ISSN: 0973-7510
E-ISSN: 2581-690X
Pseudomonas aeruginosa is inherently resistant to many drugs. It is now an emerging opportunistic pathogen of clinical relevance. The emergence of carbapenemases is another major concern. Initiation of appropriate therapy is of paramount importance thus highlighting the need of active surveillance for newer emerging resistance trends for better infection control. To study the resistance pattern of P. aeruginosa isolates obtained from lab specimens and to determine the production of ESBL and Carbapenemase among them. A hospital-based cross-sectional study was carried out in the Department of Microbiology, Government medical college Thrissur, among P. aeruginosa isolates obtained from lab specimens, from January 2018 – December 2018. 162 isolates were studied. Antimicrobial susceptibility testing was done by Kirby – Bauer disc diffusion method, extended-spectrum beta-lactamase (ESBL) production was confirmed by and phenotypic confirmatory disc diffusion test. Carbapenemase detection was done using the modified carbapenemase inactivation (mCIM) method. The obtained data was analysed. Among 162 isolates 83% were non-multidrug-resistant (MDR) strains and 17% were MDR strains. 22% of ceftazidime resistant isolates were ESBL producers. 6.2% isolates were resistant to imipenem. Among the imipenem resistant isolates, Carbapenemase production was seen in 30% isolates by mCIM test. According to our study, the most effective antibiotic against P. aeruginosa were imipenem and cefoperazone/sulbactam showing resistance in 6.3% and 6.9% isolates respectively. The diversity of antibiotic resistance mechanisms and the emergence of carbapenem resistance is a threat that limits treatment choices. This suggests the need for ongoing antimicrobial susceptibility studies in the future.
Pseudomonas aeruginosa, multidrug resistance, ESBL, carbapenemase, mCIM
P. aeruginosa is now a days considered an important cause of both hospital-acquired and community-acquired infections. It can cause significant morbidity and mortality particularly among immunocompromised individuals.
P. aeruginosa is well known for its ability to survive in varied environments. Even though P. aeruginosa is implicated in both community and hospital acquired infections, it is much more common in the latter. The other distinct properties of P. aeruginosa is that it has got very minimal nutritional requirements and the ability to form biofilms so that they can colonize several medical devices such as bronchoscopes, intravenous and urinary catheters etc.
Because of these properties, P. aeruginosa is now one among the major causes of hospital-acquired infections especially in the intensive care units.1,2 P. aeruginosa is associated with almost all infections like respiratory tract infections, otitis media, osteomyelitis, skin and soft tissue infections, keratitis, sepsis. The risk of acquiring infection includes immuno-compromised patients, especially those taking chemotherapy. The risk of these patients are raised mainly due to the underlying diseases and presence of invasive devices.1,3
Apart from intrinsic resistance to several antibiotic classes there is acquired and mutational resistance thus making the treatment even more complicated. Multiple resistance mechanisms have also been identified among the isolates.4
One of the major concerns nowadays is the development of carbapenem resistance in P. aeruginosa.5 In case of infections caused by MDR strains, carbapenems are the major choice of drugs so the treatment options get limited if there is carbapenem resistance. In such cases colistin becomes the last resort drug.6 Acquired resistance due to plasmids is also a seen in P. aeruginosa. It is particularly associated with indiscriminate antibiotic use. The rate of developing MDR P. aeruginosa strains is alarming as it creates health problems resulting in increased rates of morbidity, mortality and high health care cost.7
It is the need of the hour to develop appropriate empirical treatment for infections including P. aeruginosa infections.8 Due to resistance mechanisms the pattern also changes from time to time making it very important to know the antibiotic susceptibility of P. aeruginosa mainly for directing the empirical therapy. Hence the present study was conducted in our institution to find out the regional susceptibility profiles of P. aeruginosa with an added objective to detect the prevalence of ESBL and carbapenemase production among these isolates.
The study was done in the Department of Microbiology, Government medical college Thrissur, a hospital based cross sectional study, for a period of one year from January 2018 – December 2018 after approval and clearance from Ethical Committee.
All P. aeruginosa isolates obtained in the laboratory were included. The isolates which was repeatedly isolated from the same patient was excluded. Isolates obtained among mixed bacterial growth isolates were also excluded. All clinically relevant samples including pus, sputum, blood, csf received at the microbiology laboratory was processed according to standard procedures. Gram staining of the specimens were done to screen for pus cells and organisms. The samples was cultured on blood agar, MacConkey agar and nutrient agar. Identification of P. aeruginosa was done by colony morphology, oxidase test and various biochemical reactions like:
- Triple sugar iron showing alkaline/no change (K/NC) reaction with no gas and no H2S production
- Urea not hydrolysed
- Citrate utilized
- Nitrate reduced to nitrite which is further reduced to nitrogen gas
- Oxidative on O/F media
- Arginine hydrolysis by Moller decarboxylase test
The antimicrobial susceptibility testing was done by Kirby Bauer disc diffusion method and the results were interpreted as per CLSI 2018 guidelines.
Detection of ESBL
Screening method and Phenotypic Confirmatory Disk Diffusion test was done for detecting ESBL production.
Screening method
Zone size of ≤ 22 mm to ceftazidime disc has been screened as ESBL producers.
Phenotypic Confirmatory Disk Diffusion test for ESBL
Lawn culture of the isolate was plated on Mueller Hinton Agar (MHA) plate and tested against combination of disks viz. ceftazidime (30μg) along with ceftazidime – clavulanic acid (30/10μg). After 16-18 hours of incubation the zone sizes were measured and recorded.
ESBL production was confirmed when there is a ≥ 5mm increase in zone diameter for the antimicrobial agent test in combination with clavulanic acid when compared with it tested alone.9
Modified carbapenemase inactivation method (mCIM) for detection of Carbapenemase production
The test was done according to CLSI 2018, M100 guidelines.
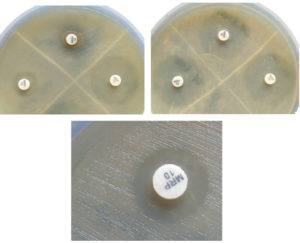
Each isolate was initially cultured overnight on blood agar. The next day, a loopful (10-μl) of bacteria was emulsified in 2 ml of trypticase soy broth (TSB). Vortex the mix for 10 to 15 s. Then meropenem disc (10-μg) was added to each tube. The suspension was then incubated at 35°C for 4 hours. Meropenem discs was then removed from each of the suspension using a 10-μl loop and place it on MHA plates which are previously inoculated with meropenem susceptible E.coli ATCC 25922 indicator strain. The MHA plates were incubated at 35°C for 18-24 hours. [Fig. 1]
Interpretation
Carbapenemase production present if there is zone diameter of 6-15 mm or colonies present within 16-18mm zone.
Carbapenemase negative – Zone diameter of ≥19mm (clear zone).
Carbapenemase indeterminate:
- Zone diameter – 16-18mm
- Zone diameter – ≥19mm and presence of minute colonies within zone
Statistical analysis
Data was analysed in Microsoft Excel version 10 after coding for different parameters and percentages were calculated.
Out of the 162 samples, P. aeruginosa was isolated in highest percentage from sputum which accounted for 29.4% followed by pus swab (23.8%). Aspirates, tissues and ear swab yielded P. aeruginosa in 10.6%, 10% and 10% cases respectively. Next highest was from tracheal aspirate (6.3%). Six out of 162 samples were urine samples which accounted for 3.8% followed by corneal scraping 3.1%. Blood samples, pleural fluid and CSF yielded P. aeruginosa in 1.9%, 1.3% and 1.3% cases respectively.
Maximum number of strains were obtained from patients belonging to age group >50 years (53.70%) with greater predominance in males. [Table 1]
Table (1):
Age and Sex distribution.
Frequency |
Percentage |
|
|---|---|---|
Sex Distribution |
||
MALE |
107 |
66.04 |
FEMALE |
55 |
33.9 |
Age Distribution |
||
<1 YEAR |
5 |
3.08 |
2-20 |
9 |
5.6 |
21-50 |
61 |
37.6 |
>50 |
87 |
53.70 |
Total |
162 |
100 |
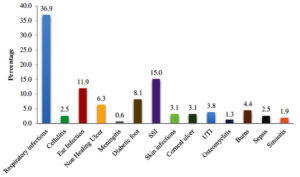
Maximum isolates were obtained from the cases with respiratory infections accounting for 59 cases i.e. 36.9%. Next highest isolation was from surgical site infections (15%) followed by ear infection (11.9%), diabetic foot (8.1%), non-healing ulcer (6.3%). Burns cases accounted for 4.4%, followed by urinary tract infections (3.8%), corneal ulcer(3.1%), skin infections (3.1%). P. aeruginosa was isolated from blood in 2.5% cases and 1.9% from sinusitis. There was 1 case (0.6%) in which P. aeruginosa was isolated from CSF in a case of meningitis [Fig. 2].
On analysing the risk factors it was found that patients with diabetes mellitus (14.4%) were more prone to P. aeruginosa infections followed by bronchiectasis (13.1%). Other risk factors included pulmonary tuberculosis (5.6%), patients on ventilator (5.6%), surgery (4.4%), on radiotherapy (4.4%), burns (4.4%), COPD (3.8%). Urinary tract catheterization (2.5%), HIV infection on anti- retroviral treatment (0.6%) and lastly pre-term baby (0.6%)
On doing antibiotic susceptibility testing the most effective antibiotic against P. aeruginosa were imipenem and cefoperazone/sulbactam with resistance in 6.3% and 6.9% isolates respectively. Least resistance was observed in piperacillin/tazobactam (10%), cefepime (11.3%), aztreonam (12.5%) and amikacin (15.6%) respectively. Analysis of susceptibility pattern showed high level of resistance among ceftazidime (25.6), gentamicin (26.9%), ciprofloxacin (33.1%), and nitrofurantoin (60% in case of urinary isolates) [Table 2].
Table (2):
Antibiotic susceptibility pattern of isolates.
| Antibiotics | Sensitive | Resistant | ||
|---|---|---|---|---|
| n | % | n | % | |
| Amikacin | 135 | 84.4 | 25 | 15.6 |
| Gentamicin | 117 | 73.1 | 43 | 26.9 |
| Ciprofloxacin | 107 | 66.9 | 53 | 33.1 |
| Ceftazidime | 119 | 74.4 | 41 | 25.6 |
| Cefoperazone/Sulbactam | 149 | 93.1 | 11 | 6.9 |
| Cefepime | 142 | 88.8 | 18 | 11.3 |
| Imipenem | 150 | 93.8 | 10 | 6.3 |
| Piperazillin/Tazobactam | 144 | 90.0 | 16 | 10.0 |
| Aztreonam | 140 | 87.5 | 20 | 12.5 |
| Nitrofurantoin (n=5 urinary isolates) | 2 | 40.0 | 3 | 60.0 |
Among 162 isolates 83% were Non-MDR strains while 17% were MDR strains. Among the 27 MDR strains maximum resistance was seen towards ciprofloxacin (96.3%), followed by gentamicin and ceftazidime (85.2% each), aztreonam (70.4%), amikacin (59.3%), cefepime (51.9%), followed by imipenem, cefoperazone/sulbactam (37.0%, 33.3% respectively) least resistance was seen towards piperacillin/tazobactam (22.2%).
Out of the 41 ceftazidime resistant isolates ESBL producing organisms accounted for 22% by double disc synergy testing testing. Among the 22% ESBL producers 55.6% were resistant to amikacin, 77.8% resistant to gentamicin, ciprofloxacin 88.9%, cefoperazone/sulbactam 55.6%, cefepime 66.7%, imipenem 33.3%, aztreonam 77.8%, and least resistance was observed to piperacillin/tazobactam 11.1%.
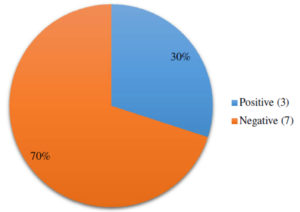
Carbapenemase production was seen among 30% of imipenem resistant isolates by mCIM method. [Fig. 3].
P. aeruginosa an important cause for opportunistic infections, now being increasingly associated with community acquired infections also. The increasing number of MDR P. aeruginosa is another major issue. It is very difficult to eradicate P. aeruginosa and moreover it is associated with greater mortality rates. The maximum number of isolates were obtained from sputum followed by pus swab. The main bulk of patients that we obtained during our study period was having respiratory infections. This may be the reason for isolating the organism in high percentage from sputum samples. We avoided the isolates to be mere colonizers of respiratory tract by isolating the organism repeatedly and also taking in to consideration the relevant clinical history. In studies done by Jakribettu sputum showed the highest Pseudomonas isolation rate.10
In this study maximum number of isolates (53.70%) were obtained in the age group of above 50 yrs and in the age group 21-50 years was 37.6%. The most common age group to get affected by P. aeruginosa is found to be between 20-50 years in several studies.11-13 The risk factors for attaining infection is much more common in this age group. Also as the age advances the natural defence mechanism of the body weakens in addition they may acquire other co-morbid conditions also and are hence more prone for this opportunistic hospital acquired infections.
Respiratory infection are found to be the most common presentation caused by P. aeruginosa in our institution, followed by other infections. Out of the 59 respiratory infection isolates majority was from pneumonia cases and most of them had risk factors like bronchiectasis, diabetes mellitus or were on ventilator. In one study by Nakamura A etal reported greater frequency of P. aeruginosa infection in diabetes mellitus.14 In another study by Zhang etal showed that diabetes mellitus was a risk factor for P. aeruginosa infection.15 India is the home for largest number of diabetic individuals and their socioeconomic conditions are poor. Diabetic foot is another complication of diabetes mellitus which can also get complicated by P. aeruginosa. Out of the five UTI cases 4 were catheterized, and these were considered as catheter associated urinary tract infections. Since P. aeruginosa can form biofilms on urinary catheters it is very important to change catheter frequently and prevent these hospital acquired infections. In this study out of the three sepsis cases two cases were from neonates. One baby was pre-term and the other baby was complicated with P. aeruginosa meningitis also which was confirmed by isolating P. aeruginosa from CSF and blood. This isolate was multi drug resistant and also a carbapenemase producer (by mCIM method). This isolate was susceptible to piperacillin/tazobactam. The baby got better by treatment except for the neurological sequelae caused due to meningitis. This case shows the importance of detecting drug resistance in P. aeruginosa.
In our study regarding the antibiotic susceptibility pattern, the most susceptible drugs were imipenem and cefoperazone/sulbactam, piperacillin/tazobactam. High level of resistance was seen towards ciprofloxacin, gentamicin, and ceftazidime and among urinary isolates to nitrofurantoin. Our institution study data shows that P. aeruginosa is becoming less susceptible to fluoroquinolones, cephalosporins, aminoglycosides especially gentamicin. Similar findings were obtained in the study done by Nithyalakshmi.16 where resistance profile of imipenem was 0%, and towards piperacillin/tazobactam (11.6%), amikacin (19.6%), ciprofloxacin 50.89% and all cephalosporins tested (28% – 38%) with cefepime (28.57%), ceftazidime (38.39%). Similar pattern was observed in other studies17-19 also which was comparable with our data. Empirical treatment that is usually started in a hospital did not include antipseudomonal coverage. The commonly used agent that has antipseudomonal coverage is ciprofloxacin but it is showing increased resistance trend. This may be the reason for getting more number of P. aeruginosa infection cases than before.
Microorganisms in hospital setups are generally resistant to multiple antibiotics due to increased usage of antibiotics. In our study among the 162 isolates 17% were MDR, and 83% were non-MDR strains which were comparable to other studies.20,21,16
Highest resistance was found to ciprofloxacin, gentamicin, followed by ceftazidime, aztreonam. While these strains showed the highest susceptibility to piperacillin/tazobactam combination. Multidrug resistance is a growing problem because of frequent use of antibiotics by primary care physicians and over the counter sale of antibiotics. This finding was similar to the study done by Gill et al.22 in which high level of resistance was seen with gentamicin (92.7%), followed by imipenem (90.2%), ciprofloxacin (87.8%) while these strains showed the highest susceptibility to piperacillin/tazobactam (36.6%) combination. Among ESBL isolates, highest resistance was obtained for ciprofloxacin 88.9%, followed by gentamicin and aztreonam (both 77.8%).
Carbapenemase detection is of great importance for preventing nosocomial transmission and in investigating outbreaks. So an attempt was made to detect carbapenemase production by mCIM method which is published in CLSI for P. aeruginosa.23 P. aeruginosa can acquire resistance to carbapenem group by a number of methods but mCIM specifically detects carbapenemase production only. In this method 30% of imipenem resistant isolates were found to be carbapenemase producers and is comparable with previous study of Lisboa et al.24
This study shows that the treatment options are very limited in carbapenemase producers and its control from spreading to other individuals and other wards is of paramount importance. Rapid detection of carbapenemase production is required for implementation of infection control practices.
mCIM method demonstrated a sensitivity of >97% and specificity of 100% for detection of various types of carbapenemases like NDM,KPC,IMI, VIM, IMP,SPM, and OXA among P. aeruginosa isolates investigated by CLSI. mCIM method is a simple, accurate method for carbapenemase detection. It does not require costly equipments or reagents, hence affordable for laboratories and hospitals.24 This method can be offered as a valuable alternative to PCR for detecting carbapenemase production in P. aeruginosa.
Limitation of the study
Being a cross sectional study, the outcome of patients after discharging from hospital was not followed up. ESBL producers among the isolates were detected by phenotypic method, the molecular methods for detection could not be done. Another limitation is that mCIM is a phenotypic method and not molecular detection of carbapenemases.
It was found that there is an increase in resistance towards commonly prescribed antimicrobial agents. The isolates showed resistance towards higher antibiotics also. In this study we found imipenem has significant action against P. aeruginosa even though carbapenem resistance is on the rise. The development of antimicrobial resistance is a natural process, which cannot be stopped but the development of resistance is accelerated when antimicrobials are misused. Recently there is progress in the development of new antibiotics but P. aeruginosa has got or can acquire mechanisms to develop resistance to these new antibiotics also. The most effective treatment strategy for P. aeruginosa is combinational therapies. The emergence of carbapenem resistance is a threat that limits treatment choices and suggests the need for ongoing antimicrobial susceptibility studies in the future.
ACKNOWLEDGMENTS
We are grateful to Late Dr Prithi Nair K, Former Professor and HOD, Department Of Microbiology, Government Medical College Thrissur for her unconditional guidance and support.
CONFLICT OF INTEREST
The authors declare that there is no conflict of interest.
AUTHORS’ CONTRIBUTION
Both the authors listed have made a substantial, direct, and intellectual contribution to the work, and approved it for publication.
FUNDING
None.
ETHICS STATEMENT
The study protocol was approved by the Institutional research committee (IRC), Govt Medical College Thrissur, Kerela, India.
AVAILABILITY OF DATA
All datasets analyzed in the study are included in the manuscript and presented as tables and Figs.
- Bonten MJ, Bergmans DC, Speijer H, Stobberingh EE. Characteristics of polyclonal endemicity of Pseudomonas aeruginosa colonization in intensive care units: implications for infection control. Am J Respir Crit Care Med. 1999;160(4):1212-1219.
Crossref - Driscoll JA, Brody SL, Kollef MH. The epidemiology, pathogenesis and treatment of Pseudomonas aeruginosa infections. Drugs. 2007;67(3):351-368.
Crossref - Kerr KG, Snelling AM. Pseudomonas aeruginosa: a formidable and ever-present adversary. J Hosp Infect. 2009;73(4):338-344.
Crossref - Mesaros N, Nordmann P, Plésiat P, et al. Pseudomonas aeruginosa: resistance and therapeutic options at the turn of the new millennium. Clin Microbiol Infect. 2007;13(6):560-578.
Crossref - Walsh TR. Clinically significant carbapenemases: an update. Curr Opin Infect Dis. 2008;21(4):367-371.
Crossref - Beno P, Krcmery V, Demitrovicova A. Bacteraemia in cancer patients caused by colistin-resistant Gram-negative bacilli after previous exposure to ciprofloxacin and/or colistin. Clin Microbiol Infect. 2006;12(5):497-498.
Crossref - Shahid M, Malik A, Sheeba. Multidrug-resistant Pseudomonas aeruginosa strains harbouring R-plasmids and AmpC β-lactamases isolated from hospitalised burn patients in a tertiary care hospital of North India. FEMS Microbiology Letters.2003;228(2):181-186.
Crossref - Kollef MH, Fraser VJ. Antibiotic resistance in the intensive care unit. Ann Intern Med. 2001;134(4):298-314.
Crossref - Laudy AE, Rog P, Smolinska-Krol K, et al. Prevalence of ESBL-producing Pseudomonas aeruginosa isolates in Warsaw, Poland, detected by various phenotypic and genotypic methods. PloS ONE. 2017;12(6):e180121.
Crossref - Jakribettu RP, Ahamed SM, Anju MM, Safeera MI, Chandran AV. Emerging biofilm producing multi-drug resistant mucoid strains of Pseudomonas aeruginosa in a Rural Medical College Hospital in North Kerala. J Microbiol Biotech Res. 2013;3(6):59-63.
- Gill JS, Arora S, Khanna SP, Kumar KH. Prevalence of multidrug-resistant, extensively drug-resistant, and pandrug-resistant Pseudomonas aeruginosa from a tertiary level intensive care unit. J Glob Infect Dis. 2016;8(4):155-159.
Crossref - Anupurba S, Bhattacharjee A, Garg A, Sen MR. Antimicrobial susceptibility of Pseudomonas aeruginosa isolated from wound infections. Indian Journal of Dermatology. 2006;51(4):286-288.
Crossref - Rashid A, Chowdhury A, Rahman SH, Begum SA, Muazzam N. Infections by Pseudomonas aeruginosa and antibiotic resistance pattern of the isolates from Dhaka Medical College Hospital. Bangladesh Journal of Medical Microbiology. 2007;1(2):48-51.
Crossref - Nakamura A, Miyake K, Misawa S, et al. Meropenem as predictive risk factor for isolation of multi drug resistant Pseudomonas aeruginosa. J Hosp Infect. 2013;83(2):153-155.
Crossref - Zhang J-F, Zhu H-Y, Sun Y-W, et al. Pseudomonas aeruginosa infection after pancreatoduodenectomy: risk factors and clinic impacts. Surg Infect. 2015;16(6):769-774.
Crossref - Nithyalakshmi J, Akhila K, Mohanakrishnan K, Sumathi G. Evaluation of resistance profile of pseudomonas aeruginosa with reference to biofilm production- An emerging challenge. IOSR Journal of Dental and Medical Sciences. 2015;14(11):13-18.
Crossref - Shahcheraghi F, Nikbin VS, Feizabadi MM. Prevalence of ESBLs genes among multidrug-resistant isolates of Pseudomonas aeruginosa isolated from patients in Tehran. Microbial Drug Resistance. 2009;15(1):37-39.
Crossref - Ibukun Aibinu TN, Odugbemi T. Occurrence of ESBL and MBL in Clinical Isolates of Pseudomonas aeruginosa From Lagos, Nigeria. The Journal of American Science. 2007;3(4):81-85.
- Peshattiwar PD, Peerapur BV. ESBL and MBL mediated resistance in Pseudomonas aeruginosa: An emerging threat to clinical therapeutics. J Clin Diagn Res. 2011;5(8):1552-1554.
- Tam VH, Chang K-T, Abdelraouf K, et al. Prevalence, resistance mechanisms, and susceptibility of multidrugresistant bloodstream isolates of Pseudomonas aeruginosa. Antimicrob Agents Chemother. 2010;54(3):1160-1164.
Crossref - Idris SN, Desa MN, Aziz MN, Taib NM. Antimicrobial susceptibility pattern and distribution of exoU and exoS in clinical isolates of Pseudomonas aeruginosa at a Malaysian hospital. Southeast Asian Journal of Tropical Medicine & Public Health. 2012;43(1):116-123.
- Gill MM, Usman J, Kaleem F, et al. Frequency and antibiogram of multi-drug resistant Pseudomonas aeruginosa. J Coll Physicians Surg Pak. 2011;21(9):531-534.
- Clinical and Laboratory Standard Institute (CLSI): Performance standards for antimicrobial susceptibility testing twenty eighth informational supplement.M100.2018.
- Lisboa LF, Turnbull L, Boyd DA, Mulvey MR, Dingle TC. Evaluation of a modified carbapenem inactivation method for detection of carbapenemases in Pseudomonas aeruginosa. J Clin Microbiol. 2018;56(1).
© The Author(s) 2021. Open Access. This article is distributed under the terms of the Creative Commons Attribution 4.0 International License which permits unrestricted use, sharing, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.