ISSN: 0973-7510
E-ISSN: 2581-690X
S. aureus Urinary Tract Infections had emergence of resistance against commonly used antibiotics and especially methicillin. The present investigation was done to study the prevalence rate and antibiotic resistance pattern of the MRSA strains isolated from immunosuppressive patients suffered from UTIs. One-hundred and twenty urine samples were collected and cultured. Those that were positive for S. aureus, were subjected to PCR and disk diffusion method. Of 120 urine sample studied, 10 samples (8.33%) were positive for S. aureus. Prevalence of MRSA among the bacterial isolates was 5.83%. MRSA strains had the highest levels of resistance against ampicillin (100%), penicillin G (100%), tetracycline (85.71%), ciprofloxacin (85.71%), amikacin (71.42%) and trimethoprim-sulfamethoxazole (71.42%). The lowest levels of resistance were seen against imipenem (14.28%) and clindamycin (28.57%). Considering the high prevalence of MRSA and its emergence for antibiotic resistance, rapid identification of infected immunosuppressive patients and their quick treatment with imipenem and clindamycin are recommended.
Methicillin resistant, Staphylococcus aureus, Antibiotic resistance, Immunosuppressive patients, Pyelonephritis.
Most urinary tract infections(UTIs) involve only the bladder and urethra. Pyelonephritis results when a UTI progresses to involve the upper urinary system including kidneys and ureters. Acute pyelonephritis is a potentially kidney and life-threatening infection that often leads to renal scarring. Acute pyelonephritis results from bacterial invasion of the renal parenchyma. Bacteria usually reach the kidney by ascending from the lower urinary tract. Bacteria may also reach the kidney via the bloodstream. Timely diagnosis and management of acute pyelonephritis has a significant impact on patient outcomes.1 Bacteria are the most important cause of pyelonephritis.
Among all bacterial agents causing UTIs, Staphylococcus aureus (S. aureus) is one of the most prevalent.2 Highly distribution of this bacterium in the hospital environment, its high resistance against hard conditions and finally occurrence of high antibiotic resistance in some of its strains caused it to be one of the most important factors of pyelonephritis. It is a gram-positive and catalase-positive coccal bacterium that is a member of the Firmicutes which is responsible for various types on infections including respiratory, urinary, skin and soft tissue, burn and blood infections.2-4
The ability of S. aureus to resist against wide range of antibiotics causing severe problems in treatment of hospital infections. Infections caused by S. aureus are treated mainly with methicillin but in recent years, increasing numbers of methicillin resistant S. aureus (MRSA) have been reported worldwide from patients with community-acquired infections.5-7 According to the available data near fifteen percent of S. aureus of hospital infections were MRSA 5-7. Previously published data revealed that MRSA strains of various types of infections and especially UTIs had a considerable levels of resistance against various types of antimicrobial agents including quinolones, aminoglycosides, macrolides, cephalosporins, sulfonamides, fluoroquinolones and tetracycline.5-9
To date, there were rare information about the epidemiology and prevalence of MRSA in the cases of UTIs in Iran. Therefore, the present investigation was carried out to study the prevalence and antibiotic resistance pattern of MRSA isolated from immunosuppressive patients suffered from pyelonephritis.
Samples and Staphylococcus aureus isolation
From May 2014 to January 2015, a total of 120 urine samples were collected from immunosuppressive hospitalized patients of educational hospitals and health centers, Iran. All of these patients were suffered from pyelonephritis. Pyelonephritis was approved using the sonographic examination.10 Midstream urine was collected in sterile condition to decrease potential bacterial, cellular and artifactual contamination. This procedure was done using the Suprapubic Aspiration (SPA).11 Urine samples were immediately transferred to the laboratory in a cooler with ice-packs.
All samples were directly cultured into 7% sheep blood agar (Merck, Darmstadt, Germany) and incubated aerobically at 37°C for 48 h. After incubation, suspicious colonies were examined by the use of morphologies compatible with Staphylococcus spp. (microscopical morphology, catalase and coagulase production). Studied colonies were cultured on Tryptic Soy Broth (TSB) (Merck, Darmstadt, Germany) and Tryptic Soy Agar (TSA) (Merck, Darmstadt, Germany). After growth, staphylococci were identified on the basis of colony characteristics, Gram staining, pigment production, hemolytic and the following biochemical reactions: catalyses activity, coagulated test (rabbit plasma), Oxidase test, glucose O/F test, resistance to bacitracin (0.04 U), mannitol fermentation on Mannitol Salt Agar (MSA) (Merck, Darmstadt, Germany), urease activity, nitrate reduction, novobiocin resistance, phosphatase, deoxyribonuclease (DNase) test and carbohydrate (xylose, sucrose, trehalose and maltose, fructose, lactose, mannose) fermentation test.12
Identification of Methicillin-resistant Staphylococcus aureus
Identification of MRSA strains was done using the PCR-based method. Bacterial strains were sub-cultured in Tryptic Soy Broth (TSB, Merck, Germany) and further incubated for 48 h at 37 oC. Genomic DNA was extracted from bacterial colonies using the DNA extraction kit (Fermentas, Germany) according to manufacturer’s instruction. Those DNA samples which were simultaneously positive for femA and mecA genes were considered as MRSA. For this purpose, the PCR method which was introduced by Jonas et al. (2002) was used (13). The PCR reactions were performed in a total volume of 25 µL, including 1.5 mM MgCl2, 50 mM KCl, 10 mM Tris-HCl (pH 9.0), 0.1% Triton X-100, 200 µM dNTPs each (Fermentas, Germany), 2.5 µL PCR buffer (10X), 25 pmoL of each primer (mecA1 (5′-GTAGAAATGACTGAACGTCCGATAA-3′) and mecA2 (5′-CCAATTCCACATTGTTTCGGTCTAA-3′) (310 bp) and femB1 (5′-TTACAGAGTTAACTGTTACC-3′) (651 bp)), 1.5 U of Taq DNA polymerase (Fermentas, Germany) and 5 µL (40-260 ng/µL) of the extracted DNA template of the MRSA isolates. The PCR cycling conditions were as follows: initial denaturation at 94°C for 4 min, followed by 30 cycles of 45 s at 94°C, 45 s at 50°C, and 60 s at 72°C, with a final extension step at 72°C for 2 min.
Antimicrobial susceptibility test
Pattern of antimicrobial resistance of MRSA isolates of the cases of pyelonephritis was studied using the simple disk diffusion technique in the Mueller–Hinton agar (Merck, Germany) medium. Instruction of the Clinical and Laboratory Standards Institute guidelines (14) was used for this purpose. Susceptibility of MRSA isolates were studied against tetracycline (30 µg/disk), cotrimoxazole (30 µg/disk), gentamycin (10 µg/disk), amikacin (30 u/disk), ampicillin (10 u/disk), ciprofloxacin (5 µg/disk), imipenem (30 µg/disk), clindamycin (2 µg/disk), penicillin G (10 u/disk), trimethoprim-sulfamethoxazole (25 µg/disk), oxacillin (1µg/disk), and erythromycin (15 µg/disk) antibiotic disks (Oxoid, UK). The plates containing the discs were allowed to stand for at least 30 min before incubated at 35°C for 24 h. The diameter of the zone of inhibition produced by each antibiotic disc was measured and interpreted using the CLSI zone diameter interpretative standards (CLSI 2012) (14). S. aureus ATCC 43300 and Escherichia coli ATCC 11775 were used as quality control organism in antimicrobial susceptibility determination.
Statistical analysis
Statistical analysis was done using the SPSS/21.0 software (SPSS Inc., Chicago, IL) and chi-square and fisher exact tests were applied for analysis. Statistical significance was regarded at a P value < 0.05.
Table 1 represents the total Prevalence of S. aureus and MRSA in the urine samples taken from immunosuppressive patients suffered from pyelonephritis. Ten out of 120 urine samples (8.33%) taken from the immunosuppressive patients suffered from pyelonephritis were positive for S. aureus. Results of the gel electrophoresis for simultaneous detection of femA and mecA genes is shown in figure 1. We found that 7 out of 10 S. aureus (70%) strains had both femA and mecA genes which were known as MRSA. There was no significant difference between the prevalence of S. aureus and also MRSA (P > 0.05).
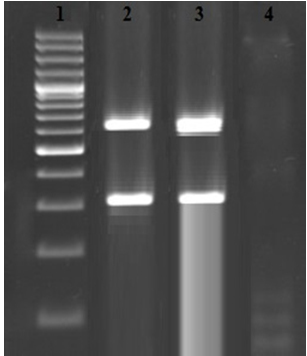
Fig. 1. Results of the gel electrophoresis for simultaneous detection of femA (651 bp) and mecA (310 bp) genes. 1: 100 bp marker (Fermentas, Germany), 2: Positive sample for mecA and femA genes, 3: Positive controls and 4: Negative control
Table (1):
Prevalence of S. aureus and MRSA in the urine samples taken from immunosuppressive patients suffered from pyelonephritis.
Types of samples |
No. samples |
Prevalence of S. aureus (%) |
Prevalence of MRSA (%) |
|---|---|---|---|
Urine |
120 |
10 (8.33) |
7 (70) |
Table 2 indicated the antibiotic resistance pattern of MRSA isolated from the urine samples of immunosuppressive patients suffered from pyelonephritis. We found that MRSA strains of our study harbored the highest levels of resistance against ampicillin (100%), penicillin G (100%), tetracycline (85.71%), ciprofloxacin (85.71%), amikacin (71.42%) and trimethoprim-sulfamethoxazole (71.42%). Prevalence of resistance against imipenem (14.28%) and clindamycin (28.57%) were low. Statistically significant differences were seen between the prevalence of resistance against ampicillin and imipenem (P = 0.011), penicillin G and imipenem (P = 0.014), ampicillin and clindamycin (P = 0.021) and ampicillin and oxacillin (P = 0.025).
Table (2):
Antibiotic resistance pattern of MRSA isolated from the urine samples of immunosuppressive patients suffered from pyelonephritis.
| Samples (No. positive) | Antibiotic resistance pattern (%) | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Tet | Cot | Gen | Amk | Amp | Cip | Imp | Cln | Pen | Tr-Su | Ox | Ert | |
| Urine (7) | 6 (85.71) | 4 (57.14) | 4 (57.14) | 5 (71.42) | 7 (100) | 6 (85.71) | 1 (14.28) | 2 (28.57) | 7 (100) | 5 (71.42) | 4 (57.14) | 5 (71.42) |
Tet: tetracycline (30 µg/disk), cot: cotrimoxazole (30 µg/disk), gen: gentamycin (10 µg/disk), amk: amikacin (30 u/disk), amp: ampicillin (10 u/disk), cip: ciprofloxacin (5 µg/disk), imp: imipenem (30 u/disk), cln: clindamycin (2 µg/disk), pen: penicillin G (10 u/disk), tr-su: trimethoprim–sulfamethoxazole (25 μg/disk), ox: oxacillin (1µg/disk), ert: erythromycin (15µg/disk).
Diabetes, acquired immune deficiency syndrome (AIDS), some kinds of surgical operation and many other conditions cause suppression of human immunity. Suppression of immunity cause quickly occurrence of infectious diseases. Presence of immunosuppressive patients in the infected environment of hospital facilitates occurrence of infectious diseases like UTIs. Dissemination of pathogenic agents which are mainly resistant against commonly used antibiotics to upper parts of the urinary system and especially kidney cause pyelonephritis which is a complex shape of UTIs.
Our results showed that 5.83% of urine samples taken from immunosuppressive patients suffered from pyelonephritis were infected with MRSA. High prevalence of resistance against various types of antibiotics including tetracycline, cotrimoxazole, gentamycin, ampicillin, ciprofloxacin penicillin G, trimethoprim-sulfamethoxazole and erythromycin is another important finding of our study. Indiscriminate and unauthorized prescription of antibiotics, inattention to the results obtained from the disk diffusion method, prescription of antibiotics based on the self-experience of medical practitioners, lack of proper disinfection of hospital environment and finally transmission of resistant pathogens from infected patients to hospital environment and also other patients are the most common reasons for the considerable prevalence of MRSA in our study. Onanuga et al. (2012)15 revealed that the prevalence of S. aureus in the urine specimens of patients suffered from UTIs in Nigeria was 33.6% which was higher than our findings. They showed that all isolates were resistant to methicillin and the prevalence of resistance against tetracycline, chloramphenicol, cotrimoxazole, gentamycin, vancomycin, cefuroxime, nitrofurantoin, ofloxacin and ciprofloxacin were 97.8%, 80.4%, 73.9%, 69.6%, 54.3%, 39.1%, 34.8% and 32.6%, respectively. Prevalence of resistance against methicillin in Australia, Jamaica, France, Spain, and USA were 21.6%, 23%, 33.6%, 30.3%, and 75%, respectively16-18. Momtaz and Hafezi (2014) 19 reported that S. aureus strainsof human clinical infections had the highest resistance against penicillin (100%), cephalothin (100%), cefazoline (100%), ceftireaxon (100%), azitromycin (62.12%), tetracycline (57.57%) and erythromycin (54.54%) which was similar to our findings. Brown et al. (2007)17 reported that the prevalence of MRSA strains was 23%. No MRSA was resistant to vancomycin and except for penicillin and to some extent co-trimoxazole (trimethoprim–sulfamethoxazole), most MSSA isolates were susceptible to nearly all antimicrobial agents.
In keeping with high prevalence of MRSA and also their considerable levels of antibiotic resistance, accurate, rapid and sensitive detection of MRSA is required especially in immunosuppressive patients which are more prone to get UTIs. Accurate application of simple disk diffusion method is another approach to reduce dissemination of MRSA. We recommended rapid identification of infected patients and their treatment with imipenem and clindamycin antibiotics.
- Morello W, La Scola C, Alberici I, Montini G. Acute pyelonephritis in children. Pediatr Nephrol. 2016; 31(8):1253-65.
- Yousefi M, Pourmand MR, Fallah F, Hashemi A, Mashhadi R, Nazari-Alam A. Characterization of Staphylococcus aureus Biofilm Formation in Urinary Tract Infection. Iran J Public Health. 2016; 45(4):485-93.
- Alavi Naeini R, Sanadgol H, Foroghani BAM, Darvishi M. Effect of Oral Rifampin in Prophylaxix of Staphylococcus Aureus Nasocarriers of Hemodialysis Patient. Annals of Military and Health Sciences Research. 2007; 4(4): 1009-1015.
- Tong SY, Davis JS, Eichenberger E, Holland TL, Fowler VG Jr. Staphylococcus aureus infections: epidemiology, pathophysiology, clinical manifestations, and management. Clin Microbiol Rev. 2015; 28(3):603-61.
- Abejew AA, Denboba AA, Mekonnen AG. Prevalence and antibiotic resistance pattern of urinary tract bacterial infections in Dessie area, North-East Ethiopia. BMC Res Notes. 2014; 7:687.
- Olona-Cabases M, Ticó-Falguera N, Ramírez-Garcerán L, Del Valle-Ortiz O, Castelló-Verdú T, García-Fernández L. Methicillin-resistant Staphylococcus aureus: a four-year experience in a spinal cord injury unit in Spain. Spinal Cord. 1996; 34(6):315-9.
- Adwan K, Abu-Hasan N, Adwan G, Jarrar N, Abu-Shanab B, Abu-Zant A. Nosocomial infection caused by methicillin-resistant Staphylococcus aureus in Palestine. Microb Drug Resist. 2005 Spring; 11(1):75-7.
- Kaur DC, Chate SS. Study of Antibiotic Resistance Pattern in Methicillin Resistant Staphylococcus Aureuswith Special Reference to Newer Antibiotic. J Glob Infect Dis. 2015; 7(2): 78–84.
- Al-Zoubi MS, Al-Tayyar IA, Hussein E, Jabali AA, Khudairat S. Antimicrobial susceptibility pattern of Staphylococcus aureus isolated from clinical specimens in Northern area of Jordan. Iran J Microbiol. 2015; 7(5):265-72.
- MacKenzie JR, Fowler K, Hollman AS, Tappin D, Murphy AV, Beattie TJ, Azmy AF: The value of ultrasound in the child with an acute urinary tract infection. Br J Urol. 1994; 74(2):240-244.
- NICE: Urinary Tract Infections in Children: Diagnosis, Treatment and Long-term Management. 2007.
- Zmantar T, Chaieb K, Ben Abdallah F, Ben Kahla-Nakbi A, Ben Hassen A, Mahdouani K, et al. Multiplex PCR detection of the antibiotic resistance genes in Staphylococcus aureus strains isolated from auricular infections. Folia Microbiol (Praha) 2008; 53(4): 357-362.
- Jonas D, Speck M, Daschner FD, Grundmann H. Rapid PCR-Based Identification of Methicillin-Resistant Staphylococcus aureus from Screening Swabs. J Clin Microbiol 2002; 40:1821–1823.
- Clinical and Laboratory Standards Institute (CLSI). Performance standards for antimicrobial susceptibility testing. Twenty-second informational supplement M100-S21. Wayne Pa 2012.
- Onanuga A, Awhowho GO. Antimicrobial resistance of Staphylococcus aureusstrains from patients with urinary tract infections in Yenagoa, Nigeria. J Pharm Bioallied Sci. 2012; 4(3): 226–230.
- Frazee BW, Lynn J, Charlebois ED, Lambert L, Lowery D, Perdreau-Remington F. High prevalence of methicillin-resistant Staphylococcus aureus in emergency department skin and soft tissue infections. Ann Emerg Med. 2005; 45(3):311-20.
- Brown PD, Ngeno C. Antimicrobial resistance in clinical isolates of Staphylococcus aureus from hospital and community sources in southern Jamaica. International Journal of Infectious Diseases, 2007; 11, 220—225.
- Voss A, Milatovic D, Wallrauch-Schwartz C, Rosdahl VT, Braveny I. Methicillin-resistant Staphylococcus aureus in Europe. Eur J Micr Infect Dis 1994; 13: 50-5.
- Momtaz H, Hafezi L. Meticillin-resistant Staphylococcus aureus isolated from Iranian hospitals: virulence factors and antibiotic resistance properties. Bosn J Basic Med Sci. 2014; 14(4):219-26.
© The Author(s) 2016. Open Access. This article is distributed under the terms of the Creative Commons Attribution 4.0 International License which permits unrestricted use, sharing, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.


