ISSN: 0973-7510
E-ISSN: 2581-690X
Ganoderma, a genus of polypore white rot fungus with some species such as G. lucidum and G. sinense have been reported with remarkable biological activities and used as traditional medicine for long. On the other hand, G. boninense is recognized as an important oil palm pathogen which causes intensive losses in oil palm industry. Hence, this study reveals the potential of antimicrobial properties of G. boninense fruiting bodies extract using different solvents. Ethyl acetate extract demonstrated a broad spectrum pathogens inhibition activity, followed by hot water > acetone > methanol > ethanol > chloroform. Ethyl acetate extract also shown the strongest growth inhibition against Proteus mirabilis (14.20 ± 0.40 mm). Lowest MIC (0.625 mg/ml) was observed in methanol extract against Coagulase-Negative Staphylococci. LC-MS analysis identified the extracts of G. boninense putatively contains natural occurring derivatives of alkaloid, fatty acid, heterocyclic compounds and glycosides. G. boninense may potential as the future drug. However, further investigation is required to understand this fungus in depth.
Ganoderma boninense, antimicrobial, natural compounds, pathogenic bacteria
New drug discovery has become a hot research area due to the emergence of antibiotic resistance bacteria. Presently, some clinical isolates such as Methicillin Resistant Staphylococcus aureus (MRSA), Penicillin Resistant Streptococcus pneumoniae (PRSP), Vancomycin Resistant Enterococci (VRE) which have become resistant to certain commonly used antibiotics are easily found in the environment1,2. The report of Global Antimicrobial Resistance Surveillance System (GLASS) rises the alarm of the seriousness in worldwide antibiotic resistance’s issue with more than 500,000 isolates showing non-susceptible to one or more of the conventional antibiotics3. According to National Surveillance of Antibiotic Resistance (NSAR) report, most of the notorious antibiotic resistant bacterial has risen since 2008 to 2016 in Malaysia4.
Hence, discovery and development of new alternative drugs to combat with the multidrug resistant bacteria remain as a challenge. Naturally occurring molecules derived from secondary metabolites formed by living organisms that are chemically and structurally diverse could further contribute knowledge and strategy to overcome such issue5. Secondary metabolites are synthesized as biochemical compounds in defense against stressful environments and invasion of predators for self-healing and rejuvenating to assist survival6. Currently, extraction of the natural compounds with antimicrobial properties such as saponins, alkaloids, sesquiterpenes, terpenoids, glycopeptide and others were used as prescription drug in numerous developed countries7,8.
Macro fungi, a group of wood-decaying fungi of the Phylum Basidomycetes have been exploited as new alternative in development of novel antibiotics due to the bioactive compounds which shown strong antimicrobial activity. In particular, Ganoderma species such as lucidum, showed good inhibition against different bacterial strains9. Ganoderma triterpenes (GTs) and polysaccharides (PS) derivatives are reported as the most common antimicrobial compounds from Ganoderma sp.10.
G. boninense is a polypore fungus in the family of Ganodermataceae. This species belongs to Ganoderma genus, which are well-known as medicinal mushrooms. On the other hand, G. boninense is a pathogen of basal stem rot (BSR) disease, a devastating disease of oil palm trees11. In the current work, different solvents were used to extract the active compounds from fruiting bodies of G. boninense to investigate the potential of antimicrobial properties.
Fungal Material
Forty-two fruiting bodies of Ganoderma were collected from infected oil palm trees in Sawit Kinabalu Oil Palm Plantation at Langkon, Sabah Malaysia. Fresh fruiting bodies were cleaned under running tap water and freeze dried using freeze dryer (ScanVac CoolSafe 110-4, Denmark). Dried fruiting bodies were stored at -80°C for further use.
Molecular Identification of G. boninense Fruiting Bodies
DNA extraction
Approximately, 100 mg sliced tissue of G. boninense fruiting bodies were powdered and homogenized with liquid nitrogen using a mortar and pestle. After that, the DNA of fruiting bodies were extracted using DNeasy plant mini kit (Qiagen, Germany) with slight modifications as described by Chong et al.12.
PCR amplification
Amplification of the fungal DNA was done on the 5.8S gene using ITS1: 5’- TCC GTA GGT GAA CCT GCG G-3’ as forward primer and ITS4: 5’- TCC GCT TAT TGA TAT GC -3’ as reverse primer. The PCR reagents (Vivantis, Malaysia) were prepared in concentration of 1 x PCR buffer, 1.5 mM MgCl2, 200 mM dNTPs, 0.4 mM forward and reverse primers, one unit of Taq polymerase and 1 ml of DNA template. The thermalcycler (GeneAtlas, Astec, Japan) was programmed for initial denaturation at 95°C for 5 min, followed by 35 cycles of denaturation at 95°C for 1 min, annealing at 53°C for 40 s and extension at 72°C for 50 s. The final extension was set at 72 °C for 10 min. The PCR product was detected by agarose gel electrophoresis with 1.5% TAE agarose gel, 70 V for 40 min.
DNA sequencing and analysis
The DNA sequencing was done by Apical Scientific Sdn Bhd at Selangor, Malaysia. The obtained sequence was trimmed and later BLAST searched for the closest matches in NCBI GenBank. Phylogenetic tree was constructed using Maximum Likelihood method with 1000 bootstraps replicate to show the relationship among sequences.
Preparation of G. boninense Extracts
Identified fruiting bodies of G. boninense were grounded using mortar and pestle with the aid of liquid nitrogen. The mortar and pestle were autoclaved, cooled and swabbed with 70% ethanol prior used. The extraction of ground powder was prepared by using maceration method. Briefly, 10 g of fine powders were soaked into 400 ml of different absolute solvents (chloroform, acetone, methanol, ethanol, ethyl acetate) at room temperature (±26°C) and shaken with 150 rpm for five days. Meanwhile, hot water extraction was prepared using water bath incubator at 90°C and 100 rpm. The supernatant was filtered through Whatman no.1 filter paper. The filtrates were then evaporated under reduced pressure using rotary evaporator (Buchi-Rotavapor R-300, Germany). Crude extracts were kept in -20°C for further use.
Microbial Culture and Inoculums Preparation
The clinical isolates were obtained from the stock culture of Genetic Laboratory, Faculty of Science and Natural Resources, Universiti Malaysia Sabah. The tested bacterial strains include eight Gram-negative bacteria (Escherichia coli, Enterobacter sp, Pseudomonas aeruginosa, Proteus mirabilis, Salmonella sp, Acinetobacter sp, Klebsiella pneumoniae, and Serratia marcescens) and four Gram-positive bacteria (Streptococcus pyogenes, S. faecalis, Coagulase negative Staphylococci, and Methicillin Resistant Staphylococcus aureus). All the bacteria were revived from glycerol stock by sub-cultured on nutrient agar (NA) and incubated overnight at 37°C. For inoculum preparation, a loopful of culture was taken from a single colony and further grown in nutrient broth (NB). The bacterial suspensions were standardized following the CLSI guidelines by adjusting the turbidity equivalent to 0.5 McFarland standard or approximately 1-2 x 108 CFU/ml13.
Antimicrobial Assay of G. boninense Fruiting Bodies Extracts
The disc diffusion method was performed to test the antimicrobial activity of each solvent extracts14. Firstly, 100 ml of bacterial inoculums containing 1-2 x 108 CFU/ml were spread over the Mueller Hinton agar (MHA) plate by sterile cotton swab. Extracts were re-dissolved in DMSO and loaded over sterile filter paper discs (6 mm in diameter) to obtain final concentration of 20mg/disc. Then, discs were placed on the top of inoculated MHA. Two control discs were used containing Chloramphenical (30 mg) and DMSO as positive and negative control respectively. The diameters of growth inhibition zones were measured after incubated for 24 h at 37°C. The experiments were performed in triplicate.
Determination of Minimum Inhibitory Concentration (MIC)
The microdilution method was used to determine the minimum inhibitory concentration (MIC) according to CLSI guidelines13. MICs were evaluated using 96 well microtiter plates. Stock solutions of the extracts were diluted in a serial of two-fold dilution, resulting in concentration ranging from 0.625-20 mg ml-1. The final concentration of bacterial suspension was adjusted to 5 x 105 CFU/ml with 50 ml of bacterial inoculum supplemented 100 ml of NB in each well. The microtiter plate was included a set of 3 controls: a column with Chloramphenical (30 mg) as positive control; a column with all solutions except the extracts; and a column of solution with DMSO as negative control. The plates were incubated at 37°C for 24 h. The absorbance was determined at 590 nm. All tests were performed in triplicate.
Metabolites Analysis of the G. boninense Crude Extracts by LC-QTOF-MS
Chromatographic separation was performed using VanquishTM Horizon UHPLC system (Thermo Fisher Scientific, USA) coupled with electrospray ionization Impact II QTOF-mass spectrometry system (Bruker Daltonics, Germany). Crude extracts (20 mL) was injected into Kinetex F5 LC column (2.1 mm x 100 mm, 2.6 mm; USA) and maintained at 30°C with flow rate of 1.0 ml min-1 with mobile phase consisted of acetonitrile (A) and 0.2% formic acid in water (B). Detection wavelength was set at 254 nm for acquiring chromatograms. The gradient elution program was used as follows: 0 min (20% A); 8 min (29% A); 25 min (29% A); 55 min (30% A); 65 min (30% A); 75 min (31% A); 90 min (65% A); 110 min (90% A); 135 min (90% A)15.
The ESI source used was negative ionization mode and deployed at 4200 V. The mass data acquisition was set to nebulizer flow at 5.0 bar, gas temperature of 300°C, drying gas flow at 12 L min-1, scan range of m/z 50-1500. The acquired data were converted into netCDFdata format (*.cdf) using Bruker Compass DataAnalysis 4.1.
Data Preprocessing and Analysis
The mass data in netCDF files were processed using an open source software tool MZmine 2.0. Typically, this software allowed raw data processing by removes of noise background. Then, peak detection was done by centroid algorithm to detect all specified data points as m/z peaks into chromatograms. After that, local minimum search was chosen for deconvolution. Prior alignment and peaks were grouped by m/z and retention time by isotope grouper. The m/z value and retention time of the fragmented ions were utilized in comparing with those from compound database, namely PubChem.
Identification of the Collected Fruiting Bodies
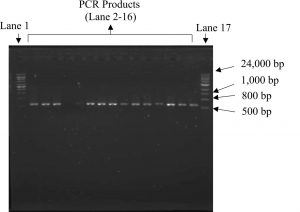
DNA extracted from the fruiting bodies were amplified using ITS1 and ITS4 primers which generates approximately 650 bp of the 5.8S rDNA gene as shown in Fig. 1. The primer pair was intended to detect the intraspecies variability in the highly reserved internal transcribed spacers (ITS) regions of fungi and basidiomycetes16. PCR produced similar size of DNA copies, nevertheless the order of nucleotide was not determined.
Fig. 1. Amplification of Ganoderma DNA isolates which produced approximately 650 bp size of PCR product using primers ITS1 and ITS4. Lane 1 and 17: 1kb DNA ladder, lane 2-16: PCR products.
Sequences obtained were subjected to BLAST search for the similarity of nucleotide sequences in NCBI database. Phylogenetic tree was constructed to study the evolutionary relationship within taxa. Table 1 showed the possible identity of Ganoderma fruiting bodies collected from field. BLAST results demonstrated the fruiting bodies possess 99% sequence similarity with Ganoderma species. The inserted nucleotide sequence showed closest homology with Ganoderma sp. BRIUMSa with highest maximum score followed by Ganoderma sp. BRIUMSb and Ganoderma boninense strain GBLS. Therefore, the closest species suggested by NCBI database matches list was G. boninense. Ganoderma sp. BRIUMSa, BRIUMSb, and BRIUMSc were belongs to the genus of Ganoderma and deposited into NCBI after isolated from Sabah, East Malaysia17 meanwhile Ganoderma boninense strain GBLS was isolated from University of Nottingham Malaysia campus, located at West Malaysia. Kok and colleagues18 suggested that the intra-species variation in fungi’s genetic could be due to the different geographical locations. Therefore, the sequence variation of basidiomata collected from field in this study possibly having lesser genetic variation with Ganoderma sp. BRIUMS compare to stain GBLS. Fig. 2 has shown that the collected basidiomata was sharing same clade with Ganoderma sp. BRIUMSa and forming a sisterly clade with Ganoderma sp. BRIUMSb with 98% bootstrap value. Both clades were closely clustered with Ganoderma boninense strain GBLS branch node with 94% bootstrap value. Hence, we can assume that Ganoderma sp. BRIUMS and collected basidiomata are mostly homologous and evolved from Ganoderma boninense strain as shown in the phylogenetic tree.
Table (1):
BLAST search results of fruiting bodies collected from Sawit Kinabalu Oil Palm Plantation shown 99% sequence identity to Ganoderma sp. BRIUMSa with highest score following by Ganoderma sp. BRIUMSb and Ganoderma boninense strain GBLS.
Accession |
Descriptions |
Max score |
Total score |
Query cover (%) |
E-value |
Percent identity (%) |
|---|---|---|---|---|---|---|
JN234427.1
|
Ganoderma sp. BRIUMSa internal transcribed spacer 1, partial sequence; 5.8S ribosomal RNA gene, complete sequence; and internal transcribed spacer 2, partial sequence |
1134 |
1134 |
97 |
0 |
99 |
JN234428.1
|
Ganoderma sp. BRIUMSb internal transcribed spacer 1, partial sequence; 5.8S ribosomal RNA gene, complete sequence; and internal transcribed spacer 2, partial sequence |
1129 |
1129 |
97 |
0 |
99 |
KF164430.1
|
Ganoderma boninense strain GBLS internal transcribed spacer 1, partial sequence; 5.8S ribosomal RNA gene and internal transcribed spacer 2, complete sequence; and 28S ribosomal RNA gene, partial sequence |
1118 |
1118 |
97 |
0 |
99 |
JN400511.1
|
Ganoderma sp. BL-94 18S ribosomal RNA gene, partial sequence; internal transcribed spacer 1, 5.8S ribosomal RNA gene, and internal transcribed spacer 2, complete sequence; and 28S ribosomal RNA gene, partial sequence |
1112 |
1112 |
97 |
0 |
99 |
JN400510.1
|
Ganoderma sp. BL-9 18S ribosomal RNA gene, partial sequence; internal transcribed spacer 2, 5.8S ribosomal RNA gene, and internal transcribed spacer 2, complete sequence; and 28S ribosomal RNA gene, partial sequence |
1112 |
1112 |
97 |
0 |
99 |
Fig. 2. Relatedness relationship of the collected fruiting bodies from the Sawit Kinabalu oil palm plantation with the five closest sequence retrieved from NCBI database. The topology was inferred by maximum likelihood. The bootstrap value is expressed in percentage and displayed next to each node. The branch length is drawn in scale with nucleotide change in unit of number of base substitutions per site.
Antimicrobial Activity of G. boninense Extracts
The zone of inhibition was recorded in Table 2. The results revealed that the fruiting bodies of G. boninense were potentially effective in suppressing microbial growth of tested pathogens with variable potency depending on the types of solvent used for extraction purpose. Solvent extraction is likely to extract the active compounds with similar polarity19. Ethyl acetate extract showed a broad variable antimicrobial properties against both Gram positive and negative bacterial strains (Enterobacter sp, P. aeruginosa, P. mirabilis, Acinetobacter sp, K. pneumoniae, S. marcescens, S. pyogenes, Coagulase negative Staphylococci, and MRSA) followed by hot water > acetone > methanol > ethanol and chloroform extract exhibited least antimicrobial activity. Ethyl acetate extract also exhibited the strongest growth inhibition against P. mirabilis (14.20 ± 0.40 mm). The results indicating solvent extracts of G. boninense fruiting bodies exerted antimicrobial properties were in agreement with previous works on other Ganoderma species. For example, solvent and aqueous extracts of G. lucidum fruiting bodies exhibited inhibitory effect against S. aureus, B. subilis, P. aeruginosa, V. cholera and S. typhi20. Moreover, extracts of G. lucidum mycelia demonstrated wide variation of bacteria inhibition for chloroform, acetone and aqueous extracts except methanol extract showed no inhibition activity against P. mirabilis, K. pneumoniae and P. aeuroginosa21. Crude extracts of G. applanatum and G. australe also exerted broad spectrum of bacteria inhibition except ethanol and water extracts of G. australe showed no inhibition activity against P.mirabilis, Actinomyces sp and P. aeruginase and K. pneumoniae respectively22. Besides Ganoderma species, various cultivated or wild macrofungi were reported for their antimicrobial activity23,24. Effect of different solvents extract of G. boninense on antimicrobial activity in this study was similar with some literatures. Wild fruiting bodies of Ganoderma collected from Causurina equiestifolia and Morinda tinctoria hard wood tree in India possess maximum and minimum inhibition activity against tested bacteria by ethyl acetate and chloroform extracts respectively25. Ameri et al.26 also reported ethyl acetate extract of G. lucidum, G. praelongum and G. resinaceum exhibited maximum inhibition activity against 30 strains of MRSA while chloroform extract was shown weak activity. However, numerous studies have shown inverse findings. Samarokoon et al.27 revealed that the chloroform fraction of G. lucidum extract from Sri Lankan showed profound antibacterial activity. Non-polar fractions of antimicrobial compounds isolated from G. lucidum were related to ergonstan-type steriods and lanostan-type triterprnoid28. All G. boninense extracts were not sensitive to E. coli, Salmonella sp (Gram negative bacteria) and S. faecalis (Gram positive bacteria). Klaus and Niksic29 also reported that aqueous extract of some G. lucidum strains showed no inhibition against Salmonella sp, E. coli, and S. aureus. Difference sensitivity of bacteria toward extracts could be related to morphological differences of bacteria cell wall. The characteristics of the cell wall and cell surface could be attributed to the permeability of the extract30. For instance, Gram positive bacterial pathogens have high resistant to treatment due to the ability of biofilm formation that creating a barrier for the penetration of extract compounds31.
Table (2):
Inhibition zone of different solvent extracts of G. boninense fruiting bodies (20 mg/disc) against several clinical isolate pathogens. Data expressed as mean SD, n = 3. Different lowercase alphabets within rows represent statistically significant difference with the different solvent extracts within same bacterial strain at p < 0.05 by One Way ANOVA Tukey’s test. Different uppercase alphabets within columns represent statistically significant difference within same solvent extracts with different bacterial strain at p < 0.05 by One Way ANOVA Tukey’s test.
| Bacterial strains | Inhibition zones (mm) | ||||||
|---|---|---|---|---|---|---|---|
| Chloramphenical (30 mg) | Chloroform | Ethyl Acetate | Acetone | Methanol | Ethanol | Hot water | |
| Gram negative pathogens | |||||||
| E. coli | 22.03 ± 0.05aAF | 0.00 ± 0.00bB | 0.00 ± 0.00bG | 0.00 ± 0.00bF | 0.00 ± 0.00bC | 0.00 ± 0.00bD | 0.00 ± 0.00b |
| E. coli (urine sample) | 22.03 ± 0.18aAG | 0.00 ± 0.00cB | 0.00 ± 0.00cG | 0.00 ± 0.00cF | 0.00 ± 0.00cC | 0.00 ± 0.00cD | 7.50 ± 0.10bBCDEFG |
| Enterobacter sp | 23.60 ± 0.08aAC | 0.00 ± 0.00dB | 11.68 ± 0.10bAC | 10.63 ± 0.06bA | 0.00 ± 0.00dC | 0.00 ± 0.00dD | 7.67 ± 0.06cBCDEFG |
| Pseudomonas aeruginosa | 21.40 ± 0.17aBCDEFG | 0.00 ± 0.00dB | 12.60 ± 0.05bAB | 8.37 ± 0.05cAE | 11.70 ± 0.09bA | 13.50 ± 0.09bA | 8.33 ± 0.14cAF |
| Proteus mirabilis | 21.30 ± 0.05aBCDEFG | 0.00 ± 0.00dB | 14.20 ± 0.40bA | 9.20 ± 0.20cAB | 0.00 ± 0.00dC | 0.00 ± 0.00dD | 8.67 ± 0.08cAC |
| Salmonella sp | 24.97 ± 0.15aA | 0.00 ± 0.00bB | 0.00 ± 0.00bG | 0.00 ± 0.00bF | 0.00 ± 0.00bC | 0.00 ± 0.00bD | 0.00 ± 0.00b |
| Acinetobacter sp | 9.97 ± 0.07aH | 0.00 ± 0.00bB | 9.83 ± 0.16aBCDEF | 7.67 ± 0.16aBCDE | 8.87 ± 0.08aB | 0.00 ± 0.00bD | 9.27 ± 0.06aAB |
| Klebsiella pneumoniae | 23.47 ± 0.10aAD | 0.00 ± 0.00cB | 10.73 ± 0.10bAE | 0.00 ± 0.00cF | 0.00 ± 0.00cC | 0.00 ± 0.00cD | 10.07 ± 0.11bA |
| Serratia marcescens | 23.47 ± 0.08aAE | 0.00 ± 0.00dB | 9.77 ± 0.04bBCDEF | 0.00 ± 0.00dF | 0.00 ± 0.00dC | 0.00 ± 0.00dD | 8.20 ± 0.03cAG |
| Gram positive pathogens | |||||||
| Streptococcus pyogenes | 18.63 ± 0.26aFG | 7.93 ± 0.04cA | 11.67 ± 0.13bAD | 8.60 ± 0.08bcAD | 0.00 ± 0.00dC | 0.00 ± 0.00dD | 8.67 ± 0.12bcAD |
| Streptococcus faecalis | 25.43 ± 0.13aA | 0.00 ± 0.00bB | 0.00 ± 0.00bG | 0.00 ± 0.00bF | 0.00 ± 0.00bC | 0.00 ± 0.00bD | 0.00 ± 0.00b |
| Coagulase neg staphylococci | 19.27 ± 0.04aFG | 0.00 ± 0.00dB | 9.17 ± 0.12cBCDEF | 8.77 ± 0.07cACD | 13.43 ± 0.18bA | 9.67 ± 0.06cB | 7.77 ± 0.06cBCDEFG |
| MRSA | 24.60 ± 0.12aAB | 0.00 ± 0.00dB | 10.23 ± 0.07bAF | 8.20 ± 0.17cBCDE | 8.80 ± 0.04bcB | 7.70 ± 0.07cC | 8.47 ± 0.06cAE |
Minimum Inhibitory Concentration (MIC) of the Effective Extracts
Results of MIC were recorded in Table 3. The MIC values obtained ranged between 0.625 mg ml-1 to 5.000 mg ml-1 with methanol extract demonstrating the highest antimicrobial activity against Coagulase neg staphylococci while hot water extracts exhibited with the lowest antimicrobial activity. Comparatively moderate activities were observed in ethyl acetate extracts exhibited MIC values of 1.250 mg ml-1 (Enterobacter sp, P. aeruginosa, P. mirabilis, S. pyogenes) and 2.500 mg ml-1 (Acinetobacter sp, K. pneumoniae, S. marcescens) against tested pathogens. Various degree of inhibitory concentration was observed for the G. boninense extracts could be due to the polarity of solvent. The extractability of secondary metabolites mainly depend on the type of solvents and methods that is used during extraction of sample. Ethyl acetate is a semi polar solvent that could dissolve broad spectrum of compounds, varying from polar to nonpolar compounds. Results of ethyl acetate extract of G. boninense showed growth inhibition of Gram positive and Gram negative pathogens indicating that the extract contains broad spectrum of active compounds and suggesting that the antimicrobial compounds are leaning toward polar-soluble. These were supported by few reports showed numerous strains of Ganoderma species and edible mushrooms exhibited broad spectrum of antibacterial properties in higher polarity extracts like ethyl acetate and methanol compare with lower polarity extracts like hexane32-34. However, the existence of low polar-soluble compounds in Ganoderma species or edible mushrooms are undeniable35,24. Preliminary phytochemical has shown that ethyl acetate soluble fraction of G. lucidum possess compounds like alkaloid, carbohydrate and glycoside, phenol, sterol, terpenoid36,37. Strong potent of inhibition zones and MIC values were observed in ethyl acetate extract of wild G. lucidum fruiting bodies against selected bacterial strains and study suggested that the antibacterial activity of solvent extracts were related to the high occurrence of total phenolics, flavonoids, alkaloids, terpenoids and glycosides compounds38. Antimicrobial compounds isolated from Ganoderma species by difference solvent polarity were reviewed by Basnet and co-workers32. Antimicrobial activity of mushrooms were reported in varied levels. Apart from solubility in solvents and extraction methods, other factors could be due to sensitivity and tolerance of the bacterial strains to mushroom crude extracts. Moreover, the production of secondary metabolites by natural resources and bioactive compositions may varied upon different environmental conditions.
Table (3):
Minimum inhibitory concentration (MIC) of the different solvent extracts of G. boninense fruiting bodies against selected bacterial strains.
| Bacterial strains | Type of solvents | MIC (mg ml-1) |
|---|---|---|
| Gram negative pathogen | ||
| E.coli (Urine sample) | Hot water | 5.000 |
| Enterobacter sp | Ethyl acetate | 1.250 |
| Pseudomonas aeruginosa | Ethanol | 1.250 |
| Proteus mirabilis | Ethyl acetate | 1.250 |
| Acinetobacter sp | Ethyl acetate | 2.500 |
| Klebsiella pneumoniae | Ethyl acetate | 2.500 |
| Serratia marcescens | Ethyl acetate | 2.500 |
| Gram positive pathogen | ||
| Streptococcus pyogenes | Ethyl acetate | 1.250 |
| Coagulase neg staphylococci | Methanol | 0.625 |
| MRSA | Ethyl acetate | 1.250 |
Crude polysaccharide of difference Ganoderma species were reported to exert broad spectrum of antimicrobial activity covering both Gram positive and Gram negative bacterial39. However, polysaccharide compounds could limit the antimicrobial activity because carbohydrates are element that help the growth of microorganism40. Further investigation on the active compound that responsible for the inhibition is necessary as crude extract contains numerous of active compounds.
Identification of the Compounds from Crude Extracts of G. boninense
Extracts with good antimicrobial activity were identified using HPLC-MS. As the result, a list of m/z chromatographic peaks were obtained and illustrated in Fig. 3 (Ethyl acetate extract), Fig. 4 (Ethanol extract) and Fig. 5 (Hot water extract).
Fig. 3. Extracted ion chromatogram (XIC) of ethyl acetate crude extract of G. boninense after peak deconvolution by local minimum search. Each individual recognized peak within the chromatogram is indicated in different colour with peak intensity and m/z value.
Fig. 4. Extracted ion chromatogram (XIC) of ethanol crude extract of G. boninense after peak deconvolution by local minimum search. Each individual recognized peak within the chromatogram is indicated in different colour with peak intensity and m/z value.
Fig. 5. Extracted ion chromatogram (XIC) of hot water crude extract of G. boninense after peak deconvolution by local minimum search. Each individual recognized peak within the chromatogram is indicated in different colour with peak intensity and m/z value
Table (4):
Putative compounds identification of ethyl acetate crude extract of G. boninense from PubChem by HPLC coupled with electrospray ionization mass spectrometry system.
Putative Identification |
Retention time |
m/z |
Peak intensity |
Compound nature |
Reference |
|---|---|---|---|---|---|
2-amino-4-chloro-3-hydroxybutanoic acid |
0.62 |
153.0211 |
1.7E6 |
Fatty acid |
Isogai et al.41 |
Cryptopleurine |
0.65 |
377.2027 |
4.5E5 |
Alkaloid |
Al-Shamma et al.42 |
6-ethoxy-2-(2-phenylethenyl)quinoline |
9.16 |
275.1291 |
2.0E5 |
Alkaloid |
Singh et al.43 |
Americine |
30.30 |
545.3056 |
1.5E5 |
Alkaloid |
Li et al.44 |
Pyroaconitine |
42.38 |
585.2982 |
5.4E5 |
Alkaloid |
Borcsa et al.45 |
(4-(Bis(2-chloroethyl)amino)phenyl)methyl hexadecanoate |
46.71 |
485.2834 |
1.5E5 |
Fatty acid |
Chandrasekaran et al.46 |
Lythranine |
73.42 |
527.2918 |
2.8E5 |
Alkaloid |
Al-Snai.47 |
Catuabine C |
77.79 |
357.1644 |
3.0E5 |
Alkaloid |
Aguiar et al.48 |
Cyano-cribrostatin IV |
77.79 |
715.3347 |
1.7E5 |
Alkaloid |
Pettit et al.49 |
4-Acetyl-4-guanidino-6-methyl(propyl)carboxamide-4,5-dihydro-2H-pyran-2-carboxylic acid |
79.32 |
341.1697 |
5.2E5 |
Phenol |
Srinivas et al.50 |
2-[(1R)-3-[di(propan-2-yl)amino]-1-phenylpropyl]-4-(hydroxymethyl)phenol; heptanoic acid |
86.46 |
471.3392 |
2.1E5 |
Fatty acid |
Ara et al.51 |
Sodium; hexadecenoic acid |
94.76 |
279.2276 |
1.5E6 |
Fatty acid |
Abubakar et al.52 |
Levofloxacin monohydrate |
94.92 |
379.1510 |
3.6E5 |
Quinolone |
Noel53 |
3-decyl-N,N-dimethyl-2H-imidazole-1-carboxamide |
99.33 |
281.2432 |
1.6E6 |
Heterocyclic compound |
Gupta et al.54 |
Dioctyltin(2+); tetradecanoate |
121.65 |
800.5725 |
1.1E5 |
Fatty acid |
Sales55 |
Based on the PubChem database search, a total of 15 compounds for ethyl acetate crude extract of G. boninense were putatively identified with previously reported with antimicrobial activity. Presented compounds presumably represented by alkaloid, quinoline, fatty acid, carboxylic acids derivatives. On the other hand, compounds of ethanol crude extract were putatively categorized into several groups, majority represented by fatty acid, alkaloid, heterocyclic compound with a few of peptide and glycoside derivative compounds as listed in Table 5. Hot water crude extract has shown the least number of compounds with only 9 compounds reported in previous studies on antimicrobial activity. Those putative identified compounds which may contribute to antimicrobial activity of G. boninense were tabulated in Table 4-6 with references.
Table (5):
Putative compounds identification of ethanol crude extract of G. boninense from PubChem by HPLC coupled with electrospray ionization mass spectrometry system
Putative identification |
Retention time |
m/z |
Peak intensity |
Compound nature |
Reference |
|---|---|---|---|---|---|
Nitrooxadiazole |
0.60 |
114.9991 |
3.5E5 |
Heterocyclic compound |
Prajapati et al.56 |
2-methylidenebutanedioic acid |
1.33 |
153.0149 |
4.6E6 |
Dicarboxylic acid |
Cordes et al.57 |
Auramycin G |
5.23 |
783.3414 |
3.5E5 |
Glycoside |
Fujiwara et al.58 |
Butyric acid, 2-bromo-4-[4-nitrophenyl]-, methyl ester |
5.66 |
300.9923 |
3.8E5 |
Fatty acid |
Fafal et al.58 |
Desoxyharringtonin |
6.10 |
515.2540 |
9.1E5 |
Alkaloid |
Mikolajczak et al.60 |
Ergovaline |
6.41 |
533.2642 |
3.1E5 |
Alkaloid |
Panaccione, et al.61 |
Deoxyharringtonine |
6.77 |
515.2536 |
4.7E5 |
Alkaloid |
Mikolajczak et al.60 |
N-[(3-cyclopentyloxyphenyl)methyl]-2-(2-propan-2-yloxy-5-propylphenoxy)ethanamine; oxalic acid |
7.18 |
501.2750 |
3.1E5 |
Fatty acid |
Kwak et al.62 |
Deoxyharringtonin |
11.91 |
515.2542 |
1.4E6 |
Alkaloid |
Mikolajczak et al.60 |
Ergocornine |
13.13 |
561.2948 |
7.0E5 |
Alkaloid |
Panaccione, et al.61 |
Leu-Nle-Uracil polyoxin C |
17.38 |
513.2382 |
2.7E5 |
Peptide |
Smith et al.63 |
Pyroaconitine |
46.14 |
585.2931 |
5.0E5 |
Alkaloid |
Borcsa, et al.45 |
amikacin |
46.38 |
585.2934 |
4.7E5 |
Aminoglycoside |
Yoshikawa et al.64 |
N-epsilon-(Octanoyl)lysyl-uracil polyoxin C |
56.60 |
541.2690 |
3.0E5 |
Glycoside |
Smith et al.63 |
Catuabine C |
78.22 |
357.1642 |
3.6E5 |
Alkaloid |
Aguiar et al.48 |
Ambruticin VS3 |
90.82 |
501.3487 |
3.0E5 |
Heterocyclic compound |
Vetcher et al.65 |
Hexadecenoic acid; linoleate |
94.78 |
279.2284 |
2.8E5 |
Fatty acid |
Canli et al.66 |
Levofloxacin monohydrate |
94.81 |
379.1514 |
2.9E5 |
Glycoside |
Noel53 |
Isooctadecanoic acid, 1,1’-(dibutylstannylene)ester |
121.83 |
800.5696 |
1.9E5 |
Fatty acid |
Fan et al.67 |
Table (6):
Putative compounds identification of hot water crude extract of G. boninense from PubChem by HPLC coupled with electrospray ionization mass spectrometry system.
Putative identification |
Retention time |
m/z |
Peak intensity |
Compound nature |
Reference |
|---|---|---|---|---|---|
2-Furanmethanethiol, 5-methyl- |
0.56 |
128.0322 |
3.8E5 |
Heteroaromatic compound |
Chen et al.68 |
Formic acid |
0.61 |
115.0003 |
1.3E6 |
Carboxylic acid |
Kovanda et al.69 |
1-benzothiophene |
0.61 |
133.0111 |
4.0E6 |
Heterocyclic compound |
Aganagowda et al.70 |
Thiazolo[5,4-d]pyrimidine-7(6H)-thione |
0.64 |
168.9781 |
3.6E5 |
Heterocyclic compound |
Habib et al.71 |
Octanoic acid |
2.65 |
187.0946 |
3.2E5 |
Fatty acid |
Huang et al.72 |
Ergovaline |
5.35 |
533.2653 |
3.6E5 |
Alkaloid |
Panaccione, et al.61 |
2-hydroxypropane-1,2,3-tricarboxylic acid |
8.70 |
517.2709 |
7.6E5 |
Carboxylic acid |
Hawkins73 |
Desoxyharringtonin |
11.51 |
515.2552 |
1.6E5 |
Alkaloid |
Mikolajczak et al.60 |
Ergocornine |
13.17 |
561.2957 |
2.9E5 |
Alkaloid |
Panaccione, et al.61 |
Numerous synthetic alkaloids using quinoline or pyridine as scaffold for development of antimicrobial drug has been exploited74,75. Nonetheless, several natural occurring alkaloid derivatives like quinoline, indole, piperazine, aptamine have been reported for their antibacterial activity76. For example, GC-MS analysis of crude extract from Plumeria alba flowers revealed compounds contributed to antimicrobial activity were alkaloid derivatives of pyridine, isoquinoline, indole, vinca and reserpine classes77. According to Ismail et al.78, fatty acids such as octadecanoic acid, dodecanoic acid, methyl ester, hexadecenoic acid were identified from chloroform extract of G. boninense fruiting bodies and exhibited growth inhibition properties against P. aeruginosa and S. aureus. Natural occurring of fatty acids such as methyl ester, tetradecenoic, hexadecenoic, octadeceadienoic extracted from G. resinaceum possess antimicrobial effects against Fusarium oxysporum, C. albicans, and S. aureus79. The presence of the compounds from three different solvent extracts of G. boninense may have contributed to the antimicrobial activity presented in the current work.
Different solvents were used to extract bioactive compounds of G. boninense fruiting body and their antimicrobial properties were examined. Ethyl acetate is a potential solvent for G. boninense extraction because of the remarkable broad spectrum of antimicrobial activity toward Gram positive and Gram negative pathogens. The extract gave the strongest inhibition to the growth of P. mirabilis. LC-MS analysis confirmed the crude extracts of G. boninense contains numerous compounds such as alkaloid, fatty acid, heterocyclic compound, peptide, and glycoside derivatives. The results obtained from this study revealed that G. boninense possess great potential with antimicrobial activities. Hence, this fungus deserves more attention in discovering their medicinal values in the future.
ACKNOWLEDGMENTS
We would like to thank Sawit Kinabalu Sdn Bhd for the assistance in samples collection for this work.
CONFLICT OF INTEREST
Authors would like to declare that there is no conflict of interest.
AUTHORS’ CONTRIBUTION
All authors participated in design of the experiments. Y.S. conducted the experiments and drafted the manuscript. K.P. supervised the project and reviewed the manuscript.
FUNDING
This study was financially supported by UMS Great Grant (GUG 0157-2/2017) and Skim Penyelidikan Bidang Keutamaan UMS (SBK 0432-2018) of Universiti Malaysia Sabah.
ETHICS STATEMENT
Not applicable.
AVAILABILITY OF DATA
All data generated and analyzed for current study were presented in form of tables and Fig.s and included in this article.
- Basak S, Singh P, Rajurkar M. Multidrug resistant and extensively drug resistant bacteria: A study. Journal of Pathogens. 2016;2016:4065603.
Crossref - Mamishi S, Moradkhani S, Mahmoudi S, Hosseinpour-Sadeghi R, Pourakbari B. Penicillin-Resistant trend of Streptococcus pneumoniae in Asia: A systematic review. Iran J Microbiol. 2014;6(4):198-210.
- World Health Organization. Global antimicrobial resistance surveillance system (GLASS) report. Early implementation 2016-2017. 2017. ISBN 978-92-4-151344-9. https://www.who.int/docs/default-source/searo/amr/global-antimicrobial-resistance-surveillance-system-(glass)-report-early-implementation-2016-2017.pdf?sfvrsn=ea19cc4a_2. Accessed 6 July, 2019
- Ministry of Health Malaysia. Malaysian action plan on antimicrobial resistance (MyAP-AMR) 2017-2021. 2017. http://www.moh.gov.my/moh/resources/Penerbitan/Garis%20Panduan/Garis%20panduan%20Umum%20(Awam)/National_Action_Plan_-_FINAL_29_june.pdf Accessed 6 July, 2019.
- Mathur S, Hoskins C. Drug development: Lessons from nature. Biomedical Reports. 2017;6(6):612-614.
Crossref - Colegate, SM, Molyneux, RJ. Bioactive natural products: detection, isolation and structure determination. 2008; pp. 421-437.CRC Press; Boca Raton, FL, USA.
Crossref - Abdallah EM. Plants: An alternative source for antimicrobials. Journal of Applied Pharmaceutical Science, 2011;01(06):16-20. https://www.japsonline.com/admin/php/uploads/118_pdf.pdf. Accessed June 12, 2020
- Thomford N, Senthebane D, Rowe A, et al. Natural products for drug discovery in the 21st century: innovations for novel drug discovery. Int J Mol Sci. 2018;19(6):1578.
Crossref - Sonawane H, Bhosle S, Garad S. Antimicrobial activity of some species of Phellinus and Ganoderma sample from Western Ghats of India. Int. J Pharm Sci Rev Res. 2012;3:1795-1799.
Crossref - Chuang CM, Wang HE, Chang CH, et al. Sacchachitin, a novel chitin-polysaccharide conjugate macromolecule present in Ganoderma lucidum: purification, composition, and properties. Pharm Biol. 2013;51:84-95.
Crossref - Chong KP, Atong M, Rossall S. The roles of syringic, caffeic and 4-hydroxybenzoic acids in Ganoderma-oil palm interaction. Asian J Microbiol Biotechnol Environ Sci. (AJMBES) 2012;14(2):157-166.
- Chong KP, Lum MS, Foong CP, et al. First identification of Ganoderma boninense isolated from Sabah based on PCR sequence homology. African Journal of Biotechnology. 2011;10(66):14718-14723.
- CLSI. Performance standards for antimicrobial disk susceptibility testing: Twenty-first informational supplement. CLSI document M100-S21. Clinical and Laboratory Standards Institute, Wayne, PA. 2011.
- Bereksi MS, Hassaine H, Bekhechi C, Abdelouahid DE. Evaluation of antibacterial activity of some medicinal plants extracts commonly used in Algerian traditional medicine against some pathogenic bacteria. Pharmacognosy Journal. 10 (3).
- Wu L, Liang W, Chen W, et al. Screening and analysis of the marker components in Ganoderma lucidum by HPLC and HPLC-MSnwith the aid of chemometrics. Molecules (Basel, Switzerland). 2017;22(4):584.
Crossref - Gardes M, Bruns TD. ITS primers with enhanced specificity for basidiomycetes- application to the identification of mycorrhizae and rusts. Molecular Ecology. 1993;2(2):113-118.
Crossref - Chong KP, Abdullah S, Ng TL. Molecular fingerprint of Ganoderma spp. from Sabah, Malaysia. Int J Agric Biol. 2013;15:1112-1118. https://agris.fao.org/agris-search/search.do?recordID=PK2014000589. Accessed June 12, 2020
- Kok S, Wong WC, Tung H, Goh YK, Goh KJ. In vitro growth of Ganoderma boninense isolates on novel palm ex- tract medium and virulence on oil palm (Elaeis guineensis) seed- lings. Malays J Microbiol. 2013;9(1):33-42.
- Zhang QW, Lin LG, Ye WC. Techniques for extraction and isolation of natural products: a comprehensive review.Chinese Medicine. 2018;13:20.
Crossref - Singh J, Gupta S, Malviya S, Ahrwar B. In-vitro evaluation of antimicrobial activity of Ganoderma lucidum. IInt J Adv Res. 2014;2(6):460-466. http://www.journalijar.com/uploads/31_IJAR-3328.pdf. Accessed June 12, 2020
- Kamble R, Venkata S, Gupte AM. Antimicrobial activity of Ganoderma lucidum mycelia. J Pure Appl Microbiol. 2011;5:1-4. https://www.researchgate.net/publication/260024799_Antimicrobial_Activity_of_Ganoderma_lucidum_Mycelia. Accessed June 12, 2020
- Jonathan SG, Awotona FE. Studies on antimicrobial potentials of three Ganoderma species. African Journal of Biomedical Research. 2010;13(2):131-139. http://hdl.handle.net/1807/54214. Accessed June 12, 2020.
- Alves ML, Ferreira IC, Martins A, Pintado M. Antimicrobial activity of wild mushroom extracts against clinical isolates resistant to different antibiotics. J Appl Microbiol. 2012;113 (2):466-475.
Crossref - Shen HS, Shao S, Chen JC, Zhou T. Antimicrobials from mushrooms for assuring food safety. Comprehensive Reviews in Food Science and Food Safety. 2017;16(2):316-329.
Crossref - Rajesh K, Dhanasekaran D. Phytochemical screening and biological activity of medicinal mushroom Ganoderma species. Malaya J Biosci. 2014;1(2):67-75.
- Ameri A, Vaidya JG, Deokule SS. In vitro evaluation of anti-staphylococcal activity of Ganoderma lucidum, Ganoderma praelongum and Ganoderma resinaceum from Pune, India. Afr J Microbiol Res. 2011;5(3):328-333.
- Samarakoon KW, Lee JH, De Silva ED, et al. Bioactivity evaluation of organic solvent extractions of Ganoderma lucidum: a Sri Lankan basidiomycete. J NSF. 2013;41(3):249-57.
Crossref - Vazirian M, Faramarzi MA, Ebrahimi SES, et al. Antimicrobial effect of the Lingzhi or Reishi medicinal mushroom, Ganoderma lucidum (higher Basidiomycetes) and its main compounds. International Journal of Medicinal Mushrooms. 2014;16(1).
- Klaus A, Niksic M. Influence of the extracts isolated from Ganoderma lucidum mushroom on some microorganisms. Zbornik Matice srpske za prirodne nauke. 2007;(113):219-226.
- Auer GK, Weibel DB. Bacterial cell mechanics. Biochemistry. 2017;56(29):3710-3724.
- Peng D. Biofilm formation of Salmonella. Microbial biofilms-Importance and Applications. 2016.
Crossref - Basnet BB, Liu L, Bai L, Liu H. Current and future perspective on antimicrobial and anti-parasitic of Ganoderma sp.: an update. Mycology. 2017;8(2):111-124.
- Chaiham M, Phutdhawong WS, Amornlerdpison D, Phutdhawong W. Antibacterial, antioxidant properties and bioactive compounds of Thai cultivated mushroom extracts against food-borne bacterial strains. Chiang Mai Journal of Science. 2018;45(4):1713-1727. http://www.thaiscience.info/Journals/Article/CMJS/10990141.pdf. Accessed June 12, 2020
- Ochoa-Zarzosa A, Vazquez-Garciduenas MS, Robinson-Fuentes VA, Vazquez-Marrufo G. Antibacterial and cytotoxic activity from basidiocarp extracts of the edible mushroom Lactarius indigo (Schw.) Fr. (Russulaceae). Afr J Pharm Pharmacol. 2011;5(2):281-288.
Crossref - Trigos A. Biologically active metabolites of the genus Ganoderma: three decades of myco-chemistry research. Revista Mexicana de Micologia. 2011;34:63-83. http://www.scielo.org.mx/pdf/rmm/v34/v34a9.pdf. Accessed June 12, 2020.
- Gowrie US, Chathurdevi G, Rani K. Evaluation of bioactive potential of basidiocarp extracts of Ganoderma lucidum. International Journal. 2014;3(1):36-46. https://ijpras.com/en/article/evaluation-of-bioactive-potential-of-basidiocarp-extracts-of-ganoderma-lucidum. Accessed June 12, 2020.
- Shamaki BU, Geidam YA, Abdulrahma F, Ogbe AO, Sandabe UK. Evaluation of phytochemical constituents and in vitro antibacterial activity of organic solvent fractions of Ganoderma lucidum methanolic extract. International Journal of Medicinal Plant Research. 2012;1(3):026-031. https://www.semanticscholar.org/paper/Evaluation-of-phytochemical-constituents-and-in-of-Bu-Ya/dee2916084bd5d70649e53dce839e42e7f2f6da3. Accessed June 12, 2020.
- Sharif S, Shahid M, Mushtaq M, Akram S, Rashid A. Wild mushrooms: a potential source of nutritional and antioxidant attributes with acceptable toxicity. Preventive Nutrition and Food Science. 2017;22(2):124.
- Skalicka-Wozniak K, Szypowski J, Los R, et al. Evaluation of polysaccharides content in fruit bodies and their antimicrobial activity of four Ganoderma lucidum (W Curt.: Fr.) P. Karst. strains cultivated on different wood type substrates. Acta Societatis Botanicorum Poloniae. 2012;81(1).
Crossref - Cushnie TT, Lamb AJ. Recent advances in understanding the antibacterial properties of flavonoids.International Journal of Antimicrobial Agents. 2011;38(2):99-107.
- Isogai A, Fukuchi N, Yamashita S, Suyama K, Suzuki A. Syringostatins, novel phytotoxins produced by Pseudomonas syringae pv. syringae. Agricultural and Biological Chemistry. 1989;53(11):3117-3119.
Crossref - Al-Shamma A, Drake SD, Guagliardi LE, Mitscher LA, Swayze JK. Antimicrobial alkaloids from Boehmeria cylindrica. Phytochemistry. 1982;21(2):485-487.
Crossref - Singh S, Kaur G, Mangla V, Gupta MK. Quinoline and quinolones: promising scaffolds for future antimycobacterial agents. J Enzyme Inhib Med Chem. 2015;30(3):492-504.
- Li XC, Cai L, Wu CD. Antimicrobial compounds from Ceanothus americanus against oral pathogens. Phytochemistry. 1997;46(1):97-102.
- Borcsa B, Fodor L, Csupor D, Forgo P, Molnar A, Hohmann J. Diterpene alkaloids from the roots of Aconitum moldavicum and assessment of Nav 1.2 sodium channel activity of aconitum alkaloids. Planta Medica. 2014;80(02/03):231-236.
- Chandrasekaran M, Senthilkumar A, Venkatesalu V. Antibacterial and antifungal efficacy of fatty acid methyl esters from the leaves of Sesuvium portulacastrum L. Eur Rev Med Pharmacol Sci. 2011;15(7):775-780. https://pubmed.ncbi.nlm.nih.gov/21780546/. Accessed June 12, 2020
- Al-Snai AE. Chemical constituents and pharmacological effects of Lithospermum officinale. IOSR Journal of Pharmacy. 2019;9(8):12-21. https://www.semanticscholar.org/paper/Chemical-Constituents-and-Pharmacological-Effects-Al-Snafi/eaa331ace9d5afa5bb3de958c2116381d99d3009. Accessed June 12, 2020
- Aguiar JS, Araujo RO, do Desterro RM, et al. Antimicrobial, antiproliferative and proapoptotic activities of extract, fractions and isolated compounds from the stem of Erythroxylum caatingae Plowman.Int J Mol Sci. 2012;13(4):4124-4140.
- Pettit GR, Knight JC, Collins JC, et al. Antineoplastic agents 430. Isolation and structure of cribrostatins 3, 4, and 5 from the republic of maldives cribrochalina species. Journal of Natural Products. 2000;63(6):793-798.
- Srinivas K, Reddy TR, Himabindu V, Reddy GM, Mohan NJ. Synthesis and antibacterial activity of novel quinoxaline-5-carboxamide derivatives. J Appl Chem. 2014;3(4):1432-1439. https://www.researchgate.net/publication/316279326_Synthesis_and_Antibacterial_Activity_of_Novel_Quinoxaline-5-Carboxamide_Derivatives. Accessed June 12, 2020
- Ara I, Bukhari NA, Solaiman D, Bakir MA. Antimicrobial effect of local medicinal plant extracts in the Kingdom of Saudi Arabia and search for their metabolites by gas chromatography-mass spectrometric (GC-MS) analysis. J Med Plant Res. 2012;6(45):5688-5694.
- Abubakar M, Majinda R. GC-MS analysis and preliminary antimicrobial activity of Albizia adianthifolia (Schumach) and Pterocarpus angolensis (DC). Medicines. 2016;3(1):3.
- Noel GJ. A review of levofloxacin for the treatment of bacterial infections. Clinical Medicine. Therapeutics, 2009;1:CMT-S28. https://doi.org/10.4137/CMT.S28.
- Gupta N, Pathak DP. Synthesis and evaluation of N-substituted imidazole derivatives for antimicrobial activity. Indian Journal of Pharmaceutical Sciences. 2001;73(6):674.
- Sales DL, Oliveira OP, Cabral MES,et al. Chemical identification and evaluation of the antimicrobial activity of fixed oil extracted from Rhinella jimi. Pharmaceutical Biology. 2015;53(1):98-103.
- Prajapati AK, Modi V. Mesogenic bent-shaped nitrooxadiazoles and thiadiazoles. Liquid Crystals. 2011;38(2):191-199. https://doi.org/10.1080/02678292.2010.535659
- Cordes T, Michelucci A, Hiller K. Itaconic acid: the surprising role of an industrial compound as a mammalian antimicrobial metabolite. Annual Review of Nutrition. 2015;35:451-473.
Crossref - Fujiwara A, Hoshino T, Tazoe M, Fujiwara M. New anthracycline antibiotics, auramycins and sulfurmycins. J Antibiot.1982;35(2):164-175.
- Fafal T, Yilmaz FF, Birincioglu SS, Hosgor-Limoncu M, Kivcak B. Fatty acid composition and antimicrobial activity of Asphodelus aestivus seeds. Human and Veterinary Medicine. 2016;8(2):103-107. https://www.semanticscholar.org/paper/Fatty-acid-composition-and-antimicrobial-activity-Fafal-Y%C4%B1lmaz/9f1066becae0ac203c892deeded06f4a9d9f5afc. Accessed June 12, 2020
- Mikolajczak KL, Powell RG, Smith Jr CR. Deoxyharringtonine, a new antitumor alkaloid from Cephalotaxus: structure and synthetic studies. Tetrahedron. 1972;28 (7):1995-2001.
Crossref - Panaccione DG. Origins and significance of ergot alkaloid diversity in fungi. FEMS Microbiology Letters. 2005;251(1):9-17.
Crossref - Kwak AM, Lee IK, Lee SY, Yun BS, Kang HW. Oxalic acid from Lentinula edodes culture filtrate: antimicrobial activity on phytopathogenic bacteria and qualitative and quantitative analyses. Mycobiology. 2016;44(4):338-342.
- Smith HA, Shenbagamurthi P, Naider F, Kundu B, Becker JM. Hydrophobic polyoxins are resistant to intracellular degradation in Candida albicans. Antimicrob Agents Chemother. 1986;29(1):33-39.
- Yoshikawa TT, Shibata, SA. In vitro antibacterial activity of amikacin and ticarcillin, alone and in combination, against Pseudomonas aeruginosa. Antimicrob Agents Chemother. 1978;13(6):997-999.
- Vetcher L, Menzella HG, Kudo T, Motoyama T, Katz L. The antifungal polyketide ambruticin targets the HOG pathway. Antimicrob Agents Chemother. 2007;51(10):3734-3736.
- Canli K, Altuner EM, Akata I, Turkmen Y, Uzek U. In vitro antimicrobial screening of Lycoperdon lividum and determination of the ethanol extract composition by gas chromatography/mass spectrometry. Bangladesh Journal of Pharmacology. 2016;11(2):389-394.
- Fan X, Wagner K, Sokorai KJ, Ngo H. Inactivation of gram-positive bacteria by novel phenolic branched-chain fatty acids. Journal of Food Protection. 2016;80(1):6-14.
- Chen C, Tong Z, Liao D, Li Y, Yang G, Li M. Chemical composition and antimicrobial and DPPH scavenging activity of essential oil of Toona sinensis (A. Juss.) Roem from China. Bioresources. 2014;9(3):5262-5278. https://ojs.cnr.ncsu.edu/index.php/BioRes/article/view/BioRes_09_3_5262_Chen_Antimicrobial_DPPH_Scavenging_Oil. Accessed June 12, 2020
- Kovanda L, Zhang W, Wei X, et al. In Vitro Antimicrobial activities of organic acids and their derivatives on several species of Gram-negative and Gram-positive bacteria. Molecules. 2019;24(20):3770.
- Aganagowda G, Thamyongkit P, Petsom A. Synthesis and antimicrobial activities of benzothiophene derivatives. Journal of the Chilean Chemical Society. 2012;57(1):1043-1047.
Crossref - Habib NS, Soliman R, El-Tombary AA, El-Hawash SA, Shaaban OG. Synthesis of thiazolo [4, 5-d] pyrimidine derivatives as potential antimicrobial agents. Arch Pharm Res. 2007;30(12):1511.
Crossref - Huang CB, Alimova Y, Myers TM, Ebersole JL. Short-and medium-chain fatty acids exhibit antimicrobial activity for oral microorganisms. Arch Oral Biol. 2011;56(7):650-654.
- Hawkins SG. Antimicrobial activity of cinnamic acid, citric acid, cinnamaldehyde, and levulinic acid against foodborne pathogens. Honors Thesis Project. University of Tennessee, Knoxville. 2014
- Brighty K, Gootz TD. The chemistry and biological profile of trovafloxacin.J Antimicrob Chemother. 1997;39 (suppl_2):1-14.
- Klimesova V, Svoboda M, Waisser K, Pour M, Kaustova J. New pyridine derivatives as potential antimicrobial agents. Il Farmaco. 1999;54(10):666-672.
- Cushnie TT, Cushnie B, Lamb AJ. Alkaloids: an overview of their antibacterial, antibiotic-enhancing and antivirulence activities. Int J Antimicrob Agents. 2014;44(5):377-386.
- Sibi G, Venkategowda A, Gowda L. Isolation and characterization of antimicrobial alkaloids from Plumeria alba flowers against foodborne pathogens. Am J Life Sci. 2014;2:1-6.
- Ismail K, Abdullah S, and Chong KP. Screening for potential antimicrobial compounds from Ganoderma boninense against selected food borne and skin disease pathogens. Int J Pharm Pharm Sci. 2014;6(2):771-774. http://eprints.ums.edu.my/9478/. Accessed June 12, 2020
- Abd El-Zaher SHF. Antimicrobial activity of lipid extracted from Gamma radiated mycelia of Ganoderma resinaceum. Egypt. J Exp Biol (Bot.). 2010;6(2):87-92. http://www.egyseb.net/fulltext/15-1430348915.pdf?1591898164. Accessed June 12, 2020
© The Author(s) 2020. Open Access. This article is distributed under the terms of the Creative Commons Attribution 4.0 International License which permits unrestricted use, sharing, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.