ISSN: 0973-7510
E-ISSN: 2581-690X
In this study, a total of 228 raw meat samples (pork: 76, beef: 76, chicken: 76) and 301 raw seafood samples (fish: 199, shrimp: 67, squid: 35) were collected randomly at traditional markets in Ho Chi Minh City (Vietnam). In meat, the ratio of Salmonella spp. was 70.61% (161/228). Among the contaminated meat samples, pork was infected with a ratio of 90.79 % (69/76) while the contamination ratios in beef and chicken were 43.42% (33/76) and 77.63% (59/76), respectively. Salmonella contamination was detected in fish (40.20%), shrimp (7.46%) and squid (17.14%). Because of sulfonamide group is used in Salmonella treatment, the study focused on sulfonamide resistance. In fresh seafood comparison, there were 32.56%, 40% and 10% Salmonella showing resistance to sulfamethoxazol in fish, squid and shrimp, respectively. In fresh meat comparison, there were 31.58%, 16.67% and 55.56% Salmonella showing resistance to sulfamethoxazol in pork, beef and chicken, respectively. Interestingly, there were 21 serovars including 19 identified serovars including S. Kentucky (8), S. Agona (2), S. Infanis (4), S. Saintpaul (1), S. Indiana (1), S. Braenderup (1), S. Potsman (2) and 2 unidentified serovars showing different phenotype to this antibiotic. Among the 21 serovars, only 23.81% strains carried both genes (sul1, sul2). For the sul1 gene, 61.9% strains were presented while sul2 occupied at a lower rate than sul1 with the rate of 52.38%. The study is very interesting and useful to go more functional analysis in sulfonamide resistance.
Salmonella, sulfonamide resistance, sul1 and sul2 genes, multiplex Polymerase chain reaction (m-PCR), serovars, raw meat, seafood
Food safety is one of the issues that we have been particularly concerned about for a long time. This is also one of significant factors for the sustainable development of socio-economic system, and public health in Vietnam. Therefore, the need for strengthened prevention, surveillance and management, of foodborne disease is extremely important at the present and future. This makes a significant contribution to economic development, poverty reduction and international integration. Salmonella spp. are the most widely distributed causes of foodborne outbreaks in human.1 They are the foodborne hazards that are responsible for the highest annual burden and the largest number of deaths globally. Globally, there were estimated that millions of people fall ill from eating contaminated food every year, resulting thousands of deaths. Salmonella spp. are found in everywhere (including water, soil, insects, animal feces, raw meats, poultry, seafood, and on factory and kitchen surfaces), distributing differently depending on geographic location.2,3 In Vietnam, more than one-third of meat samples from Ho Chi Minh City (HCMC) had Salmonella bacteria. The prevalence of Salmonella spp. in 25 g meat samples from HCMC was 71.8% (chicken), 70.7% (pork), and 62.2% (beef).
The survival of Salmonella not only depends on the water activity (aw) of the environment or food but also on other factors such as matrix composition and storage temperature. The main causes of this bacterium contamination in low-moisture foods are poor sanitation practices, substandard facilities, equipment design, and improper maintenance.4 The rise of Salmonella antibiotic resistance in food has been a global concern in recent decades because of the overuse of antibiotics for treating human and animal diseases. The inappropriate use of antibiotics in humans and animals make a great pressure on selection, leading to the presence and spread of antibiotics-resistant bacteria strains. Approximately 30 – 90% of the used antibiotics are not metabolized and exist in the environment.5 The habit of antibiotic usage in livestock production, prevention from diseases for decades is one of the main causes of increasing occurrence of antibiotic resistant Salmonella spp.6,7 In Vietnam, antimicrobials are often found in both commercial pig and poultry rations. A study estimated in-feed consumption of antimicrobials extrapolated from a retail survey of commercial feeds in 77 mg per kg of chicken produced.8 A study of medium-sized chicken farms estimated that chickens consumed 57 mg per kg of animal produced.9 However, that study was based on a small sample of 6 farms. Consequently, it is difficult to control infections, prevention and illness treatment, which gives rise to the loss of human, economy and brings about serious consequences to the society.10-13 The emergence of antibiotic resistance in bacteria raises the global problems in public health. Although there were many researches on Salmonella using the natural extracts to improve immunology and anti Salmonella activities,14-18 the traditional antibiotics were still used. One of antibiotic group has a high rate is sulfonamides. These antibiotics have bacteriostatic activity and the mechanism of action involved in the synthesis of tetrahydrofolic acid. Sulfonamide inhibits dihyrdropteroate synthetase while trimethoprim inhibits dihydrofolate reductase.19 The resistance of Salmonella to sulfonamides is due to the sul gene family. The sul1, sul2 and sul3 genes are three main genes in which the most common sul1 gene is found in the majority of serovars such as Salmonella Enteritidis, Salmonella Hadar, Salmonella Heidelberg, Salmonella Orion, Salmonella Rissen, Salmonella Agona, Salmonella Albany, Salmonella Derby, Salmonella Djugu and Salmonella Typhimurium. The sul1 gene is located on translocation genetic elements such as class I integrons or on the plasmids, however, the sul2 gene is located on the plasmids. Some of the antibiotic resistance genes of this group have been found to be dhfr and dfr located closely to sul1 and sul3 in an integron, plasmids or islands on the Salmonella genome.20
As the above state, the study aimed to evaluate the sulfonamide resistance characteristics of Salmonella strains isolated from raw meat and seafood samples at conventional markets in Ho Chi Minh City (Vietnam). The results would contribute to the science about some antibiotic resistance characteristics in Salmonella spp. isolated from food in Vietnam as well as the world.
Sample collection and preservation
228 raw meat samples (pork: 76, beef: 76, chicken: 76) and 301 raw seafood samples (fish: 199, shrimp: 67, squid: 35) were collected eventually at conventional markets in the districts of Ho Chi Minh City. 150 g -1.500 g of each sample were collected for the study. The samples were transported aseptically to the laboratory as soon as possible to be immediately analyzed. It should not let longer than 12 hours.
Salmonella spp. isolation and identification
Salmonella spp. isolation was based on ISO 6579-1:2017. To enrich bacteria for isolation, 25 g samples were placed in sterile bags (BagPage 400 mL, Interscience), added 225 mL buffer peptone water (BPW). Homogenization was performed using Stomacher method (Seward) at 230 rpm/30 seconds and incubated at 37°C in 18 ± 2 hours. Then, bacteria from cultures were spread on the selective rappaport vassiliadis medium supplied with soya (RV) and kauffmann tetrathionate novobiocin (MKTTn), then were let at 41.5°C in 24 ± 3 hours and 37°C in 24 ± 3 hours, respectively. The pure colonies were spread on xylose lysine deoxycholate (XLD) agar and mannitol lysine crystal violet brilliant green agar (MLCB) before incubated at 37°C in 24 ± 3 hours. The suspected colonies were picked up for checking Salmonella spp. by PCR using invA gene. The protocol was followed as the previous study.21 The same strains that showed many serovars with the sulfonamide resistance were used to the detect sul1 and sul2 by PCR to clarify or find out more whether sul1, sul2 or both could affect the resistance as many previous studies concluded before.
Evaluation of sulfamethoxazole susceptibility
Sterile antibiotic disks that had 6 mm-diameter were immersed in sulfonamide (Oxoid/10 μg/mL). Inhibition zones were recorded and evaluated according to the guidance of CLSI 2018 (M100-S28).22
Resistant gene detection by multiplex PCR (m-PCR)
Multiplex PCR ingredients included AmpliTaq Gold 2 UI; 0.2 mM dNTP; 1.5 mM MgCl2; 1X buffer; 0.5 μL each primer (0.625 μM); 5 μL DNA template and de-ionized water up to 25 μL.
The PCR program was set up at 95°C/10 minutes for thermal cycle. Then, PCR was performed continuously for 30 cycles. One cycle included 95°C/30 seconds, 55°C/60 seconds and 72°C/60 seconds and 72°C/7 minutes.
PCR products were run on gel agarose 1.5% containing 1 µg/mL ethidium bromide in TBE. The duration was 35 – 40 minutes in 100V and 100 mA. The primer pairs were sul1 (forward: 5’ – TCACCGAGGACTCCTTCTTC – 3’, reverse: 5’ – CAGTCCGCCTCAGCAATATC – 3’) and sul2 (forward: 5’ – CCTGTTTCGTCCGACACAGA – 3’, reverse: 5’ – C1TGTTTCGTCCGACACAGA – 3’), giving the 331 bp and 435 bp PCR products, respectively.23
Salmonella spp. Isolation and determination
Analysis methods were based on the guidelines of ISO 6579-1:2017 for detection of Salmonella spp. in raw meat samples originated from markets illustrated in Table 1.
Table (1):
The percentage of Salmonella spp. detection in raw meat samples.
| Samples | Quantity | Positive | Negative | ||
|---|---|---|---|---|---|
| Quantity | Percentage (%) | Quantity | Percentage (%) | ||
| Pork | 76 | 69 | 90.79 | 7 | 9.21 |
| Beef | 76 | 33 | 43.42 | 43 | 56.58 |
| Chicken | 76 | 59 | 77.63 | 17 | 22.37 |
| Total | 228 | 161 | 70.61 | 67 | 29.39 |
The suspected colonies of Salmonella spp. were confirmed by PCR using invA gene as a target for detection. The incorporated invA gene results revealed that among collected meat samples, there were 161 samples infected by Salmonella spp. occupying 70.61% (161/228). Pork and chicken had the approximate proportions at 90.79% (69/76) and 77.63% (59/76). The lowest proportion of contamination occurred in beef samples with the rate of 43.42% (33/76). Based on the results, there were high contamination in raw meat collected in traditional markets in Ho Chi Minh city (Vietnam). Similarly to the confirmation of suspected colonies of Salmonella spp. in meat samples, the contamination in raw seafood was also detetected and presented in Table 2.
Table (2):
The percentage of Salmonella spp. detection in raw seafood samples.
| Samples | Quantity | Positive | Negative | ||
|---|---|---|---|---|---|
| Quantity | Percentage (%) | Quantity | Percentage (%) | ||
| Fish | 199 | 80 | 40.20 | 119 | 59.80 |
| Shrimp | 67 | 5 | 7.46 | 62 | 92.54 |
| Squid | 35 | 6 | 17.14 | 29 | 82.86 |
| Total | 301 | 91 | 30.23 | 73 | 69.77 |
Antimicrobial susceptibility of Salmonella spp. strains
Salmonella spp. showed resistance to sulfonamide antibiotics in the study (Table 3 and 4). The resistance was seen in all cases of Salmonella isolated from raw meat and seafood. From Table 3, the ratios of sulfonamide resistance in pork and chicken were higher than beef samples.
Table (3):
Antimicrobial susceptibility of Salmonella spp. in raw meat.
| Pork | Chicken | Beef | Total | ||||
|---|---|---|---|---|---|---|---|
| Quantity | Percentage | Quantity | Percentage | Quantity | Percentage | Quantity | Percentage |
| 12 | 31.58 | 20 | 55.56 | 3 | 16.67 | 35 | 38.04 |
For the seafood samples, fish is a good condition for microbial growth. Therefore, there were many strains of Salmonella contaminated in fish as seen in Table 2. As the statement, Salmonella strains resist to sulfonamide more than other sources such as squid and shrimp (Table 4).
Table (4):
Antimicrobial susceptibility of Salmonella spp. in raw seafood.
| Fish | Squid | Shrimp | Total | ||||
|---|---|---|---|---|---|---|---|
| Quantity | Percentage | Quantity | Percentage | Quantity | Percentage | Quantity | Percentage |
| 14 | 32.56 | 2 | 40 | 1 | 10 | 17 | 29.31 |
The results suggested it should be careful when handling fishes to prevent contamination and resistance. In order to find out the resistance due to the same gene or other elements, the study screen some serovars and detect sulfonamide resistance genes.
Table (5):
The detection of sul1 and sul2 genes in serovars.
| Sources | Serovar | Phenotype | sul1 | sul2 | |
|---|---|---|---|---|---|
| Pork | 1 | S. Kentucky | S | + | + |
| 2 | S. Indiana | R | – | – | |
| Beef | 3 | S. Infantis | R | + | – |
| Chicken | 4 | S. Agona | R | – | + |
| 5 | S. Infantis | R | + | – | |
| 6 | S. Potsdam | R | – | + | |
| 7 | S. Kentucky | R | + | – | |
| 8 | S. Kentucky | R | + | – | |
| Fish | 9 | S. Kentucky | R | – | + |
| 10 | S. Saintpaul | R | + | + | |
| 11 | S. Braenderup | R | + | + | |
| 12 | S. Kentucky | R | + | + | |
| 13 | S. Kentucky | R | + | – | |
| 14 | S. Potsdam | R | – | + | |
| 15 | S. Kentucky | R | – | + | |
| 16 | S. Infantis | R | + | – | |
| 17 | S. Agona | R | – | – | |
| 18 | S. Infantis | S | + | – | |
| 19 | OMF:1,z6:UT | R | + | + | |
| 20 | 7:1,z6:UT | R | + | – | |
| 21 | S. Kentucky | S | – | + | |
S: Susceptible and R: Resistant
Detection of sulfonamide resistant genes
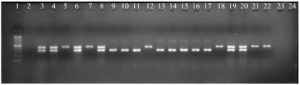
After checking sulfonamide resistance in Salmonella, the sulfonamide resistant genes were detected in serovars so that the phenotype and genotype involving resistance could be clarified partly. There were unidentified 2 serovars and 19 identified serovars of Salmonella including S. Kentucky (8), S. Infanis (4), S. Potsdam (2), S. Agona (2), S. Indiana (1), S. Saintpaul (1), S. Braenderup (1) gave the phenotype as presented in Table 5. To find out which genes involving the phenotype, PCR reaction was performed to identify sul1 and sul2. The results were presented in Fig. 1 and summarized in Table 5. Among the 19 identified serovars and 2 unidentified serovars, only 5 strains carried both genes (sul1, sul2) at the same time, accounting for 23.81% (5/21). For the sul1 gene, 13/21 strains were present (61.9%), sul2 was present at a lower rate than sul1 with the rate of 52.38% (11/21). Kentucky carried sul1 and sul2 with the highest frequency around 38.46% and 45.45%, respectively. In the study, sul1 was common in 19 serovars. However, the resistance was not only due to sul1 or sul2 involvement but also the other elements such as the plasmids, integrons or gene cassette location. Actually, the same serovar showed the phenotype and genotype difference such as there were 8 serovars belonging to S. Kentucky in which there were 2 serovars having antibiotic sensitivity while others were resistance (Fig. 1 and Table 5). Surprisingly, there was no sul1 or sul2 dectected in S. Indiana (pork) and S. Agona (fish) although they were resistant to sulfamethoxazole. The study will drive to more functional analysis in future.
From the results in Table 1, it might be that living conditions of these animals affect the situation of contamination in meat. The different dissemination in pork, chicken and beef might be due to the meat components or structure that can affect the contamination. More samples and mechanisms should be studied to aid for the contamination prevention.
The difference in Salmonella prevalence for different food groups could be caused by a variety of reasons. One of the main reasons is that the slaughter of livestock and poultry is usually carried out in unhygienic places such as the yard or slaughter tools are unsterilized knives, buckets and pots as well as the water sources are contaminated.
Furthermore, meat of cattle and poultry after slaughtering are transported by motorbike and uncovered by anything. Free sale in the market without being covered or stored for a long time along with customers who come into contact with the meat when selecting, make meat more susceptible to contaminate contaminated with Salmonella spp. by contacting with feces, skin, feathers, nails, meat cutters, trays, air, soil, water in the slaughterhouse, and from clothing, and limbs of slaughterhouse personnel. Additionally, according to Folster et al24, the difference in the prevalence of Salmonella spp. is possible that this bacterium is widespread in the environment and plays an important role in the infection between pathogens. Furthermore, Salmonella spp. distribution is very different depending on each country and region. As a result, different food sources may be contaminated with strains of Salmonella spp. with different rates.
The resistance was seen in all cases of Salmonella isolated from raw meat and seafood (Table 3 and 4). From the Table 3, the ratios of sulfonamide resistance in pork and chicken were higher than beef samples. Probably, beef was not used sulfonamide as casual as pork and chicken.
There were many strains of Salmonella contaminated in fish as seen in Table 2 with the highest resistant rate than other seafood sources such as squid and shrimp (Table 4). Probably, fish is a good condition for microbial growth that is the opportunity for antibiotic resistance. However, many mechanisms should be study more to understand deeply that is important to prevent bacterial contamination together with antibiotic resistance.
A high prevalence of sulfonamide resistance was observed in Salmonella isolated from animal products in many countries. One hundred percent sulfonamide resistance was reported in isolates originated from pigs in Thailand,25 Malaysia26 and Japan27; from broilers in Japan28 and Iran.29 High sulfonamide resistance rates were also reported in isolates originated from broilers in Japan (92.5% to 100%)30,31 and pigs (100%)25 and from a combination of poultry and swine isolates (68% to 70%) in Thailand32. A study in China found that isolates originated from broilers, ducks, geese and pigs exhibited the resistance rates of 93.8%, 20.0%, 16.7% and 52.5%, respectively.33 Resistance to sulfonamides is usually associated with the acquisition of resistance genes such as sul1, sul2 and sul3.34 Among Salmonella spp. isolates obtained from beef samples collected from retail markets in Vietnam in 2009, the resistance to sulfonamides was found in 39.7% (25/63 isolates) and 80.0% (20/25) were sul1 positive.35 Between 2007-2008, 110 isolates were obtained from meat (chicken, turkey and pork) from retail stores in Canada.36 There were 71% (78/110) showed resistance to sulphonamides. The sul1 gene was found in 5 isolates while sul2 expressed in 3 isolates and the sul3 gene was only found in one (pork) isolate. Among 88 Salmonella spp. strains isolated from retail meats and street foods in Malaysia, the sulfonamide resistant percentage was 63.6%.37 Among of these resistant strains, there were 32 strains those were positive for sul1 and sul2 while there were 5 strains carrying sul1, and 14 strains which were positive for sul2. These sulfonamide-resistance genes are commonly associated with mobile genetic elements, particularly class 1 integrons that can also relate to other antibiotic resistance. These factors facilitate the wide dissemination of sulfonamide resistance in bacteria.38
Salmonella spp. detected in raw meats and seafood collected in traditional markets in Ho Chi Minh city (Vietnam). The isolated Salmonella strains showed sulfonamide resistance in meat and seafood, but the high contamination was seen in chicken, fish and pork. The sul1 gene was common in the identified serovars but resistance was due to many elements besides sul genes such as the plasmids, integrons or gene cassette location. There was no sul1 or sul2 dectected in one serovar of isolated S. Indiana (pork) and one serovar of isolated S. Agona (fish) although they were resistant to sulfamethoxazole. This study brings out the interesting and useful points to explore the sulfonamide resistance in the future.
ACKNOWLEDGMENTS
We would like to thank scientific and technological tasks (Vietnam) for funding the study.
CONFLICT OF INTEREST
The authors declare that there is no conflict of interest.
AUTHORS’ CONTRIBUTION
THAV is proposal investigator, designed, performed experiment. THAV and NHKT did the data analysis and wrote the manuscript. NHKT did the supervision. CVH and HYH supported the research.
FUNDING
This study is supported by Scientific and Technological tasks contract No. 70/2019/HD-QPTKHCN (Vietnam).
ETHICS STATEMENT
Not applicable.
AVAILABILITY OF DATA
The data obtained in the study is available from the corresponding author and first author on reasonable request.
- Ferrari RG, Do Rosario DKA, Neto AC, Borges S, de Souza Figueiredo EE, Conte Junior CA. Worldwide Epidemiology of Salmonella Serovars in Animal-Based Foods: a Meta-analysis. Appl Environ Microbiol. 2019;85(14):e00591-19.
Crossref - Thomas Bintsis. Foodborne pathogens. AIMS Microbiol. 2017;3(3):529-563.
Crossref - Kareem SM, Al-Ezee AMM. Food poisoning (Salmonellosis). Res J Pharm Tech. 2020;13(2):529-532.
Crossref - Carrasco E, Morales-Rueda A, Garcia-Gimeno RM. Cross-contamination and recontamination by Salmonella in foods: a review. Food Res Int. 2012;45(2):545-556.
Crossref - Sarmah AK, Meyer MT, Boxall A. A global perspective on the use, sales, exposure pathways, occurrence, fate and effects of veterinary antibiotics (VAs) in the environment. Chemosphere. 2006;65(5):725-759.
Crossref - Deeb DN, Aldiab D, Sulaeman S. Effect of Storage Conditions on Bacteriological Growth in Infant Cereals. Res J Pharm Tech. 2021;14(2):667-672.
Crossref - Mousa ST, Saleh SS, Mohammed NT. Prevalence of Brucellosis and Salmonellosis in goats. Res J Pharm Tech. 2019;12(8):3690-3692.
Crossref - Cuong NV, Nhung NT, Nghia NH, et al. Antimicrobial consumption in medicated feeds in Vietnamese pig and poultry production. Ecohealth. 2016;13(3):490-498.
Crossref - Nhung NT, Nguyen HM, Cuong NV, et al. Use of colistin and other critical antimicrobials on pig and chicken farms in southern Vietnam and its association with resistance in commensal Escherichia coli bacteria. Appl Environ Microbiol. 2016;82(13):3727-3735.
Crossref - Osterblad M, Norrdahl K, Korpimaki E, Huovinen P. Antibiotic resistance. How wild are wild mammals? Nat. 2001;409(6816):37-38.
Crossref - Xi C, Zhang Y, Marrs CF, et al. Prevalence of antibiotic resistance in drinking water treatment and distribution systems. Appl Environ Microbiol. 2009;75(17):5714-5718.
Crossref - Swaroop VK, Mukherjee A, Sharma S, Osborne WJ. Isolation and characterization of drug resistant Salmonella typhi from sewage water. Res J Pharm Tech Feb. 2015;8(2):167-171.
Crossref - Bidhendi SM, Mojtahedi A, Alaei F. Identification and Serotyping of Salmonella Strains from poultry by PCR-RFLP in Shiraz, Iran. Res J Pharm Tech. 2018;11(6):2591-2594.
Crossref - Kalia P, Kumar NR, Harjai K. Preventive effect of honey bee propolis on Salmonella enterica serovar Typhimurium infected BALB/c mice: A Hematological Study. Res J Pharm Tech. 2020;13(7):3389-3393.
Crossref - Adak S, Chakraborty D, Maji HS, et al. Comparison of the antimicrobial activity of the phyto-constituents obtained from the stem bark and leaf extracts of Phyllanthus emblica L. against different strains of Staphylococcus aureus and Salmonella typhi. Res J Pharmacol Pharmacodynamics. 2018;10(2):53-60.
Crossref - Abdulazeem L, Jassim Y. The Effect of yolk immunoglobulin and heat killed Sallmonella Typhi on rabbits. Res J Pharm Tech. 2018;11(6):2503-2506.
Crossref - Shiyan S, Arifin A, Amriani A, Herlina, Pratiwi G. Immunostimulatory activity of ethanol extract from Calotropis gigantea L. Flower in rats against Salmonella typhimurium infection. Res J Pharm Tech. 2020;13(11):5244-5250.
- Fatmawati S, Dzariyani LR, Estri WMMR, Erryana M, Dewanti WT, Rifa’i M. Fermented ethanolic extract of moringa oleifera leaves with Lactobacillus plantarum FNCC 0137 as immunomodulators on Salmonella typhi infected mice. Res J Pharm Tech. 2020;13(12):5777-5782.
Crossref - Mascaretti OA. Bacteria versus antibacterial agents: an integrated approach. Washington, D.C. 2003.
Crossref - Antunes P, Machado J, Peixe L. Characterization of antimicrobial resistance and class 1 and 2 integrons in Salmonella enterica isolates from different sources in Portugal. J Antimicrob Chemother. 2006;58(2):297-304.
Crossref - Vu THA, Huu NN, Ly HD, Tu NHK. Detection of Salmonella spp. in feed and their antibiotic susceptibility for alternative therapy. J Appl Pharm Sci. 2016;6(5):18-21.
Crossref - CLSI. Performance Standards for Antimicrobial Susceptibility Testing. 28th ed. CLSI supplement M100. Wayne, PA: Clinical and Laboratory Standards Institute; 2018.
- Chen S, Zhao S, White DG, et al. Characterization of multiple antimicrobial resistant Salmonella serovars isolated from retail meats. Appl Environ Microbiol. 2004;70(1):1-7.
Crossref - Folster JP, Campbell D, Grass J, et al. Identification and Characterization of Multidrug-Resistant Salmonella enterica Serotype Albert Isolates in the United States. Antimicrob Agents Chemother. 2015;59(5):2774-2779.
Crossref - Sanpong P, Theeragool G, Wajjwalku W, Amavisit P. Characterization of multiple antimicrobial resistant Salmonella isolated from pig farms in Thailand. Kasetsart J Nat Sci. 2010;44(4):643-651.
- Choe DW, Hassan L, Loh TC. The prevalence of antimicrobial resistant Salmonella spp. and the risk factors associated with their occurrence in finisher pigs in Seberang Perai, Malaysia. Pertanika J Trop Agric Sci. 2011;34(2):303-310.
- Dahshan H, Chuma T, Shahada F, et al. Characterization of antibiotic resistance and the emergence of AmpC producing Salmonella Infantis from pigs. J Vet Med Sci. 2010;72(11):1437-1442.
Crossref - Shahada F, Sugiyama H, Chuma T, Sueyoshi M, Okamoto K. Genetic analysis of multidrug resistance and the clonal dissemination of beta-lactam resistance in Salmonella Infantis isolated from broilers. Vet Microbiol. 2010;140(1-2):136-141.
Crossref - Rahmani M, Peighambari SM, Svendsen CA, Cavaco LM, Agerso Y, Hendriksen RS. Molecular clonality and antimicrobial resistance in Salmonella enterica serovars Enteritidis and Infantis from broilers in three Northern regions of Iran. BMC Vet Res. 2013;9:66.
Crossref - Chuma T, Miyasako D, Dahshan H, et al. Chronological change of resistance to β-Lactams in Salmonella enterica serovar Infantis isolated from broilers in Japan. Front Microbiol. 2013;4:113.
Crossref - Shahada F, Chuma T, Tobata T, Okamoto K, Sueyoshi M, Takase K. Molecular epidemiology of antimicrobial resistance among Salmonella enterica serovar Infantis from poultry in Kagoshima, Japan. Int J Antimicrob Agents. 2006;28(4):302-307.
Crossref - Khemtong S, Chuanchuen R. Class 1 integrons and Salmonella genomic island 1 among Salmonella enterica isolated from poultry and swine. Microb Drug Resist. 2008;14(1):65-70.
Crossref - Pan ZM, Geng SZ, Zhou YQ, et al. Prevalence and antimicrobial resistance of Salmonella sp. isolated from domestic animals in Eastern China. J Anim Vet Adv. 2010;9(17):2290-2294.
Crossref - Antunes P, Machado J, Peixe L. Dissemination of sul3 containing elements linked to class 1 integrons with an unusual 3′ conserved sequence region among Salmonella isolates. Antimicrob Agents Chemother. 2007;51(4):1545-1548.
Crossref - Chau TT, Campbel JI, Galindo CM, et al. Antimicrobial drug resistance of Salmonella enterica serovar Typhi in asia and molecular mechanism of reduced susceptibility to the fluoroquinolones. Antimicrob Agents Chemother. 2007;51(12):4315-4323.
Crossref - Aslam M, Checkley SL, Avery B, et al. Phenotypic and genetic characterization of antimicrobial resistance in Salmonella serovars isolated from retail meats in Alberta, Canada. Food Microbiol. 2012;32(1):110-117.
Crossref - Thong KL, Modarressi S. Antimicrobial resistant genes associated with Salmonella from retail meats and street foods. Food Res Int. 2011;44(9):2641-2646.
Crossref - Antunes P, Machado J, Sousa JC, Peixe L. Dissemination of sulfonamide resistance genes (sul1, sul2, and sul3) in Portuguese Salmonella enterica strains and relation with integrons. Antimicrob Agents Chemother. 2005;49(2):836-839.
Crossref
© The Author(s) 2021. Open Access. This article is distributed under the terms of the Creative Commons Attribution 4.0 International License which permits unrestricted use, sharing, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.