ISSN: 0973-7510
E-ISSN: 2581-690X
The incidence of methicillin-resistant Staphylococcus aureus (MRSA) having an increased rate of fluoroquinolone resistance is currently noted in Egypt necessitating the implication of new strategies to restrain the rapid spread of this resistance. The approach of repurposing could present a solution to this problem. In the current study, the efficacy of the antifungal agent, caspofungin, in increasing the susceptibility of MRSA isolated from Alexandria Main University Hospital (AMUH) to a novel fluoroquinolone, zabofloxacin, was investigated. A percentage of 30.3% of tested isolates were found to be resistant to zabofloxacin. Upon treatment with subinhibitory concentration of caspofungin, these isolates had their zabofloxacin minimum inhibitory concentration values decreased by a range varying from 2- to 32-fold. Proteomic approach was performed to provide insights into the involved mechanism of caspofungin sensitization. The profiles of cellular proteins generated by electrophoretic techniques varied between treated and untreated MRSA samples with polymorphism values ranging from 82.4 to 94.1%. Homology was detected between 1,3-beta-glucan synthase (the fungal target enzyme of caspofungin) and SdrM, a multidrug efflux pump in S. aureus. This homology was elaborated by immunoblotting analysis which showed downregulation of SdrM efflux pump proteins. The percentage of reduction in the SdrM proteins expression level varied from 18 to 61%. Caspofungin succeeded in sensitizing zabofloxacin-resistant MRSA clinical isolates by blocking the action of SdrM efflux pump and inhibiting its extrusion. Further research is urgently required to endorse the repositioning of caspofungin as an agent used in combination with zabofloxacin in the management of MRSA infections with higher efficiency.
Caspofungin, efflux pump proteins, MRSA, repurposing, western blot, zabofloxacin.
Despite the development of novel antibiotics and the significant revolution in infection prevention measures, MRSA remains one of the most threatening and hazardous modern pathogens related with elevated rates of morbidity and mortality1. It is believed that MRSA possesses several features that render its eradication extremely complicated. Being an unpredictable and versatile microorganism with a high potential for genetic adaptation and for developing epidemic strains, this pathogen continues to endanger thousands of patients worldwide1.
The prevalence of MRSA has been noted currently in Egypt. In 2017, the occurrence rate of MRSA within hospitalized patients in Cairo and Alexandria, the two major cities in Egypt, was declared to reach 50-82%2. In 2018, Elshimy et al. reported a MRSA incidence rate of 44% across various laboratories of Egyptian hospitals3. Consequently, finding strategies to stop further dissemination of MRSA in hospital settings has become a necessity that could be achieved by the provision of both innovative antimicrobial agents and auxiliary facets of care, including infectious disease consultation and source control1.
Fluoroquinolones are broad-spectrum antimicrobials exerting their action mainly through inhibition of DNA gyrase and topoisomerase IV, thus, hindering the process of DNA supercoiling and killing the bacteria4. They are still considered the ultimate option for MRSA infections in patients with vancomycin tolerance problem5. Zabofloxacin (ZAB), a novel member of fluoroquinolones, has recently gained attention due to its considerable activity against Streptococcus pneumonia, methicillin-resistant coagulase-negative staphylococci, and MRSA6. Nevertheless, our previous study aiming at its evaluation as a novel fluoroquinolone not launched in Egyptian hospital settings yet, depicted a resistance rate of 37.9% among 116 MRSA clinical isolates. This disturbing situation indicates that the resistance of MRSA, even towards new antimicrobial agents, is developing expeditiously and necessitates new strategies to restrain the rapid spread of this resistance7.
The approach of screening and repurposing of molecules of completely disparate clinical indications has seized renovated interest, as reported by a number of latest studies8. Among the suggested drugs used in combination with other antibiotics for the eradication of MRSA infections are: the antirheumatic auranofin, the organoselenium compound ebselen, the anthelmintic ivermectin, and anticancer agents clomiphene and 5-fluoro-2’-deoxyuridine9,10. The repurposing strategy was applied as well to hinder the bacterial virulence instead of bacterial viability, where naftifine, an antifungal agent, was stated to suppress staphyloxanthin biosynthesis, an essential virulence determinant of S. aureus responsible for its protection from the host oxidant killing activity9. Similarly, caspofungin (CAS), an echinocandin agent, was repurposed by Siala et al. in combination with moxifloxacin or delafloxacin to abolish another virulence factor of S. aureus; its biofilm-forming ability10. In the current study, the repurposing strategy was explored where the efficacy of the antifungal CAS in increasing the susceptibility of MRSA clinical isolates to the novel fluoroquinolone, ZAB, was tested. In addition, proteomic analysis was performed to investigate the underlying mechanism of CAS sensitization of MRSA isolates to ZAB.
Clinical isolates and bacterial identification
Eighty clinical isolates of MRSA were obtained from the inpatient routine laboratory of AMUH from January 2019 to July 2019. The isolates belonged to the following clinical specimens: bronchoalveolar lavages (n = 48), blood (n = 12), urine (n = 10), pus (n= 7) and wound swabs (n= 3). Standard methicillin-sensitive S. aureus (MSSA) strain (ATCC 25923), and standard MRSA strain (ATCC 43300) were used as quality control strains. Identification was performed by conventional techniques and the biochemical characteristics were confirmed by Dry Spot Staphytect Plus (Oxoid, UK).
Antibiotics
Caspofungin diacetate (CAS) purchased from pharmaceutical markets as Cancidas® (Merck & Co., Inc., USA), oxacillin (OXA) (Sigma-Aldrich, Germany), and zabofloxacin powder (ZAB) granted by Dongwha Pharm. Co., Ltd. (Seoul, Korea) were used in this study.
Phenotypic detection of methicillin resistance
Modified Kirby-Bauer disc diffusion method with a cefoxitin disc (FOX, 30μg, Oxoid, UK) and the agar dilution method with OXA were used to confirm methicillin resistance in accordance with the Clinical and Laboratory Standards Institute guidelines11. In FOX disc diffusion assay, the turbidity of an overnight suspension was adjusted to 0.5 McFarland, swabbed on Mueller-Hinton agar (Oxoid, UK), and FOX discs were placed on the surface of the agar. The plates were incubated at 37°C for 18 h; and the diameters of inhibition zones were recorded. Isolates with an inhibition zone diameter of ≤21 mm were considered to be methicillin resistant. For OXA agar dilution assay, overnight bacterial cultures at 0.5 McFarland turbidity were inoculated onto the surface of Mueller-Hinton agar medium containing 6µg/ml OXA and supplemented with 4% NaCl, then incubated at 37°C for 24 h. The growth of more than one colony was considered an indication of OXA resistance according to CLSI11.
Detection of mecA gene by polymerase chain reaction (PCR)
The presence of a 286-base pair (bp) PCR product corresponding to mecA gene was confirmed using the following forward (5’-TGCTATCCACCCTCAAACAGG-3’) and reverse (5’-AACGTTGTAACCACCCCAAGA-3’) primers12 in a thermocycler (Thermo Fisher Scientific, Waltham, M.A., USA.). A positive control (ATCC 33591) elaborating mecA gene was included. After an initial denaturation step at 93°C for 5 min, samples were subjected to 35 cycles of denaturation at 93°C for 1 min, annealing at 55°C for 1 min and extension at 72°C for 1 min. A final extension step was performed at 72oC for 5 min. The resulting PCR products were separated by electrophoresis on a 2% agarose gel containing ethidium bromide (0.2 mg/ml), using a 100-bp ladder (Fermentas, USA) as DNA molecular weight marker.
Antimicrobial susceptibility testing
Minimum inhibitory concentrations (MICs) of ZAB and CAS were determined by the microtiter broth dilution method in accordance with CLSI guidelines13. A volume of 100µl of each tested isolate at 5.0 x 105 CFU/ml were incubated with 100µl of serial dilutions of ZAB or CAS in Mueller-Hinton broth (MHB, Oxoid, UK) at 37°C for 24 h in 96-well microtiter plates. An antibiotic-free well was set as a growth control, and well containing MHB only was used as a negative control. MICs were determined by deducing the optical density at 630 nm (OD630) using a microtiter plate reader and the lowest concentration of the antibiotic that inhibited growth was considered MIC.
Determination of the antibiotic-resistance modifying activity of CAS against ZAB-resistant MRSA clinical isolates
The MIC of ZAB against ZAB-resistant isolates was determined in the presence of subinhibitory concentration of CAS by microtiter broth dilution method as described above. Wells containing 100µl of each tested isolate at 5.0 x 105 CFU/ml were supplemented with 50µl of CAS at a final concentration of 16µg/ml and 50µl of serial dilutions of ZAB. The lowest concentration of ZAB in the presence of CAS that prevented growth was recorded as MIC and the degree of CAS sensitization of tested isolates for ZAB was calculated from fold decrease values in ZAB MIC14.
Proteomic studies
Extraction of whole cell proteins
An overnight culture of 7 selected isolates in MHB incubated without or with 16µg/ml of CAS were centrifuged (6000 rpm, 10 min), the pellets were washed 3 times with phosphate-buffered saline, and harvested by centrifugation (6000 rpm, 10 min). TriFastTM (Peqlab, VWR, USA) for the simultaneous isolation of RNA, DNA and protein was used as per manufacturer’s instructions. Briefly, after the precipitation of RNA and DNA, the proteins were precipitated from the ethanol/phenol supernatant with isopropanol, centrifuged (12000 rpm, 4°C, 10 min), washed 3 times with 0.3 M guanidinium hydrochloride in 95% ethanol, harvested, vortexed with 100% ethanol, stored for 20 min at room temperature and centrifuged (7500 rpm, 4°C, 5 min). Protein pellets were dried, dissolved in 1% sodium dodecyl sulphate (SDS), centrifuged, and the supernatant was stored at -20°C. The total protein fraction concentration was determined according to Bradford method15 based on the interaction between protein and Coomassie Brilliant Blue G250 (CBBG-250, Sigma) by recording the absorbance at 595 nm with bovine serum albumin as standard.
Sodium dodecyl sulphate polyacrylamide gel electrophoresis (SDS-PAGE)
Whole cell proteins were analysed by SDS-PAGE according to Laemmli16. Protein samples of 30μg were loaded on polyacrylamide gels and separated based on their molecular weight in a Biolab gel apparatus (V20-CDCY, Canada). Electrophoresis was performed with a buffer system (25 mM Tris, 186 mM glycine, 0.1% SDS) at 75 V through a stacking gel (4% polyacrylamide) and a separating gel (13% polyacrylamide) at 125 V during approximately 2 h. The gels were then stained by 2 h incubation in 0.1% Coomassie Brilliant Blue G-250, then destained in a solution of glacial acetic acid, methanol, and water mixed at a of ratio 1:3:6. A protein ladder ranging from 10 to 200 kDa (Invitrogen, USA) was used as SDS-PAGE molecular weight marker.
Image analysis
The gels were analysed using the gel documentation system (Geldoc-it, UVP, England) and TotalLab software analysis (www.totallab.com). The software was used to analyze the protein profiles of untreated samples and to compare them to the profiles of the treated samples with 16µg/ml of CAS. The protein changes as well as protein pattern maps were analysed from 3 independent biological replicates and 3 technical replicates.
Western blot analysis
Total protein extract (30μg) of each untreated and 16µg/ml CAS treated samples were run in SDS-PAGE as described earlier. The separated proteins were transferred onto nylon membrane (Hybond™, GE Healthcare, 0.45µm pore size) as described by electrophoretic transfer method of Towbin et al.17, using a Bio-Rad Mini TransBlot® cell (100 mA, 90 min). Membrane was blocked in Tris-buffered saline with tween solution (TBST, 50 mM Tris-HCl, pH 8.0, 150 mM NaCl, and 0.1% Tween-20) containing 5% non-fat dry milk for 1 h at room temperature. The blots were then incubated with primary antibody (recombinant anti-1,3-beta-glucan synthase antibody, catalog no. MOB-2, Creative Biolabs, USA; at 1:1000 dilution) for overnight at 4°C. Next day, membrane was rinsed in a rotatory shaker, in TBST for 5 times, 10 min each, and incubated for 1 h at room temperature with secondary antibody (horseradish peroxidase, HRP, catalog no. 32106, Thermo Fisher Scientific, USA; at 1:4000 dilution). Blots were developed by chemiluminescence, scanned using ChemiDoc scanner, and the densitometric intensity of each band was quantified by the TotalLab software analysis.
Statistical analysis
Data were expressed as mean ± S.D. of triplicates. One-way ANOVA followed by the Bonferroni testing through Prism 3 GRAPHPAD computer program was performed for multi-variable comparisons. Differences were considered significant at p-value < 0.05.
Confirmation of methicillin resistance
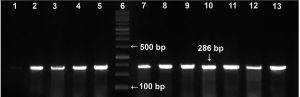
Before reaching our laboratory, the clinical isolates collected in the routine laboratory of AMUH have passed through an identification procedure by being inoculated on mannitol salt agar supplemented with 4µg/ml FOX as a part of their adopted protocol for MRSA diagnosis18. In our facility, the confirmation of methicillin resistance was accomplished by subjecting the tested isolates to 2 phenotypic tests; the FOX disc diffusion and the OXA agar dilution methods, as well as the gold standard of MRSA identification, the molecular detection of mecA gene. Fig. 1 shows a representative agarose gel with PCR amplification of mecA gene bands appearing at 286 bp. From the 80 isolates received, 76 were confirmed to possess mecA gene and thus were identified as MRSA isolates. The 4 isolates which didn’t harbour mecA gene were excluded from this study. Among the detection methods used in our laboratory, the FOX disc diffusion method was able to identify 74/76 of MRSA isolates with a 97.4% sensitivity while the OXA agar dilution method identified 71/76 of the MRSA isolates with a specificity of 75% (Table 1).
Table (1):
Comparison of two phenotypic tests used for MRSA isolates detection versus the molecular detection method by PCR amplification of mecAgene.
| Phenotypic tests | mecA positive (n= 76) |
mecA negative (n=4) | Sensitivity % | Specificity % | ||
|---|---|---|---|---|---|---|
| True +ve | False -ve | True-ve | False +ve | |||
| FOX DD | 74 | 2 | 2 | 2 | 97.4 | 50 |
| OXA AD | 71 | 5 | 3 | 1 | 93.4 | 75 |
FOX DD:cefoxitin disc diffusion test; OXA AD: oxacillin agar dilution assay.
Fig. 1. Representative agarose gel (2%) viewing PCR amplification of mecA gene (286 bp). Lane 6: DNA molecular weight marker (100-bp ladder). Lane 1: methicillin-sensitive S. aureus (MSSA strain, ATCC 25923, negative control); lane 2: positive control (MRSA, ATCC 43300); lanes 3–5 and lanes 7-13: representative MRSA isolates from the study.
MIC determination
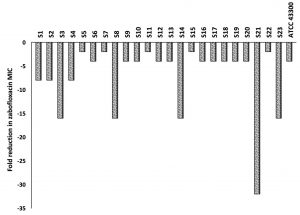
The microtiter broth dilution MICs were determined for 76 MRSA clinical isolates as well as for the two control strains (MSSA; ATCC 25923 and MRSA; ATCC 43300) against ZAB and CAS. The MIC range for ZAB varied between 0.03 and 4µg/ml with an MIC90 of 2µg/ml, while for CAS, the MIC50 and MIC90 reached 32µg/ml and 64µg/ml, respectively. A percentage of 30.3% (n=23) of the clinical isolates were resistant to ZAB (Table 2). These 23 isolates, in addition to the standard MRSA (ATCC 43300), were included for further investigations. The MIC of ZAB was calculated in the presence of a subinhibitory concentration of CAS (16µg/ml) and the degree of CAS sensitization was calculated from fold decrease values in ZAB MIC as presented in Fig. 2. All of the tested isolates showed a decrease in their ZAB MIC values when used in combination with CAS and the fold decrease varied between 2- to 32-fold. More than 78% of the tested isolates had their ZAB MIC values decreased by 4-fold or more upon incubation with 16µg/ml CAS (Fig. 2 and Table 3).
Table (2):
MIC distribution of zabofloxacin and caspofungin determined by broth microdilution method for 76 MRSA clinical isolates.
| Antimicrobial agent | MIC (µg/ml) | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0.03 | 0.06 | 0.125 | 0.25 | 0.5 | 1 | 2 | 4 | 8 | 16 | 32 | 64 | |
| Zabofloxacin | 12 | 9 | 19 | 7 | 5 | 1 | 15 | 8 | 0 | 0 | 0 | 0 |
| Caspofungin | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 46 | 30 |
The bold figure indicates the CLSI breakpoint for zabofloxacin: 2µg/ml.
Table (3):
In vitro activity of zabofloxacin alone and combined with caspofungin (16µg/ml) against 23 MRSA clinical strains isolated from different specimens.
| MRSA isolate | MIC (µg/ml) | |||
|---|---|---|---|---|
| Code | Clinical source | ZABa | CAS | ZAB + 16µg/mlCAS |
| S1 | BAL | 2 | 32 | 0.25 |
| S2 | BAL | 2 | 64 | 0.25 |
| S3 | pus | 2 | 64 | 0.125 |
| S4 | blood | 4 | 64 | 0.5 |
| S5 | BAL | 2 | 32 | 1 |
| S6 | urine | 4 | 64 | 1 |
| S7 | wound | 2 | 64 | 1 |
| S8 | BAL | 2 | 64 | 0.125 |
| S9 | BAL | 2 | 32 | 0.5 |
| S10 | pus | 2 | 32 | 1 |
| S11 | blood | 2 | 64 | 1 |
| S12 | urine | 2 | 64 | 1 |
| S13 | urine | 2 | 64 | 0.5 |
| S14 | urine | 2 | 64 | 0.125 |
| S15 | BAL | 2 | 32 | 1 |
| S16 | BAL | 2 | 64 | 0.5 |
| S17 | BAL | 2 | 64 | 0.5 |
| S18 | wound | 2 | 64 | 0.5 |
| S19 | BAL | 2 | 64 | 0.5 |
| S20 | BAL | 2 | 64 | 0.5 |
| S21 | urine | 4 | 64 | 0.0625 |
| S22 | pus | 2 | 64 | 1 |
| S23 | urine | 2 | 32 | 0.125 |
| ATCC 43300 (MRSA) | 0.5 | 32 | 0.125 | |
aCLSI breakpoint for zabofloxacin is 2µg/ml.BAL: bronchoalveolar lavage; ZAB: zabofloxacin; CAS: caspofungin.
Fig. 2. Fold reduction in zabofloxacin MIC when combined with 16µg/ml of caspofungin tested against MRSA clinical isolates, S1 to S23, and the standard MRSA, ATCC 43300.
Proteomic analysis
To provide insights into the involved mechanism of CAS sensitization, the effect of subinhibitory concentration of CAS (16µg/ml) on cellular protein extracts of 7 MRSA isolates was explored by proteomic approaches. These 7 isolates (S1, S6, S7, S8, S14, S21 and S23) were selected to display different fold reduction in ZAB MIC varying from 2- to 32-fold reduction (Fig. 2).
Protein variation between control and treated samples with CAS
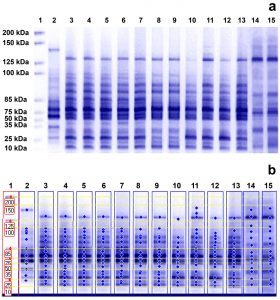
The profiles of cellular proteins generated by SDS-PAGE diverged between treated and untreated MRSA samples with differences observed in the molecular weight (kDa) of the obtained bands, their intensity, their fractionation, the display of new bands (unique bands appearing only in treated samples), and the missing of others (non-unique bands that existed in control samples and were lost from treated samples)19. (Fig. 3 a
and b).
Fig. 3. Whole cell protein fingerprinting patterns (a) and the molecular weight computerized detection of the polypeptide bands (b) generated by SDS-PAGE for 7 MRSA isolates. Lane 1 represents the protein marker; lanes 3, 5, 7, 9, 11, 13, and 15 represent the protein profiles of untreated isolates S1, S6, S7, S8, S14, S21 and S23, respectively; and lanes 2, 4, 6, 8, 10, 12, and 14 represent the protein profiles of these isolates treated with 16µg/ml of caspofungin, respectively.
Table (4):
SDS-PAGE protein analysis for the untreated and treated samples with 16µg/ml of caspofungin for 7 MRSA clinical isolates.
| MRSA isolates | Control Sample | Treated Sample | Monomorphic bands | Polymorphic bands | % Polymorphism | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| No. of bands | Molecular weight range (kDa) | No. of bands | Molecular weight range (kDa) | No. of bands | Molecular weight (kDa) | % | Unique bandsa | Non-unique bandsa | ||||
| No. | % | No. | % | |||||||||
| S1 | 16 | 133.6 – 17.2 | 12 | 133.3 – 24.2 | 1/16 | 80.5 | 6.3 | 11/16 | 68.8 | 4/16 | 25 | 93.8 |
| S6 | 17 | 133.2 – 14.8 | 17 | 133.6 – 13.6 | 3/17 | 80.5, 70.4, 52.2 | 17.6 | 14/17 | 82.4 | – | – | 82.4 |
| S7 | 17 | 132.1 – 18.3 | 17 | 133.2 – 16.0 | 1/17 | 22.4 | 5.9 | 16/17 | 94.1 | – | – | 94.1 |
| S8 | 17 | 132.6 – 13.6 | 15 | 132.6 – 13.6 | 2/17 | 132.6, 13.6 | 11.8 | 13/17 | 76.5 | 2/17 | 11.8 | 88.2 |
| S14 | 19 | 141.5 – 14.8 | 13 | 129.8 – 12.4 | 2/19 | 27.6, 20.4 | 10.5 | 11/19 | 57.9 | 6/19 | 31.6 | 89.5 |
| S21 | 17 | 131.3 – 11.2 | 16 | 131.3 – 11.2 | 2/17 | 131.3, 11.2 | 11.8 | 14/17 | 82.4 | 1/17 | 5.9 | 88.3 |
| S23 | 14 | 140.5 – 16.0 | 14 | 138.3 – 14.8 | 2/14 | 80.8, 70.4 | 14.3 | 12/14 | 85.7 | – | – | 85.7 |
aUnique bands are referring to new bands that appeared due to treatment while non-unique bands are bands that disappeared due to treatment with16 µg/ml of caspofungin.
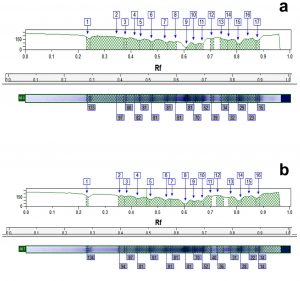
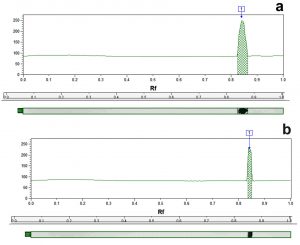
The results in Table 4 illustrate that the whole cell protein content of control MRSA samples was fractionated by electrophoretic forces into different band number varying from 14 to 19 polypeptide bands with molecular weights ranging from 141.5 to 11.2 kDa. Fig. 4 is a representative example of a dendogram for isolate S6 showing the electrophoretic control (a) and treated (b) banding patterns which were used for the computerized detection of the occurrence of monomorphic bands (bands having identical molecular weight and retention factor, Rf, as in control) between each control and its treated pair. The maximum number of bands in control samples (19 bands) was observed in isolate S14 (molecular weights ranging from 141.5 to 14.8 kDa), while for treated samples the maximum number reached 17 bands in both S6 and S7 samples. Monomorphic bands were detected among the 7 MRSA isolates with different proportions. The percentage of occurrence of these monomorphic bands ranged from 5.9 to 17.6% with highest percentage being detected in isolate S6 (17.6%) (Table 4).
Fig. 4. Representative dendogram viewing the protein pattern for S6 MRSA isolate control (a) and treated (b) sample with 16µg/ml of caspofungin used for the computerized detection of the occurrence of monomorphic bands.
Treatment with CAS resulted in the disappearance of certain bands (non-unique bands) in samples S1, S8, S14 and S21 while samples S6, S7 and S23 had an equivalent number of bands before and after treatment. SDS-PAGE gave rise to a total of 91 unique polypeptide bands, varying in molecular weight, number, and band intensity within the treated samples (the molecular weights of the unique bands and their band intensity percent are provided in the Supplementary File: Tables 1-S to 7-S). The emergence of these unique bands is an indication of protein alteration due to CAS. The maximum unique bands’ number was 16 with a percentage of 94.1%, observed in isolate S7, compared to a minimum number of 11 observed in isolates S1 and S14, with percentages of 68.8% and 57.9%, respectively. The polymorphism values generated by SDS-PAGE analysis were high ranging between 82.4 and 94.1% (Table 4).
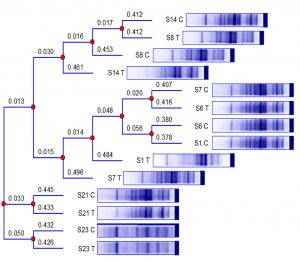
A phylogenetic tree was constructed to confirm the variations in the protein profiles obtained as a result of treatment with 16µg/ml of CAS (Fig. 5). The branch lengths of the phylogenetic tree clearly demonstrate that the protein profile of each isolate has differed after its exposure to subinhibitory concentration of CAS. The protein profile of control isolate of S14 is shown to possess similarity to the treated profile of S8, and both of them were related to the profile of the untreated isolate of S8. The previously mentioned cluster had resemblance to the protein profile of the treated isolate S14. Analogy was exhibited by the protein profile of control isolate S7 and the profile of treated S6. Likewise, resemblance was displayed between protein profiles of both control isolates S6 and S1. The latter cluster was related to the protein pattern of treated S1 and the resultant summative cluster to the protein profile of treated isolate S7. Interestingly, the profiles of control and treated isolates S21 and S23 remained related in pairs as displayed by the branch lengths of the phylogenetic tree (Fig. 5).
Fig. 5. Phylogenetic tree constructed for the protein profiles of 7 untreated MRSA clinical isolates and those subjected to overnight treatment with 16µg/ml of caspofungin. The numbers above the branch points represent the branch lengths. Codes, “C” and “T” refer to the control and treated samples, respectively, of the protein profiles of S1, S6, S7, S8, S14, S21, and S23 MRSA isolates.
Western blot assay
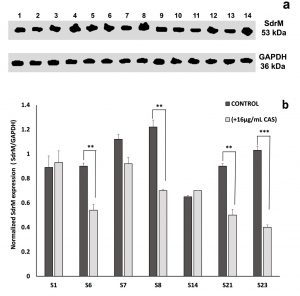
To examine whether the enhancement of ZAB penetration induced by CAS might be associated with an alteration in the efflux pump mechanism of the treated MRSA isolates, western blot assay was performed using the recombinant anti-1,3-beta-glucan synthase antibody. In fungi, CAS prevents the inclusion of uridine diphosphate (UDP)-glucose into 1,3-beta-glucan, thus blocking the function of 1,3-beta-glucan synthase20. A BLAST analysis exhibited that the fungal 1,3-beta-glucan synthase shares 33.3% identity with SdrM, a multidrug efflux pump in S. aureus belonging to the major facilitator superfamily (MFS). On the basis of this similarity, anti-1,3-beta-glucan synthase antibody was used to quantitate the effect of CAS on SdrM in 7 selected MRSA isolates by immunoblotting assay. A representative dendogram viewing the densitometric intensity computerized analysis of SdrM efflux pump proteins in MRSA isolate S23 is shown in Fig. 6 for control (a) and CAS treated (b) samples. A constant expression level of the loading control (GAPDH, 36 kDa) was observed for all tested samples (Fig. 7a). Following normalization against GAPDH (anti-GAPDH antibody, 6C5, Thermo Fisher Scientific, USA), the probing results showed relative downregulation in SdrM efflux pump proteins upon CAS treatment in isolates: S6, S7, S8, S21, and S23 when compared to the control samples (Fig. 7b). The percentage of reduction in the level of expression of SdrM proteins was highly significant (p-value < 0.001) for isolate S23 and was equivalent to 61%. Isolates S6, S8, and S21 showed a significant downregulatory effect (p-value < 0.01) with percentages of reduction equal to 40, 43 and 45%, respectively. The downregulation was not significant for isolate S7 reaching 18% reduction. No obvious differences in the expression level of SdrM proteins between treated and control samples of S1 and S14 isolates were observed (Fig. 7 b).
Fig. 6. Representative dendogram showing the densitometric intensity computerized analysis of SdrM efflux pump protein in MRSA isolate S23 control (a) and treated sample with 16µg/ml of caspofungin (b).
Fig. 7. Effect of caspofungin on SdrM efflux pump protein (53 kDa) in 7 MRSA clinical isolates. (a) Western blot analysis for untreated samples (control), lanes: 2, 4, 6, 8, 10, 12, and 14 and in the presence of 16µg/ml caspofungin (treated), lanes: 1, 3, 5, 7, 9, 11, and 13 representing isolates S1, S6, S7, S8, S14, S21, and S23, respectively. (b) The corresponding SdrM protein expression values for isolates S1, S6, S7, S8, S14, S21, and S23 following calibration against GAPDH. All data are mean ± SD of triplicates. The error bars represent SDs. The p-values indicate significance where ** p<0.01 and *** p<0.001.
In Egypt, the increasing rate of fluoroquinolone resistance among MRSA isolates is alarmingly recognized, contributing to a situation that could be described as countrywide. In a prospective cohort study, conducted in 12 hospitals in Egypt from 2005 to 2013, the rate of ciprofloxacin resistance reached 80% among MRSA isolates21. In Cairo, 60% of MRSA strains isolated from 211 patients suffering from pneumonia were found to be resistant to ciprofloxacin, while in Alexandria, MRSA isolated from different AMUH’s departments showed a rate of 74% of ciprofloxacin resistance18,22. Fluoroquinolones are still being prescribed by physicians in different Alexandria University hospitals to treat MRSA infections as per Infectious Diseases Society of America, IDSA, guidelines followed in these hospitals23,24. In our previous work, we showed that ZAB could be considered as an option for MRSA infections due to its efficacy encompassing older fluoroquinolones7. However, a percentage of 30.3% of MRSA isolated from AMUH were found to be resistant to ZAB in the current study (Table 2). This percentage of resistance, although being lower than that of ciprofloxacin reported across Egypt, necessitates the implementation of an innovative strategy widely discussed by researchers nowadays; the repurposing strategy. Guided by Siala et al. studies which emphasised the potentiating effect of CAS in destroying the biofilm matrix of S. aureus by inhibiting bacterial N-acetylglucosamine transferase10, in the current work, we investigated the sensitizing effect of CAS on ZAB in an attempt to increase ZAB efficacy against ZAB-resistant MRSA isolates. All of the ZAB-resistant isolates showed a decrease in their MIC values when combined with subinhibitory concentration of CAS (16µg/ml) and the fold reduction varied from 2- to 32-fold (Fig. 2 and Table 3). Proteomic approaches were applied to study the involved mechanism of CAS sensitization using cellular protein extracts of 7 MRSA isolates (S1, S6, S7, S8, S14, S21 and S23). Prominent qualitative and quantitative variations in the protein profiles of untreated and treated MRSA isolates were observed using SDS-PAGE (Fig. 3 a and b). These alterations included changes in the number of bands, their molecular weights, their intensities, along with the addition or the deletion of certain bands resulting in elevated percentages of polymorphism ranging from 82.4 to 94.1% (Table 4). The bands generated by SDS-PAGE analysis express the end-products of transcriptional and translational activities of active structural genes. Therefore, SDS-PAGE is considered to be a successful tool providing information about gene expression under stress due to a certain treatment25, or gene mutations caused by this treatment. Differences in the polypeptide band sizes might be attributed to the occurrence of point mutations in the relevant genes, creating stop codons that lead to production of longer or shorter polypeptide chains25. The intensity of polypeptide bands varied among treated and untreated samples (as shown in the Supplementary File: Tables 1-S to 7-S) and this might be due to changes in the regulatory gene expression, which modulated the rate of transcription of a specific structural protein, an observation consolidated by other researchers26. The appearance of unique bands, as a result of CAS treatment, may be interpreted by the fragmentation of polypeptide bands due to gene replication following a frameshift mutation. Shikazono et al. related the appearance of unique bands to the possible structural variations in DNA including transpositions, additions or deletions, giving rise to different amino acid sequences and accordingly, a different protein27. Contrastingly, the loss of protein bands following CAS treatment might be explained by several rationales. Cleavage of certain peptide bonds with the appearance of shorter polypeptide chains, deletion of the corresponding genes, or the cross linking of a particular polypeptide chain leading to the denaturation of protein, are among these explanations27. The constructed phylogenetic tree confirmed the variations in the protein profiles obtained as a result of treatment with 16µg/ml of CAS and underlined the existence of high level of polymorphism which may have emerged from from the inclusion or elimination of amino acids in mutated bands, or from changes in the sequence of these amino acids28 (Table 4 and Fig. 5).
Running a BLAST analysis, we found that β-1-3-glucan synthase, the fungal target enzyme of CAS, presented 33.3% identity to one of the members of S. aureus MFS efflux pumps, the SdrM efflux pump29. Similarity between fungal 1,3-beta-glucan synthase and another member of staphylococcal multidrug resistance (MDR) efflux pumps belonging to the adenosine-triphosphate (ATP)-binding cassette, (ABC) superfamily, has been reported in other bacteria, as well30. The SdrM efflux pump was shown to promote an energy-dependent efflux of hydrophilic fluoroquinolones31. Being a hydrophilic fluoroquinolone with a log P value of -0.89, ZAB might act as a possible substrate to SdrM efflux pump which extruded this fluoroquinolone, contributing to the resistance noticed among the tested isolates in this study. The resistance to fluoroquinolones in S. aureus has been generally related to mutations in topoisomerase IV and in DNA gyrase thus decreasing the affinity of these cellular targets to the drug32. Nevertheless, the fact that all of the ZAB-resistant isolates showed a decrease in their MIC values when combined with subinhibitory concentration of CAS (Fig. 2 and Table 3), implies the involvement of another mechanism of resistance. This fact was emphasized by western blot analysis using anti-1,3-beta-glucan synthase antibody, where treatment with CAS resulted in downregulation of SdrM efflux pump proteins in 5/7 tested isolates when compared to the untreated samples. The percentage of reduction in the level of expression of SdrM proteins varied between 18 to 61% with values being significant in 4/5 tested isolates (Fig. 7 a and b). Since the mode of action of CAS is by non-competitive inhibition of the enzyme 1,3-beta-glucan synthase which was found to be homologous to the staphylococcal SdrM efflux pump protein, accordingly, CAS was capable of inhibiting SdrM efflux pumps in ZAB-resistant isolates, preventing the extrusion of ZAB by this pump and consequently, restoring its antimicrobial activity against MRSA isolates. We are fully aware that the administration of CAS to patients with no fungal burden could jeopardize their normal flora. However, a preceding example of an antifungal agent being repurposed for the treatment of MRSA infections due to its potent inhibitory action on the staphylococcal enzyme diapophytoene desaturase (CrtN) is rising hope. Naftifine, an antifungal agent has already found its way in the preclinical phase with a new application for MRSA treatment and awaits submission to the Chinese FDA in the near future9.
In conclusion, this study identified a new bacterial target for the antifungal CAS. It was proved that CAS succeeded in restoring the antimicrobial activity of ZAB and sensitizing ZAB-resistant MRSA clinical isolates by blocking the action of SdrM efflux pump and inhibiting the expel of ZAB. Further research for the development of structurally-related agents to CAS with minimal antifungal activity is urgently required to endorse the repurposing of CAS as an agent used in combination with ZAB in the management of MRSA infections with higher efficiency.
Additional file: Additional Table 1S- 7S.
ACKNOWLEDGMENTS
The authors acknowledge Dongwha Pharm. Co., Ltd., Seoul, Korea, for kindly supplying zabofloxacin powder used in this study.
CONFLICT OF INTEREST
The authors declare that they have no conflict of interest.
AUTHORS’ CONTRIBUTION
All authors listed have made a substantial, direct, and intellectual contribution to the work, and approved it for publication.
FUNDING
None.
ETHICS STATEMENT
This article does not contain any studies with human participants or animals performed by any of the authors.
AVAILABILITY OF DATA
All datasets generated and analysed within this study are included in the manuscript and the Supplementary Files.
- Turner NA, Sharma-Kuinkel BK, Maskarinec SA, Eichenberger EM, Shah PP, Carugati M, Holland TL, Fowler VG. Methicillin- resistant Staphylococcus aureus: an overview of basic and clinical research. Nat Rev Microbiol, 2019; 17: 203-218.
Crossref - Abouelfetouh A. The status of methicillin resistance among Egyptian Staphylococcus aureus isolates: an overview. Infect Disord Drug Targets, 2017; 17: 67-69.
Crossref - Elshimy R, Khattab RA, Zedan H, Hosny AS, Elmorsy TH. Study on prevalence and genetic discrimination of methicillin-resistant Staphylococcus aureus (MRSA) in Egyptian hospitals. Afr J Microbiol Res, 2018; 12: 629-646.
Crossref - Kocsis B, Domokos J, Szabo D. Chemical structure and pharmacokinetics of novel quinolone agents represented by avarofloxacin, delafloxacin, finafloxacin, zabofloxacin and nemonoxacin. Ann Clin Microbiol Antimicrob, 2016.
Crossref - Hashem RA, Yassin AS, Zedan HH, Amin MA. Fluoroquinolone resistant mechanisms in methicillin-resistant Staphylococcus aureus clinical isolates in Cairo, Egypt. J Infect Dev Ctries, 2013; 7: 796-803.
Crossref - Park HS, Kim HJ, Seol MJ, Choi DR, Choi EC, Kwak JH. In vitro and in vivo antibacterial activities of DW-224a, a new fluoronaphthyridone. Antimicrob Agents Chemother, 2006; 50: 2261-2264.
Crossref - Mohamed NM, Zakaria AS, Edward EA, Abdel-bary A. In vitro and in vivo activity of zabofloxacin and other fluoroquinolones against MRSA isolates from a university hospital in Egypt. Pol J of Microbiol, 2019; 68: 59-69.
Crossref - Younis W, Thangamani S, Seleem MN. Repurposing non-antimicrobial drugs and clinical molecules to treat bacterial infections. Curr Pharm Des, 2015; 21: 4106-4111.
Crossref - Crunkhorn S. Antifungal agent blocks MRSA. Nature Reviews Drug Discovery, 2016.
Crossref - Siala W, Kucharikova S, Braem A, Vleugels J, Tulkens P M, Mingeot-Leclercq M, Van Dijck P, Van Bambeke F. The antifungal caspofungin increases fluoroquinolone activity against Staphylococcus aureus biofilms by inhibiting N-acetylglucosamine transferase. Nature communications, 2016.
Crossref - Clinical and Laboratory Standards Institute (CLSI). 2018. Performance standards for antimicrobial susceptibility testing: approved standard twenty-seventh edition M100-S28, Wayne, PA, USA.
- Mahadevan K. Methicillin-resistant Staphylococcus aureus; identification from clinical isolates and analysis of antibiotic resistance pattern. Aust J of Sci and Technol, 2017; 1: 17-20.
- Clinical and Laboratory Standards Institute (CLSI). 2012. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically: approved standard ninth edition M07-A9, Wayne, PA, USA.
- Fankam AG, Kuiate JR, Kuete V. Antibacterial and antibiotic resistance modifying activity of the extracts from allanblackia gabonensis, combretum molle and gladiolus quartinianus against Gram-negative bacteria including multi-drug resistant phenotypes. BMC Complementary and Alternative Medicine, 2015.
Crossref - Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem, 1976; 72: 248-254.
Crossref - Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature, 1970; 227: 680-685.
Crossref - Towbin H, Staehelin T, Gordon J. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedure and some applications. Proc Natl Acad Sci U S A, 1979; 76: 4350-4354.
Crossref - Sadaka SM, El-Ghazzawy EF, Harfoush RA, Meheissen MA. Evaluation of different methods for the rapid diagnosis of methicillin-resistance in Staphylococcus aureus. Afr. J. Microbiol. Res., 2009; 3: 49-55.
- Abdelhaliem E, Al-Huqai AA. Detection of protein and DNA damage induced by elevated carbon dioxide and ozone in Triticum aestivum L. using biomarker and comet assay. Genet Mol Res, 2016.
Crossref - Douglas CM. Fungal beta(1,3)-D-glucan synthesis. Med Mycol, 2001; 39:55-66.
Crossref - Abdel-Maksoud M, El-Shokry M, Ismail G, Hafez S, El-Kholy A, Attia E, Talaat M. Methicillin-resistant Staphylococcus aureus recovered from healthcare- and community-associated infections in Egypt. Int J Bacteriol, 2016.
Crossref - Helal Z, Radwan S, El-Tayeb W. Involvement of PVL producing MRSA in Pneumonia patients, Cairo, Egypt. Arch of Clin Microbiol, 2012; 3: 1-7.
- Lambert M. IDSA guidelines on the treatment of MRSA infections in adults and children. Am Fam Physician, 2011; 84: 455-463.
- Soliman G S, Abu-Youssef R M, Saleib B F, El-Moughazi A M, Zaki A. Awareness of World Health Organization methicillin-resistant Staphylococcus aureus guidelines at Alexandria University hospitals. East Mediterr Health J, 2013; 19: 622-628.
Crossref - Mondini L, Noorani A, Pagnotta M A. Assessing plant genetic diversity by molecular tools. Diversity, 2009; 1: 19-35.
Crossref - AL-Huqail A A, Abdelhaliem E. Evaluation of genetic variations in maize seedlings exposed to electric field based on protein and DNA markers. BioMed Res Intern, 2015.
Crossref - Shikazono N, Suzuki C, Kitamura S, Watanabe H, Tano S, Tanaka A. Analysis of mutations induced by carbon ions in Arabidopsis thaliana. J Exp Bot, 2005; 56: 587-596.
Crossref - Galani S, Naz F, Soomro F, Jamil I, Zia-ul-hassan, Azhar A, Ashraf A. Seed storage protein polymorphism in ten elite rice (Oryza sativa L.) genotypes of Sindh. Afr J Biotechnol, 2011; 10: 1106-1111.
- Costa SS, Viveiros M, Amaral L, Couto I. Multidrug efflux pumps in Staphylococcus aureus: an update. Open Microbiol J, 2013; 7: 59-71.
Crossref - Beauvais A, Bruneau JM, Mol PC, Buitrago MJ, Legrand R, Latge JP. Glucan synthase complex of Aspergillus fumigatus. J Bacteriol, 2001; 183: 2273-2279.
Crossref - Yamada Y, Hideka K, Shiota S, Kuroda T, Tsuchiya T. Gene cloning and characterization of SdrM, a chromosomally-encoded multidrug efflux pump, from Staphylococcus aureus. Biol Pharm Bull, 2006; 29: 554-556.
Crossref - Hooper DC. Mechanisms of fluoroquinolone resistance. Drug Resist Updat, 1999; 2: 38-55.
Crossref
© The Author(s) 2020. Open Access. This article is distributed under the terms of the Creative Commons Attribution 4.0 International License which permits unrestricted use, sharing, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.