ISSN: 0973-7510
E-ISSN: 2581-690X
This study aimed to optimize the medium compositions and cultural conditions for improved chitinase production by a potential strain of Bacillus isolated from the marine environment and determine the antifungal activity of its chitinase against plant pathogenic fungi. Five potential isolates were cultured for chitinase production by submerged fermentation using colloidal chitin in a liquid medium. In this study, chitinase activity was determined by measuring reducing sugars, which were determined by the 3,5-dinitrosalicylic acid (DNS) assay. The most potential isolate, B26, showed similarity to Bacillus paramycoides based on the 16S rRNA gene sequence. The maximum chitinase production was achieved at 6.52±0.02 U/mL after 72 h of incubation in a medium containing 2% squid pen powder, supplemented with 0.5% sodium nitrate and 2% NaCl, with an initial pH of 7. It was observed that the optimization of cultural conditions resulted in 2.83 times higher chitinase production than an unoptimized medium. The antifungal activity of crude chitinase against phytopathogenic fungi was evaluated by a well-diffusion method. The chitinase of B. paramycoides B26 effectively inhibited the growth of Fusarium solani TISTR 3436 (83.4%) and Penicillium chrysogenum TISTR 3554 (80.12%).
Antifungal, Bacillus paramycoides, Chitinase, Squid Pen
Marine products, including shrimp, crab, and squid, have high global market demand. Remarkably, the expansion of seafood industries contributes to tons of shellfish waste that contains up to 58% chitin.1 Therefore, the treatment of chitin waste plays a significant role in ecological sustainability while also increasing the economic value of by-product utilization.2 Bioconversion of chitin into value-added products such as N-acetylglucosamine (NAG), chitosan, and chitinase has been proposed as an effective method of reducing waste because it is both helpful and environmentally sustainable.3
Chitinase (EC 3.2.1.14) is an enzyme that degrades chitin, a polymer found in the cell walls of fungi, crab shells, and insect cuticles.4 As noted previously, chitin is present in fungal cell walls; therefore, it can be decomposed by chitinolytic microorganisms.5,6 Global fungal pathogen spread has a significant impact on animal and plant biodiversity. Approximately 8,000 fungal and Oomycetes species are linked to plant diseases.7 Natural fungicides, such as chitinases, have the potential to replace chemical fungicides in plant biocontrol.8 Marine microorganisms are considered a source of novel enzymes and useful secondary metabolites.9-11 In contrast to terrestrial microbes, marine organisms have produced many novel molecules with diverse structural and functional features, representing a reservoir of new bioactive compounds with high biocontrol potential. For this reason, marine microbes are being explored to find new beneficial microorganisms and novel chemical entities as sources for new lead compounds. Chitinase-producing bacteria from the marine have attracted great attention because of their functional and industrial applications.9,12
A recent study shows that chitinase from marine Exiguobacterium antarcticum DW2 has a wide pH range, good thermostability, and low Km values, indicating chitin degradation capacity.13 Chitinases from the marine Pseudoalteromonas flavipulchra DSM 14401T have the potential to directly degrade natural crystalline chitin to produce N,N’-diacetylchitobiose.14 Furthermore, chitinase from the marine Streptomyces anulatus CS242 has been shown to possess antifungal activity against Aspergillus niger.15 Marine chitinases have unique properties such as higher pH, salt tolerance, and cold-adapted than terrestrial chitinases that may serve specific biotechnological applications. A receptor-inducer system regulates chitinase production in microorganisms. Thus, the composition of the culture medium can affect chitinase production.16 The nitrogen source and fermentation conditions impact chitinase synthesis in marine Bacillus sp. R2.17 Based on these, the present study aims to optimize chitinase production by marine-associated Bacillus paramycoides under submerged fermentation and determine its antifungal activity against nine pathogenic fungi. Alternaria alternata, Aspergillus flavus, A. fumigatus, A. niger, Cladosporium cladosporioides, Fusarium solani, Mucor ruber, Penicillium chrysogenum, and Rhizopus stolonifer were used in this study. Most of these fungi cause disease in crops, other organisms, or as allergens in humans.7,18
Preparation of Microorganisms
Bacillus strains isolated from sand and seawater were obtained from the Microbiology Laboratory, Department of Science, Faculty of Science and Technology, Prince of Songkla University, Pattani Campus, Thailand. The selection of five isolates used in this study was based on their chitinase and antifungal activity, as explained in the previous experiment.19 Bacterial isolates were grown in nutrient agar (HiMedia, India) enriched with 3% NaCl and incubated at 30°C for 24 h. The nine pathogenic fungi were obtained from the Thailand Institute of Scientific and Technological Research (TISTR). The fungal strains were Alternaria alternata TISTR 3435, Aspergillus flavus TISTR 3041, A. fumigatus TISTR 3108, A. niger TISTR 3130, Cladosporium cladosporioides TISTR 3285, Fusarium solani TISTR 3436, Mucor ruber TISTR 3006, Penicillium chrysogenum TISTR 3554, and Rhizopus stolonifer TISTR 3016. The fungi were grown on potato dextrose agar (PDA) (HiMedia, India) at 27°C for 5 days before use.
Preparation of Colloidal Chitin Medium
Colloidal chitin was prepared according to the method as described by Chandrasekaran 20 with minor modifications. As a medium for chitinase production, a colloidal chitin medium containing (g/L): yeast extract (HiMedia) 0.5; MgSO4. 7H2O 0.3; KH2PO4 1.36; (NH4)2. SO4 1; NaCl 30; and colloidal chitin (1% w/v) was used in this study. The initial pH of the culture medium was adjusted to 7.0±0.2. The medium was autoclaved at 121°C for 15 min.
Extracellular Chitinase Activity
The inoculum was prepared by adding one loop of culture grown for 24 h to 100 mL of nutrient broth supplemented with 3% NaCl in a 250 mL Erlenmeyer flask and incubated at 30°C for 24 h while stirring at 180 rpm. Four milliliters of inoculum (OD600nm = 0.5) was added to 100 mL of colloidal chitin medium in a 250 mL Erlenmeyer flask and incubated at 30°C for 72 h while stirring at 180 rpm. The fermentation broth was centrifuged for 10 min at 10,000 rpm at 4°C in an EppendorfTM Model 5810R (Germany). The obtained supernatant was used to determine chitinase activity.
Chitinase Assay
The chitinase activity was determined by the amount of reducing sugar (N-acetylglucosamine) produced during the hydrolysis of the colloidal chitin. The reducing sugar was determined by modifying the DNS method.21 One milliliter of the supernatant was added to 1 mL of 1% colloidal chitin in 0.1 M citrate phosphate buffer (pH 5) and incubated for 1 h at 37°C. The reaction mixture was centrifuged at 10,000 rpm at 4°C for 5 min. One milliliter of the mixture supernatant was mixed with 1 mL of the 3,5-dinitrosalicylic acid reagent (Merck, Germany) and heated for 5 min in a boiling water bath. The samples and blank absorbance were measured at 540 nm using a Multiskan™ GO Microplate Spectrophotometer (Thermo Scientific™, USA). One unit of chitinase activity was defined as the amount of enzyme required to generate 1 mol of reducing sugar per hour under the experimental conditions.
Identification of Chitinolytic Bacillus Isolate B26
Genomic DNA extraction was performed using phenolic-chloroform extraction.22 The 16S rRNA gene was amplified with universal primers (forward o27F and reserve o1492R) and performed using GeneAmp PCR (PerkinElmer, USA). The amplified samples were separated on a 1% agarose gel containing ethidium bromide (0.5 μg/mL) with a DNA ladder marker (GeneRulerTM 1 kb, Fermentas, USA). PCR products with a length of around 1,500 bp were purified and sequenced at the Bioneer sequencing company in South Korea. The results were compared with the GenBank database using the BLAST program.
Optimization of Chitinase Production
Effect of Substrates and Concentrations on Chitinase Production
The different substrates, including colloidal chitin, shrimp shell powder (SSP), crab shell powder (CSP), and squid pen powder (SPP), at a concentration of 1%, were used for chitinase production. Furthermore, the best substrate was evaluated at various concentrations (0.5, 1, 2, and 3%) to determine the optimal substrate concentration for chitinase production.
Effect of Additional Carbon Sources on Chitinase Production
In this experiment, the best substrate and its concentration from the previous experiment were used instead of colloidal chitin. Furthermore, various additional carbon sources (1%), including glucose, lactose, maltose, and N-acetylglucosamine, were added to the medium. A control experiment without an additional carbon source was also involved. The following procedure was identical to the one described previously.
Effect of Additional Nitrogen Sources on Chitinase Production
In this experiment, the best medium compositions from previous experiments were applied. Various additional nitrogen sources (1%), including sodium nitrate (NaNO3), ammonium nitrate (NH3NO4), peptone, and yeast extract, were evaluated for chitinase production. A control experiment without an additional nitrogen source was also involved. The following procedure was identical to the one described previously. Furthermore, the best additional nitrogen source was evaluated at various concentrations (0.5, 1, 2, and 3%) to determine the optimal nitrogen concentration for chitinase production.
Effect of NaCl Concentrations and Initial pH on Chitinase Production
In this experiment, the best medium compositions from previous experiments were applied. Various NaCl concentrations (0, 1, 2, 3, and 4%) were evaluated for chitinase production. The following procedure is identical to the one described previously. Furthermore, the initial pH of the medium (5, 6, 7, 8, and 9) was applied to determine the optimal initial pH for chitinase production. The following procedure was identical to the one described previously.
Effect of Incubation Time on Chitinase Production
The best medium compositions and cultural conditions from previous experiments were evaluated for different incubation times. The following procedure was identical to the one described previously. The crude chitinase was prepared for antifungal activity.
Antifungal Activity of Crude Chitinase
The crude chitinase from the optimization process was evaluated for antifungal activity against plant pathogenic fungi using a well-diffused method. A fungal plug with a diameter of 5 mm was removed from the 5-day-old culture and centered on Petri plates containing potato dextrose agar. Four wells were created, each with a diameter of 1 cm and located at an equal distance from the center of the plate. Equal aliquots (150 µL) of supernatant from crude chitinase, boiled crude chitinase (negative control), uninoculated culture (negative control), and 2 ug/mL of amphotericin B (positive control) were placed in these wells and incubated at 30°C for 5 days. The percentage of mycelial growth inhibition was measured using the formula below.23
Mycelial growth inhibition (%) = [R1 – R2 / R1] X 100
R1 denotes the distance of the fungal growth radius (mm) measured from the fungal colony’s center to the negative control well, and R2 represents the fungal growth radius (mm) distance to the sample. The growth of the test fungal strains was considered inhibited when the inhibition percentage exceeded 20%.
Statistical Analysis
All experiments were performed in triplicates. The results were statistically analyzed using variance analysis (ANOVA) and Bonferroni correction. The letter label variation in the figure showed a significant difference of P< 0.05.
Screening of Chitinase-producing Bacillus Isolates
Five Bacillus isolates were evaluated for chitinase production in the colloidal chitin medium. As shown in Table 1, isolate B26 showed the most potent strain with chitinase activity of 0.753 U/mL. The results were consistent with a previous study 19 that isolate B26 showed the highest chitinase activity observed by clear-zone formation on the colloidal chitin agar. Nevertheless, no significant difference in the chitinase production by the Bacillus isolates was observed. However, our previous study proved that isolate B26 had more antagonistic activity against pathogenic fungi than the other isolates tested in this experiment.19
Table (1):
Chitinase production by chitinolytic Bacillus isolates in the colloidal chitin medium.
Isolates |
Chitinase activity (U/mL) |
|---|---|
B07 |
0.735±0.056 |
B10 |
0.722±0.005 |
B19 |
0.742±0.027 |
B20 |
0.748±0.083 |
B26 |
0.753±0.007 |
Data is represented by the mean and standard deviation. The P-value of all the samples is not statistically significant.
Identification of Isolate B26
Isolate B26 was identified using 16S rRNA gene sequencing and found 99.47% identical to sequences of Bacillus paramycoides. A study of the morphological, physiological, and biochemical characteristics of isolate B26 was conducted to substantiate this finding.19
Effect of Substrates on Chitinase Production
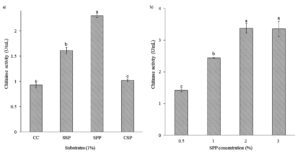
Isolate B26 was evaluated to produce chitinase on various substrates (Figure 1a). Squid pen powder (SPP) as a substrate gave significantly higher chitinase activity (2.3 U/mL) than other substrates. Different concentrations of SPP were evaluated to achieve maximum chitinase production. The increased chitinase activity of 3.38 U/mL was obtained in the culture medium containing 2% SPP and slowly decreased afterwards (Figure 1b).
Figure 1. Effect of (a) substrates and (b) substrate concentrations on chitinase production by isolate B26 under submerged fermentation
Effect of Additional Carbon Sources on Chitinase Production
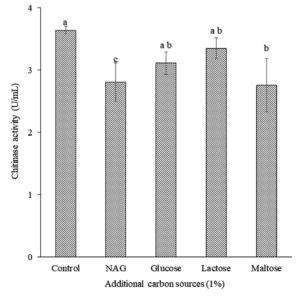
Additional carbon sources, namely glucose, lactose, NAG, and maltose, were evaluated to obtain the maximum chitinase production by isolate B26. Interestingly, isolate B26 produced the maximum chitinase production (3.65 U/mL) in the SPP medium without an additional carbon source (Figure 2), significantly higher than the SPP with NAG and maltose addition. A decrease in chitinase production by Bacillus isolate B26 in SPP medium with NAG, maltose, lactose, and glucose was observed.
Figure 2. Effect of additional carbon sources on chitinase production by isolate B26 in an optimized medium under submerged fermentation
Effect of Additional Nitrogen Sources on Chitinase Production
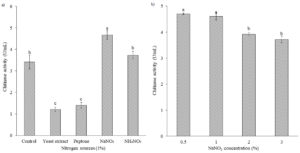
The addition of nitrogen sources (1%) to the culture medium, which consisted of yeast extract (0.05%) and (NH4)2SO4 (0.1%) as its original nitrogen sources, was conducted to evaluate their effects on chitinase production. As an inorganic nitrogen source, sodium nitrate was the most favorable nitrogen source for chitinase production (4.67 U/mL), significantly compared with other nitrogen sources (Figure 3a). In contrast, lower chitinase production was found in peptone or yeast extract. Different concentrations of NaNO3 were further evaluated to achieve maximum chitinase production. The results showed that the isolate B26 produced chitinase maximally at 0.5% NaNO3 (4.07 U/mL) (Figure 3b).
Figure 3. Effect of (a) nitrogen sources and (b) NaNO3 concentrations on chitinase production by isolate B26 in an optimized medium under submerged fermentation
Effects of NaCl Concentrations and Initial pH on Chitinase Production
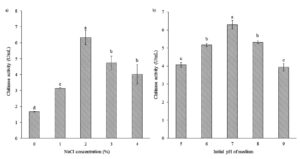
Various NaCl concentrations (0, 1, 2, 3, and 4%) affected chitinase production by Bacillus isolate B26 (Figure 4a). The results showed that isolate B26 produced chitinase maximally at 2% NaCl with chitinase activity of 6.34 U/mL. The optimum medium was evaluated at different initial pH for chitinase production (Figure 4b). The highest chitinase activity (6.3 U /mL) was observed at pH 7. The culture medium with an initial pH below and above pH 7 significantly reduced chitinase production.
Figure 4. Effect of (a) NaCl concentration and (b) initial pH of the medium on chitinase production by isolate B26 in an optimized medium under submerged fermentation
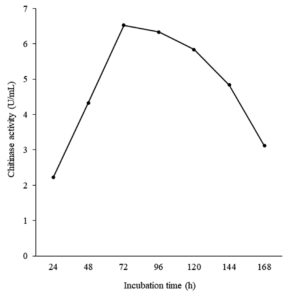
Effect of Incubation Time on Chitinase Production
The effect of incubation time on chitinase production by Bacillus isolate B26 was conducted (Figure 5). Maximum enzyme production was observed at 72 h of incubation with an enzyme activity of 6.52 U/mL. After that, its production decreased because it had a longer incubation time.
Figure 5. Effect of incubation time on chitinase production by isolate B26 in an optimized medium under submerged fermentation
Antifungal Activity of Crude Chitinase
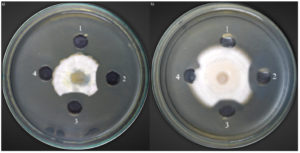
The crude chitinase from the optimized medium of Bacillus isolate B26 was evaluated for antifungal activity against nine fungi. The results of the fungal growth on the plate assay are shown in Table 2. Only two fungal strains, F. solani TISTR 3436 and P. chrysogenum TISTR 3554 were sensitive to crude chitinase. The fungal growth inhibition by crude chitinase was 83.4% and 80.12% for F. solani TISTR 3436 and P. chrysogenum TISTR 3554. The negative control well showed no fungal growth inhibition, but the others showed a crescent of inhibition around the wells due to antifungal chitinase (Figure 6).
Table (2):
Antifungal activity of crude chitinase of isolate B26 against fungi.
Fungi |
Mycelial growth inhibition (%) |
|---|---|
Alternaria alternata TISTR 3435 |
0 |
Aspergillus flavus TISTR 3041 |
0 |
Aspergillus fumigatus TISTR 3108 |
0 |
Aspergillus niger TISTR 3130 |
0 |
Cladosporium cladosporioides TISTR 3285 |
8.67±5.49 |
Fusarium solani TISTR 3436 |
83.4±2.9 |
Mucor ruber TISTR 3006 |
4.64±0.43 |
Penicillium chrysogenum TISTR 3554 |
80.12±4.5 |
Rhizopus stolonifer TISTR 3016 |
0 |
The data is represented by the mean and standard deviation.
Bacillus isolate B26, a potent isolate producing chitinase, has DNA sequence similarity to B. paramycoides, a novel strain in the member group of B. cereus.24 Medium optimization revealed that SPP as a substrate significantly affected the chitinase production by Bacillus isolate B26. A similar finding was reported that Streptomyces thermocarboxydus produced the highest chitinase activity when cultured in a medium containing SPP.25 These results correspond to some studies that showed that the chitin content in squid pens26 was higher than in shrimp and crab shells.27 The β–chitin found in squid pens is less crystalline and more degradable. Therefore, chitinolytic bacteria degrade β–chitin faster than α-chitin in shrimp and crab shells, which have a more stable structure.28 Most bacteria have been reported to produce the highest chitinase activity when colloidal chitin is present in the medium.29,30 However, researchers reported the highest bacterial chitinase activity in a medium containing shrimp shells.31 This finding indicates that chitinase production by bacteria depends on the culture medium and enzyme producer characteristics.
The effect of substrate concentrations on chitinase production by isolate B26 showed that 2% of SPP was optimal. This finding was similar to Serratia marcescens DSM 30121T, which produces the maximum chitinase in 2% of carbon sources.32 Additionally, we observed that NAG was not beneficial as a substrate for chitinase production. A similar result was also found in B. thuringiensis R176.33 Another study showed that lactose and maltose decreased chitinase production, while chitin gave the highest enzyme production.34 Isolate B26 produced the maximum chitinase when cultured in a medium containing the sole substrate. However, the medium suppressed its chitinase production with additional carbon sources. In other words, B. paramycoides genes that encode chitinase are controlled by carbon catabolite repression.35 However, adding different carbon sources in the culture medium, namely fructose and starch, increased chitinase production by Streptomyces pratensis36 and Aeromonas hydrophila,29 respectively.
The effects of different nitrogen sources revealed that inorganic nitrogen sources were the best for chitinase production by isolate B26. The same findings were found in B. thuringiensis33 and Cohnella sp. A01.34 Furthermore, 0.5% sodium nitrate was suitable for chitinase production by Bacillus isolate B26. Nitrate assimilation is a good nitrogen source for Bacillus species. However, nitrate utilization requires a reductive pathway with nitrate and nitrite reductase enzyme participation.37 As a result, sodium nitrate may be an excellent alternative for industrial-scale chitinase production.
Additionally, in this present study, a reduction in chitinase production was found in a medium with a complex nitrogenous source addition, such as yeast extract and peptone. Adding yeast extract as an additional nitrogen source will increase its concentration in the medium. In studying the a-amylase synthesis by B. amyloliquefaciens,38 in uncontrolled pH fermentations, increasing the yeast extract concentration to 5 g/L decreased the pH significantly. As a result, the enzyme synthesis is completely suppressed. The medium contains yeast extract that is reported to cause nitrogen catabolite repression in Saccharomyces cerevisiae during wine fermentation.39
Optimum chitinase activity was observed when the medium was supplemented with 2% NaCl. Comparable results were obtained with 2.5 to 3% NaCl by Bacillus sp. R2 isolated from the marine sample.17 The sodium ion is implicated in pH homeostasis. Every living cell tends to expel high sodium ions from its cytoplasm.40 Furthermore, the most favorable conditions for chitinase production were observed at pH 7. Our previous study supported these findings regarding the effect of NaCl concentration and the initial pH on the growth of isolate B26.19 A comparable study reported that chitinase is maximally produced at pH 7.5 by Bacillus sp. R2.17 The initial pH of the medium influenced enzyme biosynthesis due to the ionization of the proton gradient at the enzyme’s catalytic site.41
The duration of incubation affected the growth rate and pattern of enzyme production. After 72 h, the production began to decline, possibly because the microorganism’s growth had reached a point where it could no longer maintain a stable growth rate in conjunction with the availability of nutrient resources. Similar findings have also been found for Bacillus sp. HU1.42 A comparable study reported that Massilia timonae produced the most chitinase after 36 h 43 and 48 h by A. punctata HS6 and A. hydrophila HS4.29
Crude chitinase from isolate B26 showed the most effective inhibition against P. chrysogenum and F. solani. Moreover, the 5-min boiled crude chitinase showed no fungal growth inhibition because of the denaturation of the enzyme when heated, leading to the loss of enzyme activity. The antifungal chitinase produced by Bacillus has been proven in numerous studies. The crude and purified chitinase of B. pumilus showed inhibition against F. solani and A. niger.44 Furthermore, purified chitinase of B. cereus inhibited the growth of F. oxysporum and Pythium ultimum.45 Due to its natural origin, each has its unique properties. The polymer found in fungi is structurally like chitin, which is found in other organisms. However, unlike arthropod exoskeletons, fungal chitin is associated with other polysaccharides.46 Therefore, the subsite structure in the binding cleft must affect chitinolytic ability. This explains why the chitinase did not affect other fungi.
Medium composition and conditions significantly increase the chitinase production of B. paramycoides B26. We can conclude that chitinase production increased 2.83-fold after media improvement using squid pen powder as a substrate. The crude chitinase showed inhibitory activity against F. solani TISTR 3436 and P. chrysogenum TISTR 3554. This study suggests that B. paramycoides B26 may benefit chitin waste utilization and control of plant pathogenic fungi.
ACKNOWLEDGMENTS
The authors would like to thank Graduate School of Prince of Songkla University for the fund which made this important study viable and effective.
CONFLICT OF INTEREST
The authors declare that there is no conflict of interest.
AUTHORS’ CONTRIBUTION
All authors listed have made a substantial, direct, and intellectual contribution to the work and approved it for publication.
FUNDING
This study was supported by Thailand’s Education Hub for ASEAN Countries (TEH-AC) scholarship (067) and a research fund from graduate school, Prince of Songkla University.
DATA AVAILABILITY
All datasets generated or analyzed during this study are included in the manuscript.
ETHICS STATEMENT
This article does not contain any studies with human participants or animals performed by any authors.
- Subramanian K, Sadaiappan B, Aruni W, et al. Bioconversion of chitin and concomitant production of chitinase and N-acetylglucosamine by novel Achromobacter xylosoxidans isolated from shrimp waste disposal area. Sci Rep. 2020;10(1):11898.
Crossref - Narayan B, Velappan SP, Zituji SP, Manjabhatta SN, Gowda LR. Yield and chemical composition of fractions from fermented shrimp biowaste. Waste Manag Res. 2010;28(1):64-70.
Crossref - Suresh P, Sachindra N, Bhaskar N. Solid state fermentation production of chitin deacetylase by Colletotrichum lindemuthianum ATCC 56676 using different substrates. J Food Sci Technol. 2011;48(3):349-356.
Crossref - Rathore AS, Gupta RD. Chitinases from bacteria to human: properties, applications, and future perspectives. Enzyme Res. 2015;2015:791907.
Crossref - Cheba BA, Zaghloul TI. Bacillus Sp. R2 chitinase: substrate specificity, shelf-life stability, and antifungal activity. Procedia Manuf. 2020;46:879-884.
Crossref - Sheikh HMA, Hamshary OIM, Khattab AE-NAE-H. Molecular identification, characterization and improvement of a chitinase producing Bacillus strain showing significant control against some dermatophytic fungi. J Pure Appl Microbiol. 2022;16(1):643-654.
Crossref - Fisher MC, Gurr SJ, Cuomo CA, et al. Threats posed by the fungal kingdom to humans, wildlife, and agriculture. MBio. 2020;11(3):e00449-00420.
Crossref - Veliz EA, Martinez-Hidalgo P, Hirsch AM. Chitinase-producing bacteria and their role in biocontrol. AIMS Microbiol. 2017;3(3):689-705.
Crossref - Paulsen SS, Andersen B, Gram L, Machado H. Biological potential of chitinolytic marine bacteria. Mar Drugs. 2016;14(12):230.
Crossref - Karthikeyan A, Joseph A, Nair BG. Promising bioactive compounds from the marine environment and their potential effects on various diseases. J Genet Eng Biotechnol. 2022;20(1):1-38.
Crossref - Buatong J, Rukachaisirikul V, Sangkanu S, Surup F, Phongpaichit S. Antifungal metabolites from marine-derived Streptomyces sp. AMA49 against Pyricularia oryzae. J Pure Appl Microbiol. 2019;13(2):653-665.
Crossref - Jahromi ST, Barzkar N. Marine bacterial chitinase as sources of energy, eco-friendly agent, and industrial biocatalyst. Int J Biol Macromol. 2018;120(Part B):2147-2154.
Crossref - Fu X, Guo Y, Jin Y, Ma M. Bioconversion of chitin waste using a cold-adapted chitinase to produce chitin oligosaccharides. LWT. 2020;133:109863.
Crossref - Ren X-B, Dang Y-R, Liu S-S, et al. Identification and characterization of three chitinases with potential in direct conversion of crystalline chitin into N, N′-diacetylchitobiose. Mar Drugs. 2022;20(3):165.
Crossref - Mander P, Cho SS, Choi YH, et al. Purification and characterization of chitinase showing antifungal and biodegradation properties obtained from Streptomyces anulatus CS242. Arch Pharm Res. 2016;39(7):878-886.
Crossref - Warda EA, Abeer A, Eman RH, Mahmoud AS, Ahmed I. Applications of Plackett-Burman and central composite design for the optimization of novel Brevundimonas diminuta KT277492 chitinase production, investigation of its antifungal activity. Braz Arch Biol Technol. 2016;59:e16160245.
Crossref - Cheba BA, Zaghloul TI, El-Mahdy AR, El-Massry MH. Effect of nitrogen sources and fermentation conditions on Bacillus sp. R2 chitinase production. Procedia Manuf. 2018;22:280-287.
Crossref - Pfavayi LT, Sibanda EN, Mutapi F. The pathogenesis of fungal-related diseases and allergies in the African population: the state of the evidence and knowledge gaps. Int Arch Allergy Immunol. 2020;181(4):257-269.
Crossref - Kurniawan E, Panphon S, Leelakriangsak M. Potential of marine chitinolytic Bacillus isolates as biocontrol agents of phytopathogenic fungi. IOP Conf Ser Earth Environ Sci. 2018;217:012044.
Crossref - Chandrasekaran R, Revathi K, Nisha S, Kirubakaran SA, Sathish-Narayanan S, Senthil-Nathan S. Physiological effect of chitinase purified from Bacillus subtilis against the tobacco cutworm Spodoptera litura Fab. Pestic Biochem Physiol. 2012;104(1):65-71.
Crossref - Miller GL. Use of dinitrosalicylic acid reagent for determination of reducing sugar. Anal Chem. 1959;31(3):426-428.
Crossref - Cheng H-R, Jiang N. Extremely rapid extraction of DNA from bacteria and yeasts. Biotechnol Lett. 2006;28(1):55-59.
Crossref - Farag AM, Abd-Elnabey HM, Ibrahim HA, El-Shenawy M. Purification, characterization and antimicrobial activity of chitinase from marine-derived Aspergillus terreus. Egypt J Aquat Res. 2016;42(2):185-192.
Crossref - Liu Y, Du J, Lai Q, et al. Proposal of nine novel species of the Bacillus cereus group. Int J Syst Evol Microbiol. 2017;67(8):2499-2508.
Crossref - Tran TN, Doan CT, Nguyen AD, Wang S-L. The isolation of chitinase from Streptomyces thermocarboxydus and its application in the preparation of chitin oligomers. Res Chem Intermed. 2019;45(2):727-742.
Crossref - Cuong HN, Minh NC, Van Hoa N, Trung TS. Preparation and characterization of high purity β-chitin from squid pens (Loligo chenisis). Int J Biol Macromol. 2016;93(Part A):442-447.
Crossref - Synowiecki J, Al-Khateeb NAAQ. The recovery of protein hydrolysate during enzymatic isolation of chitin from shrimp Crangon crangon processing discards. Food Chem. 2000;68(2):147-152.
Crossref - Jang MK, Kong BG, Jeong YI, Lee CH, Nah JW. Physicochemical characterization of a-chitin, β-chitin, and g-chitin separated from natural resources. J Polym Sci Part A-1: Polym Chem. 2004;42(14):3423-3432.
Crossref - Kuddus M, Ahmad I. Isolation of novel chitinolytic bacteria and production optimization of extracellular chitinase. J Genet Eng Biotechnol. 2013;11(1):39-46.
Crossref - Jankiewicz U, Brzezinska MS. Purification, characterization, and gene cloning of a chitinase from Stenotrophomonas maltophilia N4. J Basic Microbiol. 2015;55(6):709-717.
Crossref - Brzezinska MS, Walczak M, Lalke-Porczyk E, Donderski W. Utilization of shrimp-shell waste as a substrate for the activity of chitinases produced by microorganisms. Pol J Environ Stud. 2010;19(1). http://www.pjoes.com/Utilization-of-Shrimp-Shell-Waste-as-a-Substrate-r-nfor-the-Activity-of-Chitinases,88370,0,2.html. Accessed June 20, 2022.
- Lamine B, Lamine B, Bouziane A. Optimisation of the chitinase production by Serratia marcescens DSM 30121T and biological control of locusts. J Biotechnol Biomater. 2012;2(3):3-7.
Crossref - Chaiharn M, Lumyong S, Hasan N, Plikomol A. Solid-state cultivation of Bacillus thuringiensis R 176 with shrimp shells and rice straw as a substrate for chitinase production. Ann Microbiol. 2013;63(2):443-450.
Crossref - Aliabadi N, Aminzadeh S, Karkhane AA, Haghbeen K. Thermostable chitinase from Cohnella sp. A01: isolation and product optimization. Braz J Microbiol. 2016;47(4):931-940.
Crossref - Vinuselvi P, Kim M-K, Lee S-K, Ghim C-M. Rewiring carbon catabolite repression for microbial cell factory. BMB Rep. 2012;45(2):59-70.
Crossref - Shivalee A, Lingappa K, Mahesh D. Influence of bioprocess variables on the production of extracellular chitinase under submerged fermentation by Streptomyces pratensis strain KLSL55. J Genet Eng Biotechnol. 2018;16(2):421-426.
Crossref - Lin JT, Stewart V. Nitrate assimilation by bacteria. Adv Microb Physiol. 1997;39:1-30.
Crossref - Alam S, Hong J, Weigand WA. Effect of yeast extract on alpha-amylase synthesis by Bacillus amyloliquefaciens. Biotechnol Bioeng. 1989;33(6):780-785.
Crossref - Beltran G, Novo M, Rozes N, Mas A, Guillamon JM. Nitrogen catabolite repression in Saccharomyces cerevisiae during wine fermentations. FEMS Yeast Res. 2004;4(6):625-632.
Crossref - Wood JM. Bacterial responses to osmotic challenges. J Gen Physiol. 2015;145(5):381-388.
Crossref - Romano G, Costantini M, Sansone C, Lauritano C, Ruocco N, Ianora A. Marine microorganisms as a promising and sustainable source of bioactive molecules. Mar Environ Res. 2017;128:58-69.
Crossref - Dai D, Hu W, Huang G, Li W. Purification and characterization of a novel extracellular chitinase from thermophilic Bacillus sp. Hu1. Afr J Biotechnol. 2011;10(13):2476-2485. https://www.ajol.info/index.php/ajb/article/view/93176. Accessed June 10, 2022.
- Faramarzi M, Fazeli M, Yazdi MT, et al. Optimization of cultural conditions for production of chitinase by a soil isolate of Massilia timonae. Biotechnology. 2009;8(1):93-99.
Crossref - Gurav R, Tang J, Jadhav J. Novel chitinase producer Bacillus pumilus RST25 isolated from the shellfish processing industry revealed antifungal potential against phyto-pathogens. Int Biodeterior Biodegradation. 2017;125:228-234.
Crossref - Chang W-T, Chen C-S, Wang S-L. An antifungal chitinase produced by Bacillus cereus with shrimp and crab shell powder as a carbon source. Curr Microbiol. 2003;47(2):0102-0108.
Crossref - Zarei M, Aminzadeh S, Zolgharnein H, et al. Characterization of a chitinase with antifungal activity from a native Serratia marcescens B4A. Braz J Microbiol. 2011;42(3):1017-1029.
Crossref
© The Author(s) 2022. Open Access. This article is distributed under the terms of the Creative Commons Attribution 4.0 International License which permits unrestricted use, sharing, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.