococcus aureus strains were most resistance to Penicillin G (90.62%), Ampicillin (81.25%) and Methicillin (71.87%).
Multi drug resistant pathogens in food is alarming. Milk and milk based beverages are an essential part of diet in south Asia. Significance of maintaining hygiene during their preparation and storage is vital. The current study indicates poor hygiene and management of the food processing in some cases. Antibiotic resistance pattern is also important to identify the correct antibiotic immediately during the medical treatment. Public awareness is crucial to avoid unhygienic foods.
ACKNOWLEDGMENTS
The R & D project of Institute of Food Science and Technology (IFST), Bangladesh Council of Scientific and Industrial Research (BCSIR) provided the funding of this study. The authors are grateful to IFST and BCSIR authority as well.
CONFLICT OF INTEREST
The authors declare that there are no conflict of interest.
AUTHORS’ CONTRIBUTION
SP planned the study and supervised the research work. KWH performed the whole investigation. NT did the data analysis and methodology portions. KWH wrote down the original draft and all the authors reviewed it.
FUNDING
This study was funded by R & D project of Institute of Food Science and Technology (IFST), Bangladesh Council of Scientific and Industrial Research (BCSIR).
ETHICS STATEMENT
Not applicable.
AVAILABILITY OF DATA
All datasets generated or analyzed during this study are included in the manuscript.
- Griffiths MW. The microbiological safety of raw milk. Improving the safety and quality of milk. 2010;27-63.
Crossref - Giacometti F, Bonilauri P, Albonetti S, et al. Quantitative Risk Assessment of Human Salmonellosis and Listeriosis Related to the Consumption of Raw Milk in Italy. J Food Prot. 2015;78:13-21.
Crossref - Anderson M, Hinds P, Hurditt S, Miller P, McGrowder D, Alexander-Lindo R. The microbial content of unexpired pasteurized milk from selected supermarkets in a developing country. Asian Pac J Trop Biomed. 2011;1(3):205-211.
Crossref - Angulo FJ, LeJeune JT, Rajala-Schultz PJ. Unpasteurized milk: a continued public health threat. Clin Infect Dis. 2009;48(1):93-100.
Crossref - Reta MA, Addis AH. Microbiological quality assessment of raw and pasteurized milk. Int J Food Sci Microbiol. 2015;2(6):087-091.
- MacDonald LE, Brett J, Kelton D, Majowicz SE, Snedeker K, Sargeant JM. A systematic review and meta-analysis of the effects of pasteurization on milk vitamins, and evidence for raw milk consumption and other health-related outcomes. Journal of Food Protection. 2011;74(11):1814-1832.
Crossref - Amagliani G, Brandi G, Schiavano GF. Incidence and role of Salmonella in seafood safety. Food Research International. 2012;45(2):780-788.
Crossref - FDA Circular No.2013-010. Revised Guidelines For The Assessment of Microbiological Quality of Processed Food. 2013.
- Lim JY, Yoon JW, Hovde CJ. A brief overview of Escherichia coli O157: H7 and its plasmid O157. J Microbiol Biotechnol. 2010;20(1):5-14.
Crossref - De Buyser ML, Janin F, Dilasser F. Contamination of ewe cheese with Staphylococcus aureus: study of an outbreak of food poisoning. Zentralblatt fur Bakteriologie, Mikrobiologie und Hygiene. I. Abteilung: Supplemente. 1985.
- Miwa N, Kawamura A, Masuda T, Akiyama M. An outbreak of food poisoning due to egg yolk reactionnegative Staphylococcus aureus. Int J Food Microbiol. 2001;64:361-366.
Crossref - Fetsch A, Contzen M, Hartelt K, et al. Staphylococcus aureus food-poisoning outbreak associated with the consumption of ice-cream. Int J Food Microbiol. 2014;187:1-6.
Crossref - Garnier L, Valence F, Mounier J. Diversity and control of spoilage fungi in dairy products: an update. Microorganisms. 2017;5(3):42.
Crossref - Filtenborg O, Frisvad JC, Thrane U. Moulds in food spoilage. Int J Food Microbiol. 1996;33(1):85-102.
Crossref - in’t Veld JHH. Microbial and biochemical spoilage of foods: an overview. Int J Food Microbiol. 1996;33(1):1- 18.
Crossref - Westall S, Filtenborg O. Spoilage yeasts of decorated soft cheese packed in modified atmosphere. Food Control. 1998;15:243-249.
Crossref - Kure CF, Skaar I, Brendehaug J. Mould contamination in production of semi-hard cheese. International Journal of Food Microbiology. 2004;93(1):41-49.
Crossref - Beletsiotis E, Guikas D, Kalantzi K. Incorporation of microbiological and molecular methods in HACCP monitoring scheme of molds and yeasts in a Greek dairy plant: A case study. Proc Food Sci. 2011;1:1051- 1059.
Crossref - Temelli S, Anar S, Sen C, Akyuva P. Determination of microbiological contamination sources during Turkish white cheese production. Food Control. 2006;17:856- 861.
Crossref - Oyugi E, Buys EM. Microbiological quality of shredded Cheddar cheese packaged in modified atmospheres. Int J Dairy Technol. 2007;60(2):89-95.
Crossref - Mazumder R, Abdullah A, Ahmed D, Hussain A. High Prevalence of blaCTX-M-15 Gene among Extended- Spectrum β-Lactamase-Producing Escherichia coli Isolates Causing Extraintestinal Infections in Bangladesh. Antibiotics. 2020;9(11):796.
Crossref - FAW/WHO. Risk management and food safety, FAW Food and Nutrition paper number 65, Food and Agriculture Organization Rome, Italy. 1997.
- Cappuccino JG, Sherman N. Microbiology: A laboratory manual 10 th Edition, 2005.
- Marjan S, Das KK, Munshi SK, Noor R. Drug-resistant bacterial pathogens in milk and some milk products. Nutrition & Food Science. 2014;44(3):241-248.
Crossref - Silliker JH, Elliott RP, Bryan FL, Christian JHB, Clark DS. Microbial Ecology of Foods, 1980.
- Feroz F, Senjuti J, Tahera J, Das K, Noor R. Investigation of microbiological spoilage and demonstration of the anti-bacterial activity of the major imported fruits within Dhaka Metropolis. Stamford Journal of Microbiology. 2015;4(1):1-4.
Crossref - Das JS, Feroz F, Tahera J, Das KK, Noor R. Assessment of microbiological contamination and the in vitro demonstration of the anti-bacterial traits of the commonly available local fruit blend within Dhaka Metropolis. J Pharmacog Phytoche. 2014;3(1):73-77.
- Sultana T, Rana J, Chakraborty SR, Das KK, Rahman T, Noor R. Microbiological analysis of common preservatives used in food items and demonstration of their in vitro anti-bacterial activity. Asian Pac J Tropi Dis. 2014;4(6):452-456.
Crossref - Rahman F, Noor R. Prevalence of pathogenic bacteria in common salad vegetables of Dhaka Metropolis. Bangladesh Journal of Botany. 2012;41(2):159-162.
Crossref - FDA Bacteriological Analytical Manual. 18th Ed., AOAC, Washington, DC. 2005.
- Salfinger Y, Tortorello ML. Compendium of Methods for the Microbiological Examination of Foods, 5th Ed., American Public Health Association, Washington, D.C. 2015.
Crossref - Wehr HM, Frank JH. Standard Methods for the Microbiological Examination of Dairy Products, 17th, Ed., APHA Inc., Washington, D.C. 2004.
- Jagessar RC, Mars A, Gones G. Selective antimicrobial roperties of leaf extract against various microorganisms using disc diffusion and agar well diffusion method.
J Nat Sci. 2008;6(2):24-38. - Das M, Sabuj AAM, Haque ZF, et al. Characterization of Staphylococcus aureus isolated from human dental infection. African Journal of Microbiology Research. 2019;13(14):273-278.
Crossref - Hudzicki J. Kirby-Bauer disk diffusion susceptibility test protocol. American Society for Microbiology. 2009.
- Santos RMD, Santos FLMD, Ramacciato JC, Junqueira JLC. Evaluation of antimicrobial contamination and resistance to Staphylococcus aureus collected from radiographic materials used in dentistry. RGO. Revista Gaúcha de Odontologia (Online). 2012;60(4):467-477.
- Kumar PA. Bacterial resistance to antimicrobial agents and microbiological quality among Escherichia coli isolated from dry fishes in southeast coast of India. Roum Biotechnol Lett. 2008;13(6):3984-3989.
- Jamal JB, Akter S, Uddin MA. Microbiological quality determination of pasteurized, UHT and flavoured milk sold in Dhaka, Bangladesh. Stamford Journal of Microbiology. 2018;8(1):1-6.
Crossref - Malek M, Akter J, Ahmed T, Uddin MA. Isolation and quantification of microorganisms from some common milk products within Dhaka city, Bangladesh. Stamford Journal of Microbiology. 2015;5(1):13-17.
- Biva NA, Ismail I, Azad F, Rifat MA, Alim SR. Microbiological quality of some common dairy beverages available in Dhaka University campus of Bangladesh. Cogent Food & Agriculture. 2019;5(1):1707054.
Crossref
© The Author(s) 2021. Open Access. This article is distributed under the terms of the Creative Commons Attribution 4.0 International License which permits unrestricted use, sharing, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.