ISSN: 0973-7510
E-ISSN: 2581-690X
The aim of the present study was to isolate antagonistic actinomycetes from marine sediment collected from the Red Sea coast at Hurghada city and the Suez Gulf. A total of 16 actinomycete isolates were obtained in October 2018 and their antagonistic activities were evaluated against Aeromonas hydrophila, Vibrio damsela, Staphylococcus aureus ATCC6538, Bacillus subtilis ATCC 6633, Pseudomonas aeruginosa ATCC 9027, Salmonella typhimurium ATCC 14028, Escherichia coli ATCC 19404 and Candida albicans ATCC 10231 by using the agar well diffusion method. Among the 16 isolates, 14 (87.5%) isolates exhibited antimicrobial activity against most of the tested pathogens. The most potent isolate was identified by 16S rRNA sequence analysis as Streptomyces sp. and designated Streptomyces sp. MK388207. The experimental design of Plackett-Burman was implemented to optimize the culture conditions for antimicrobial agent(s) production by the most powerful isolate against C. albicans ATCC 10231 when grown in starch nitrate broth and the data revealed that negative (–) levels of KNO3, K2HPO4 and pH and positive (+) levels of starch, MgSO4.7H2O, FeSO4 and incubation period supported the production of the antimicrobial agent(s). Growth under the optimized culture conditions led to a 1.4-fold rise in antimicrobial activity. The ethyl acetate extract of Streptomyces sp.MK388207 was subjected to gas-liquid chromatography mass spectrometry (GC MS) and revealed the presence of six fractions with the major component being phenol, 2,4-bis (1,1 dimethylethyl). The findings of this study suggested that the antagonistic marine Streptomyces, in particular Streptomyces sp MK388207, the antibacterial compounds produced by this isolate, could be used as antibiotics that could have future applications in the pharmaceutical industry.
Antimicrobial, Actinomycetes, Marine sediment, Optimization.
Antibiotic resistant pathogenic bacteria seriously threaten global health and food security and occur at an alarming rate in many parts of the world1. Moreover, approximately 700,000 people worldwide die yearly because of drug-resistant contagion2. The search for novel antibiotics, especially in less explored habitats, is the only way to address this problem. Microorganisms derived from different marine sources produce potential antimicrobial compounds that be used for the treatment of many contagious illnesses and that could replace conventional drugs3. As the rate of novel bio-active compounds discovered from terrestrial actinomycetes declines over time, a great deal of attention was focused on screening actinomycetes from various environments for their ability to produce new secondary metabolites. Research have shown that marine-isolated Actinomycetes are metabolically active and have adapted to marine life4. Actinobacteria are the most prominent bioactive compound producers; they produce approximately two-thirds of the all-natural antibiotics used in medicine and agriculture. Among them, the genus Streptomyces has produced the majority of these compounds with wide range of pharmaceutical activities, such as antimicrobial, anticancer and antiviral activities5.
The biosynthesis of secondary metabolites by microorganisms depends on their growth conditions. Therefore, alterations in nutritional and physical factors during the incubation process could optimize the production of bioactive compounds by microbial isolates6. Statistical optimization methodologies such as surface methodology and the Plackett-Burman experimental design are used to design the nutritional conditions and remove the determination of the single-factor optimization practicability by optimizing all of the affecting parameters collectively7. However, bioactive actinomycetes from Suez Gulf are less explored.
In this study we aimed to isolate actinomycetes from marine soils of Suez Gulf and Hurghada with wide spectrum antimicrobial activity against various pathogenic bacteria. The optimal culture conditions for maximum antimicrobial agent production by the most potent isolate wed determined by using the Plackett-Burman experimental design.
Sample collection and processing
Sediment samples were collected from the Suez Gulf, Hurghada city, National Institute and Oceanography Beach, located at 27o 17’03″N 33o 46’15″E, using sterile techniques. Samples were collected and serially named S1 and S2, and then they were transported to the microbiology laboratory and stored at 4°C. To remove gravel and debris, the collected samples were blended altogether and passed through a 2 mm sifter. At that point each samples was dried overnight at 27°C to dispose of bacterial development8.
Isolation of marine actinomycetes
The isolation of marine actinomycetes was carried out on starch nitrate agar medium prepared with 50% seawater and containing the following (g/mL): starch, 20; K2HPO4, 0.5; KNO3, 1; MgSO4 7H2O, 0.5; FeSO4, 0.01; and agar, 15. The pH of the medium was adjusted to 7 before sterilization and myco-statin (500 µL/L) was supplemented after sterilization. From each sediment sample, approximately 1 gm was taken and mixed separately with 9 mL of sterile sea water. Five milliliters of the suspension were added to 100 mL of sterile starch nitrate agar medium. Approximately 20 mL of medium was poured into each sterile plate without air bubbles. The plates were incubated at 30°C for 7 days. Actinomycete colonies were selected and purified by streaking on the same medium. The purified isolates were grown in starch nitrate broth and stored in 25% glycerol at -20°C 9.
Screening for antibacterial activity
To test the antimicrobial potential of the actinomycete isolates, each isolate was inoculated into a 250 mL Erlenmeyer flask containing 100 mL starch nitrate broth (pH 7) at 30°C for 7 days under shaking conditions. To obtain the cell free supernatant, the culture broth was centrifuged at 8,000 rpm for 10 min and the antimicrobial activity of the supernatant was assessed by the agar well diffusion method. Sterile Mueller-Hinton agar plates were separately swabbed with the following pathogens that were used as test organism: Aeromonas hydrophila, Vibrio damsela, Staphylococcus aureus ATCC6538, Bacillus subtilis ATCC 6633, Pseudomonas aeruginosa ATCC 9027, Salmonella typhimurium ATCC 14028, Escherichia coli ATCC 19404 and Candida albicans ATCC. A sterile cork borer was used to make 6mm diameter wells in each plate. Approximately 100µL of the supernatant of each actinomycete isolate was loaded into each well against each of the test organisms. The plates were incubated at 37°C for 24 h. The antimicrobial activity was assessed by evaluating the inhibition zones diameter10. Only isolates with broad-spectrum activity were selected for further investigations.
Molecular taxonomy and phylogenetic analysis
A single colony of the most potent isolate was grown in starch nitrate liquid medium for 7 days. The growth was harvested and genomic DNA was extracted with the genomic DNA extraction protocol of the Gene JET genomic DNA Puri cation Kit (Fermentas). PCR was preformed using universal primers (16S 27F and 16S 1492R). The PCR product was then purified using a gel/DNA extraction kit. The 16S rRNA gene obtained was submitted to a DNA sequencing facility, the genetic laboratory at Color Company, Egypt (2018). The sequence was compared to the database available at the National Center for Biotechnology Information database (NCBI) via a BLAST search. A Phylogenetic tree was constructed using MEGA software version 6.0.
Optimization of culture conditions
The experimental design of Plackett Burman11 was used to show the comparative significance of medium components in antimicrobial agents manufacturing by isolate Streptomyces sp. MK388207 (M12) against C.albicans ATCC 10231 in liquid culture. Seven autonomous variables (Table 1) in eight combinations were organized according to the Plackett-Burman design matrix (Table 2); row no. 9 represents the basal control. For each variable, high (+1) and low (-1) levels were tested. All trials were performed in triplicate, and each experiment was repeated twice, with the mean considered as the response. The primary impact of each variable was determined by the equation below:
Exi = (SMi+ – SMi-) / N
where Exi is the variable main effect, Mi+ and Mi- are the radii of the clear zones around each well in the trials, where the independent variables were present in high and low concentrations, respectively, and N is the number of trials divided by 2. using Microsoft Excel 2007 was used to calculate the statistical t-values for equal unpaired samples for the determination of variable significance.
Verification experiment
Verification experiments were conducted in parallel using the anticipated optimized medium against the basal condition medium to confirm the outcomes acquired from the Plackett-Burman design statistical analysis. Antimicrobial agent output was determined by evaluating the average diameter (mm) of the inhibition area.
Effect of temperature on the production of antimicrobial agents
To study the effect of temperature on the production of antimicrobial agents, isolate Streptomyces sp. MK388207 (M12) was inoculated into 100 mL of the culture formula obtained from the experimental design and incubated at different temperatures (20, 25, 30, 35, 40 and 50°C) for 7 days. Supernatant from each experiment was tested using the method of agar well diffusion agai nst C.albicans ATCC 10231 12.
Extraction of bioactive compounds
The most potent isolate Streptomyces sp. MK388207 (M12) was grown in the verified medium as a production medium. The culture was centrifuged at 10,000 rpm for 20 min and the supernatant obtained was extracted with an equal volume of ethyl acetate by shaking for one hour for complete extraction. The organic phase was separated and evaporated with a heating mantle at 40°C to achieve dry powder13.
Characterization of the purified bioactive compounds
Cmass spectrometry (GC-MS) analysis
A Trace GC1310-ISQ mass spectrometer (Thermo Scientific, Austin, TX, USA) with a direct capillary column TG–5MS (30 m x 0.25 mm x 0.25 לm movie thickness) was used to determine the chemical structure of the crude extract. Initially, the column oven temperature was held at 50°C and then increased to 230°C by 5°C / min. The temperature was raised by 30°C / min to the final temperature of 290°C and kept for 2 min. The temperatures of the injector and MS transfer line were maintained at 250 and 260°C respectively; helium was used as a carrier gas at a steady flow rate of 1 mL / min. The solvent delay was 3 min and 1µL diluted specimens were automatically injected using a divided mode Autosampler AS1300 combined with GC. EI mass spectra were collected in full scan mode at 70 eV voltages of ionization over the range of m / z 40–1000. The temperature of the ion source was set at 200°C. Comparing their retention times (RTs) and mass spectra with the WILEY 09 and NIST 11 mass spectral databases, the elements were recognized. (Fungi Center, Azhar University, Cairo, 2019).
Infrared spectroscopy (IR)
In the Central Laboratory of Al-Azhar University, Cairo Branch, IR-spectral assessment was examined. One mg sample and 300 mg KBr (IR grade) were correctly blended for this purpose14. Using Ultraviolet Visible Spectrophotometer evaluation, the purified bioactive compounds were defined by recording the absorption peak.
Isolation and screening of marine actinomycetes
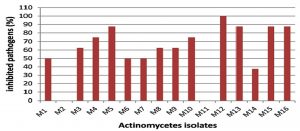
The marine environment is an excellent habitat for the discovery of new bioactive agents useful for the treatment of human illness. Microorganisms derived from this environment have provided new therapeutic agents. Among them, the most active producers are actinomycetes15. In this study, sixteen morphologically different marine actinomycete isolates were obtained from marine sediment collected from the Suez Gulf, Hurghada city using starch nitrate agar medium. The nutritional conditions of the starch medium allow the growth of actinomycetes and suppress the growth of other bacteria and fungi16. All isolates were screened for antagonistic activity against different pathogens. Among them, 14 isolates (87.5%) exhibited antimicrobial activity (Fig. 1).
However, several previous studies reported lower percentages of antagonistic isolates among actinomycetes isolated from marine environments17-19. Only 2 isolates (12.5%) did not exhibit any antimicrobial activity against any of the pathogens tested. This could be attributed to the nutritional or cultivation conditions which seemed to be inadequate for the formation of antimicrobial compound(s) by these isolates; it is expected that alterations in these conditions could induce antimicrobial activities20. Among the sixteen isolates, isolate M12 exhibited marked antimicrobial activity against all selected species of both gram negative and gram-positive bacteria and C. albicans ATCC 10231 and the maximum diameter of the inhibition zone was recorded against C. albicans ATCC 10231 (Fig. 1 and Table 3). Broad-spectrum antimicrobial activity is widespread among marine actinomycetes16, 21. Moreover, antagonistic activity against Candida species has been frequently reported22. These results suggest that isolate M12 could be a promising source for the production of bioactive compound(s) against clinically important Candida spp as well as both gram negative and gram-positive bacterial species.
Molecular taxonomy and phylogeny of the most potent isolate
The BLAST search of the 16S rDNA sequence of the most potent isolate (M12) showed maximum similarity (99%) with Streptomyces sp. and the phylogenetic tree was constructed (Figure 2). Based on the molecular taxonomy and phylogeny the strain was identified as Streptomyces sp. and deposited in the GenBank as Streptomyces sp. MK388207.
Optimization of antimicrobial agent(s) production by Streptomyces sp. MK388207 (M12)
Statistical experimental designs are the strongest techniques for the fast screening of important variables from a multivariable scheme and for the reduction of mistakes in determining the impact of distinct parameters7,23,24. The Plackett-Burman design was employed to evaluate the significant effect of the different components of starch nitrate medium on the production of antimicrobial agents from Streptomyces sp. MK388207 (M12) against C. albicans ATCC 10231. The components of starch nitrate medium selected for the production of experimental antimicrobial agents, in addition to other cultural factors including pH and incubation period were each examined at two levels (-, +) as shown in Table 1. The Placket-Burman design presented in Table 2 was applied to examine the importance of these factors with respect to the production of antimicrobial agent(s). The inhibition zone diameters (mm) obtained by experimental trials (in triplicate) were found to be in the range of 14-24 mm. Statistical analysis of the data (t-value) showed that, among the medium components, starch was the most significant independent variable that affected the suppression of C. albicans ATCC 10231. Moreover, MgSO4.7H2O and FeSO4 showed also considerable effect on C. albicans with respect to the production of antimicrobial agent(s) (Table 4).
Table (1):
Levels of independent variables in the Plackett-Burman experiment.
| Variable | Level (g/L) | ||
|---|---|---|---|
| -1 | 0 | +1 | |
| Starch | 10 | 20 | 30 |
| KNO3 | 0.5 | 1 | 1.5 |
| K2HPO4 | 0.25 | 0.5 | 0.75 |
| MgSO4.7 H2O | 0.25 | 0.5 | 0.75 |
| FeSO4 | 0.005 | 0.01 | 0.015 |
| Incubation period (days) | 5 | 7 | 9 |
| pH | 6 | 7 | 8 |
Table (2):
The Plackett-Burman design for 7 independent variables against Candida albicans ATCC10231.
| Trial no. | Factors | Inhibition zone (mm) | ||||||
|---|---|---|---|---|---|---|---|---|
| Starch | KNO3 | K2HPO4 | MgSO47H2O | FeSO4 | IP | pH | ||
| 1 | -1 | -1 | -1 | 1 | 1 | -1 | 1 | 16 |
| 2 | 1 | -1 | -1 | -1 | -1 | 1 | 1 | 14 |
| 3 | -1 | 1 | -1 | -1 | 1 | 1 | -1 | 0 |
| 4 | 1 | 1 | -1 | 1 | -1 | -1 | -1 | 20 |
| 5 | -1 | -1 | 1 | 1 | -1 | 1 | -1 | 0 |
| 6 | 1 | -1 | 1 | -1 | 1 | -1 | -1 | 16 |
| 7 | -1 | 1 | 1 | -1 | -1 | -1 | 1 | 0 |
| 8 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 24 |
| 9 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 20 |
Table (3):
Antimicrobial activity of the marine actinomycete isolates.
| Inhibition zone (mm) | ||||||||
|---|---|---|---|---|---|---|---|---|
| Isolate | A.hydrophila | V.damsela | S.aureus | B.subtilis | P.aeruginosa | S.typhimurium | E.coli | C.albicans |
| M1 | 0.0 | 10.0 | 0.0 | 12.0 | 0.0 | 8.0 | 16.0 | 0.0 |
| M2 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 |
| M3 | 18.0 | 16.0 | 10.0 | 0.0 | 0.0 | 0.0 | 10.0 | 12.0 |
| M4 | 14.0 | 12.0 | 0.0 | 0.0 | 10.0 | 12.0 | 14.0 | 10.0 |
| M5 | 8.0 | 10.0 | 12.0 | 8.0 | 12.0 | 10.0 | 8.0 | 0.0 |
| M6 | 18.0 | 12.0 | 0.0 | 0.0 | 16.0 | 0.0 | 10.0 | 0.0 |
| M7 | 12.0 | 0.0 | 0.0 | 0.0 | 0.0 | 16.0 | 10.0 | 12.0 |
| M8 | 0.0 | 10.0 | 16.0 | 16.0 | 16.0 | 10.0 | 0.0 | 0.0 |
| M9 | 16.0 | 0.0 | 15.0 | 0.0 | 0.0 | 12.0 | 16.0 | 8.0 |
| M10 | 16.0 | 12.0 | 8.0 | 0.0 | 12.0 | 0.0 | 10.0 | 12.0 |
| M11 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 |
| M12 | 18.0 | 16.0 | 18.0 | 14.0 | 16.0 | 18.0 | 16.0 | 20.0 |
| M13 | 10.0 | 12.0 | 12.0 | 0.0 | 8.0 | 10.0 | 10.0 | 12.0 |
| M14 | 12.0 | 16.0 | 0.0 | 10.0 | 0.0 | 0.0 | 0.0 | 0.0 |
| M15 | 0.0 | 16.0 | 14.0 | 10.0 | 10.0 | 8.0 | 15.0 | 12.0 |
| M16 | 16.0 | 16.0 | 18.0 | 0.0 | 15.0 | 10.0 | 8.0 | 15.0 |
A. hydrophila: Aeromonas hydrophila, V. damsela : Vibrio damsela, S. aureus: Staphylococcus aureus, B. subtilis: Bacillus subtilis, P.aeruginosa: Pseudomonas aeruginosa, S. typhimurium: Salmonella typhimurium, E. coli: Escherichia coli, C. albicans: Candida albicans
Table (4):
Statistical analyses of the Plackett-Burman experimental results.
Variable |
Main effect |
t-value* |
|---|---|---|
Starch |
7.25 |
2.485 |
KNO3 |
-0.25 |
-0.085 |
K2HPO4 |
-1.25 |
-0.4285 |
MgSO4.7H2O |
3.75 |
1.2857 |
FeSO4 |
2.75 |
0.9428 |
pH |
-1.75 |
-0.6 |
Incubation period |
2.25 |
0.7714 |
*t-value significant at the 1% level = 3.70
t-value significant at the 5% level = 2.446
t-value significant at the 10% level = 1.94
t-value significant at the 20% level =1.372
Standard t-values were obtained from the Statistical Methods 25
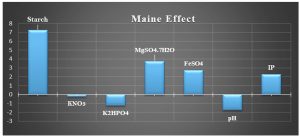
The mean values of the main impact on the inhibition zone diameter of the factors examined were calculated and shown in Fig. 3.
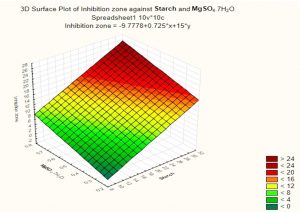
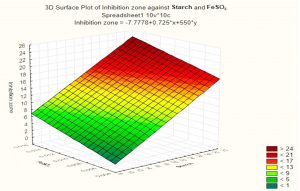
Based on these results, negative (–) levels of KNO3, K2HPO4 and pH as well as positive (+) levels of starch, MgSO4.7H2O, FeSO4 and incubation period promoted the production of the antimicrobial agent(s) formed by Streptomyces sp. MK388207 (M12). Moreover, the t-value represented in Table 4 supports this observation. The interaction of MgSO4.7H2O and FeSO4 increased the inhibition zone diameter. This strategy checked the validity of the design applied. The basal medium versus the optimized medium was evaluated using a verification experiment. The interaction effects of the starch concentration with the MgSO4.7H2O concentration and the FeSO4 concentration with respect to the inhibition zone diameter of the produced antimicrobial agent(s) based on the Plackett-Burman results are described in three-dimensional graphs (Fig. 4 (a), (b).
Fig. 4a. The interaction of starch concentration with FeSO4 with respect to inhibition zone diameter of antimicrobial agent based on the Plackett Burman results.
Fig. 4b. The interaction of starch concentration with MgSO4.7H2O with respect to inhibition zone diameter of antimicrobial agent based on the Plackett Burman results.
Verification experiment
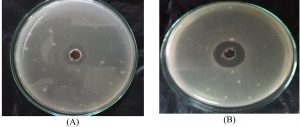
A nearly optimal manufacturing medium was developed as follows: (g/L) starch 30; KNO3 0.5; K2HPO4 0.25; MgSO47H2O 1.5FeSO4; 0.015 and pH 8 for a 5-day incubation period according to the information acquired. A verification experiment has been used to compare the predicted optimum levels of the independent variables and the underlying conditions. Cultivation of Streptomyces sp.MK388207 (M12) in the verified medium adjusted to pH 8 for 5 days resulted in a 1.4 fold increase in the inhibition zone diameter compared to the basal conditions (Table 5 and Fig. 5). Hassan and his team reported that, increasing pH concentrations and declining concentrations of peptone, beef extract and culture volume led Pseudoalteromonas piscicida to enhance antagonistic activity by about 1.2 fold7. Abd-Elnaby and others stated that, high pH values and a large inoculum size caused an approximately 1.3-fold increase in the antimicrobial efficacy of Streptomyces parvus26.
Table (5):
A verification experimental: antagonistic activity of Streptomyces MK388207 (M12) grown in basal versus verified media against Candida albicans ATCC 10231.
Medium |
Inhibition zone (mm) |
|---|---|
Basal medium |
20 |
Verified medium |
28 |
Fig. 5. Inhibition zone diameter of Streptomyces MK388207 (M12) supernatant grown on basal medium (A) compared with the verified medium (B) against Candida albicans ATCC10231.
Effect of temperature on the production of antimicrobial agents from isolate Streptomyces sp. MK388207 (M12)
The effect of temperature on the growth of isolate Streptomyces MK388207 (M12) using verified medium and the production of antagonistic agents against C. albicans ATCC10231 were evaluated. The actinomycete isolate grew well ranging from 25 to 40°C. The highest antagonistic activities were observed in incubated at 35°C (Table 6). Aliero and his team reported that, the minimum temperature for antagonistic activity production from three actinomycete isolates KBRWDSa3 (RF), KBMWDSb6 (M6) and BRWDSc (SP) was 25°C while the maximum temperature was 35°C12. Higher antimicrobial activity was observed when cultures were grown at 30-35°C.
Table (6):
Effect of the incubation temperature of Streptomyces MK388207 (M12) on the production of antimicrobial agents against Candida albicans ATCC 10231.
Inhibition zone (mm) |
Temperature °C |
|---|---|
22 |
25 |
27 |
30 |
29 25 |
35 40 |
Chemical characterization of the ethyl acetate extract
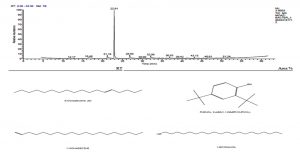
Ethyl acetate an efficient solvent for the extraction of bioactive compounds from actinomycete cultures27-29. The GC-MS chromatogram of ethyl acetate crude extracts of Streptomyces sp. K388207 (M12) showed different bioactive compounds as shown in Fig. 6.
Fig. 6. GC-MS chromatogram and structure formulas of compounds of the bioactive compounds extracted from Streptomyces sp. strain MK388207 (M12).
The active principles with their RT, molecular formula and molecular weight (MW) of Streptomyces sp. strain MK388207 (M12) are presented in Table 7.
Table (7):
Components detected in ethyl acetate crude extract of Streptomyces sp. MK388207 strain (M12).
No. |
R T Value |
Compound |
Molecular Formula |
Molecular Weight |
Peak Area (%) |
|---|---|---|---|---|---|
1 |
21.19 |
5-Octadecene, (E)- |
C18H36 |
252 |
5.48 |
2 |
22.91 |
Phenol,2,4-bis (1,1 dimethylethyl)- |
C14H22O |
206 |
63.39 |
3 |
26.79 |
1-nonadecene |
C19H38 |
266 |
5.31 |
4 |
32.09 |
1-Eicosanol |
C20H42O |
298 |
3.79 |
5 |
36.83 |
1-Docosene |
C22H44 |
308 |
2.49 |
6 |
43.14 |
Podocarp-7-en-3-one, 13á-methyl-13-vinyl- |
C20H30O |
286 |
1.67 |
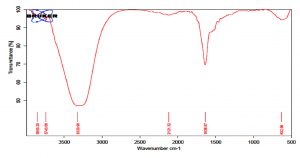
The GC MS patterns revealed peaks that represented (5-octadecene, (E-phenol,2,4-bis (1,1 dimethylethyl; 1-Nonadecene; 1-Eicosanol; 1-Docosene and podocarp-7-en-3-one, 13ב-methyl-13-vinyl-). The antibacterial, antifungal and anti-inflammatory activities of phenol, 2,4-bis (1,1-dimethylethyl)- have previously been reported30,31. 1-nonadecene extracted from the Streptomyces sp. TN 256 strain exhibited antibacterial activity and potent antifungal activity against C. albicans32. 5-octadecene, 1-hexadecene and 1-nonadecene were obtained from Streptomyces sp. TN 272, a novel terrestrial species isolated from soil samples of Tunisian regions33. In another study, Naragani and his team reported that, 5-octadecene and 1-nonadecene from Streptomyces cheonanensis VUK-A which was isolated from a mangrove have antibacterial and antifungal activities34. 1-Eicosanol produced by Streptomyces sp. DOSMB-A107 isolated from mangrove sediments has been shown to have antimicrobial activity35. Two non-bioactive compounds reported in the present study (1-docosene and podocarp-7-en-3-one, 13a-methyl-13-vinyl-) previously reported35,36. In addition, FTIR spectrum of ethyl acetate extracts were used to determine organic functional groups. Ethyl acetate crude extracts of Streptomyces sp. K388207 (M12) represented absorption at 3743.86 cm-1 showing O-H group, 3320.98 cm-1 showing N-H amino group, 2121.32 cm-1 showing N=N=N azide group, 1638.97 cm-1 showing C=N imine group and 632.96 cm-1 showing C-Br bromine group (Fig. 7).
Fig. 7. FTIR spectrum of the extracts bioactive compounds isolated from Streptomyces sp. MK388207 strain (M12).
Parthasarathi and his team (2012) reported that, FTIR spectrum of S. hygroscopicus BDUS 49 which isolated from marine environment locations of Bigeum Island, South West coast of South Korea, showed two absorption peaks in the region of 3500 and 1730 cm-1. The spectrum shows that OH and potentially lactone carbonyl group are present in the compound. Another two absorption peaks showed in the region of 1700 (carbonyl) and between 1610 and 1630 cm-1. This peak indicates that the compound had carbonyl and alkenes (C=C) group. The absence of carboxylic acid (COOH) and ester (COOR) alkynes was confirmed by the lack of bands in the region of 1670-1674 and 1700-750 cm-1 respectively37. In summary, we could show that the marine sediment collected from the Red Sea coast at Hurghada city and the Suez Gulf is an excellent source of antagonistic actinomycetes. Most of the obtained isolates exhibited antimicrobial activity against most of the tested pathogens. Among them, Streptomyces sp. MK388207 was chosen as the most powerful strain and the cultivation conditions were optimized for maximum antimicrobial production using the experimental design of Plackett-Burman. Gas-liquid chromatography mass spectrometry (GC-MS) analysis of the ethyl acetate crude extract of Streptomyces sp. MK388207 revealed the presence of some previously reported bioactive agents and two other compounds with no known activity. Further studies are needed to purify and elucidate the bioactive compounds.
The Actinobacteria class is a very important bacterial group with significant value as a prolific antibiotic producer and other therapeutic compounds. Our study showed that Suez Gulf, Hurghada City sediment samples contained a variety of actinomycetes that may inhibit the growth of some test microorganisms and Candida albicans. The strain Streptomyces sp. MK388207 was selected as the most potent strain exhibited strong antimicrobial activity. The results of the present work clearly demonstrate the effectiveness of experimental design of Plackett-Burman in optimizing medium composition and improving the growth of Streptomyces sp. MK388207. Its usefulness in designing an efficient fermentation process for the production of the bioactive compounds by Streptomyces sp. MK388207 to be used for pharmaceutical industries. In the future study, complete identification of the bioactive compounds extracted will be tracked.
ACKNOWLEDGMENTS
None.
CONFLICT OF INTEREST
The authors declare that there is no conflict of interest.
AUTHORS’ CONTRIBUTION
LSA carried out the statistical experiment analysis. NMF wrote the manuscript. All authors read and approved the manuscript for publication.
FUNDING
None.
ETHICS STATEMENT
Hereby, I Moaz Mohamed Hamed consciously assure that for the manuscript: Antimicrobial activity of marine actinomycetes and the optimization of culture conditions for the production of antimicrobial agent(s) the following is fulfilled:
1) This material is the authors’ own original work, which has not been previously published elsewhere.
2) The paper is not currently being considered for publication elsewhere.
3) The paper reflects the authors’ own research and analysis in a truthful and complete manner.
4) The paper properly credits the meaningful contributions of co-authors and co-researchers.
5) The results are appropriately placed in the context of prior and existing research.
6) All sources used are properly disclosed (correct citation). Literally copying of text must be indicated as such by using quotation marks and giving proper reference.
7) All authors have been personally and actively involved in substantial work leading to the paper, and will take public responsibility for its content.
AVAILABILITY OF DATA
The data that support the findings of this research are available from the corresponding author, Dr. Moaz M. Hamed, upon reasonable request.
- W.H. Organization, World malaria report 2014, World Health Organization, 2015.
- C. Privault, J. O’neill, J.-M. Renders, V. Ciriza, Y. Hoppenot, G. Bauduin, A. Fucs, Y. Deng, G. Gerard and M. Knibiehly, Journal, 2016.
- E.J. Gudioa, E.C. Fernandes, A.I. Rodrigues, J.A. Teixeira and L.R. Rodrigues, Frontiers in Microbiology, 2015; 6: 59.
- S. Valli, S.S. Suvathi, O. Aysha, P. Nirmala, K.P. Vinoth and A. Reena, Asian Pacific Journal of Tropical Biomedicine, 2012; 2: 469-473.
Crossref - S.J. Naine, C.S. Devi and V. Mohanasrinivasan, Brazilian Archives of Biology and Technology, 2015; 58: 198-207.
Crossref - A. Rajnisz, A. GU PIEL, M. Postek, J. Ziemska, A. Laskowska, D. Rabczenko and J. Solecka, Polish Journal of Microbiology, 2016; 65, 51-61.
Crossref - S.W.M. Hassan, N.A. El Sersy, A.E.-S. Abdelwahab and M. A.-R. Ali, Journal of Applied Pharmaceutical Science Vol., 2017; 7: 084-093.
- S. Valli, S.S. Suvathi, O. Aysha, P. Nirmala, K.P. Vinoth and A. Reena, Asian Pacific Journal of Tropical Biomedicine, 2012; 2: 469.
Crossref - P. Ellaiah, T. Ramana, K. Raju, P. Sujatha and A.U. Sankar, Asian Journal of Microbiology Biotechnology and Environmental Sciences, 2004; 6: 53-56.
- P.N. Waithaka, F.B. Mwaura, J.M. Wagacha, E.M. Gathuru and B.M. Githaiga, Cell Bio., 2017, 6: 13.
- R.L. Plackett and J.P. Burman, Biometrika, 1946; 305-325. https://doi.org/10.1093/biomet/33.4.305 A.A. Aliero, A.S. Adam, I. Ntulume, A.I. Bagudo, M.C. Ondieki, S.A. Jega, J. Odda and M.A. Okech, Current Trends in Biotechnology & Pharmacy, 2018; 12.
- R. Balachandar, N. Karmegam, M. Saravanan, R. Subbaiya and P. Gurumoorthy, Microbial Pathogenesis, 2018; 121: 155-165. https://doi.org/10.1016/j.micpath.2018.05.027
- K. Bayoub, T. Baibai, D. Mountassif, A. Retmane and A. Soukri, African Journal of Biotechnology, 2010; 9: 4251-4258.
- S. Pooja, T. Aditi, S.J. Naine and C.S. Devi, Frontiers in Biology, 2017; 12: 280-289.
Crossref - S. Ramesh, India Dissertation, University of Madras, Chennai, India, 2009.
- S. Ramesh and N. Mathivanan, World Journal of Microbiology and Biotechnology, 2009; 25: 2103-2111.
Crossref - Z.A. Zainal Abidin, N. Abdul Malek, Z. Zainuddin and A.J.K. Chowdhury, Frontiers in Life Science, 2016: 9: 24-31.
Crossref - Y. Retnowati, L. Sembiring, S. Moeljopawiro, T. S. Djohan and E.S. Soetarto, Biodiversitas Journal of Biological Diversity, 2017; 18: 1453-1461.
- M. Oskay, International Journal of Agriculture & Biology, 2011, 13.
- M. Jayaprakashvel, Journal of Modern Biotechnology, 2012; 1: 1-7.
- C. Spadari, T. Antunes, R. Teixeira, E. Minotto, A. M. Fuentefria, D. Sand and S. Van, Revista Brasileira de Biociencias, 2013; 11.
- A. Saraniya and K. Jeevaratnam, Brazilian Journal of Microbiology, 2014; 45: 81-88.
Crossref - A. Zeinab, M. Aly, A. Faiza and E. Fatma, Int. J. Curr. Microbiol. App. Sci., 2015; 4: 1082-1096.
- G. W. C. Snedecor and G. William, Statistical Methods/George W. Snedecor and William G. Cochran, 1989.
- H. Abd-Elnaby, G. Abo-Elala, U. Abdel-Raouf, A. Abd-elwahab and M. Hamed, Biotechnology & Biotechnological Equipment, 2016; 30: 180-191.
Crossref - T. Ahsan, J. Chen, X. Zhao, M. Irfan and Y. Wu, AMB Express, 2017; 7: 54.
Crossref - O.H. El Sayed, M.M. Asker, M.A. Swelim, I.H. Abbas, A.I. Attwa and M.E. El Awady, Journal of Genetic Engineering and Biotechnology, 2016; 14: 161-168.
Crossref - H. Rante, R. Yulianty, N. Djide, R. Burhamzah and M. Prasad, 2017.
- A.C.F. Amaral, L.A. Gomes, J.R.d.A. Silva, J.L.P. Ferreira, A.d.S. Ramos, M.d.S.S. Rosa, A.B. Vermelho and I.A. Rodrigues, BioMed research international, 2014; 2014.
Crossref - T. Chandrasekar, M.R.K. Rao, R.V. Kumar, K. Prabhu, S. Nandha Kumar and D. Divya, J. Chem. Pharm. Res., 2015; 7: 124-136.
- S. Smaoui, F. Mathieu, L. Elleuch, Y. Coppel, G. Merlina, I. Karray-Rebai and L. Mellouli, World Journal of Microbiology and Biotechnology, 2012; 28: 793-804.
Crossref - L. Elleuch, K.A. Shaaban, M.S. Abdel-Aziz, A. Chakchouk, M.M. Nagia, L. Mellouli and M. Shaaban, African Journal of Microbiology Research, 2012; 6: 2202-2210.
- K. Naragani, U. Mangamuri, V. Muvva, S. Poda and R. K. Munaganti, Int J Pharm Pharm Sci., 2016; 8: 53-57.
- R. Baskaran, P. Mohan, M. Madanan, A. Kumar and M. Palaniswami, 2015.
- M. Padma, S. Ganesan, T. Jayaseelan, S. Azhagumadhavan, P. Sasikala, S. Senthilkumar and P. Mani, Journal of Drug Delivery and Therapeutics, 2019; 9: 85-89.
Crossref - S. Parthasarathi, S. Sathya, G. Bupesh, R.D. Samy, M.R. Mohan, G. S. Kumar, M. Manikandan, C. Kim and K. Balakrishnan, World J. Fish Mar. Sci., 2012; 4: 268-277.
© The Author(s) 2019. Open Access. This article is distributed under the terms of the Creative Commons Attribution 4.0 International License which permits unrestricted use, sharing, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.