Proteus mirabilis is a specific opportunistic pathogen of many infections including urinary tract infections (UTIs). Risk factors are linked with the acquisition of multidrug-resistant (MDR) to 3 or more classes of antimicrobials) strains. The resistance in extended-spectrum alpha-lactamase is rare, but the rising resistance in extended-spectrum beta-lactamase (ESBL) producing strains is a matter of concern. β-lactamases and antibiotic modifying enzymes mainly constitute the ESBLs resistance mechanism by hydrolyzing the antibiotics. Mutation or Porin loss could lead to the reduced permeability of antibiotics, enhanced efflux pump activity hindering the antibiotic access to the target site, antibiotic failure to bind at the target site because of the target modification, and lipopolysaccharide mutation causing the resistance against polymyxin antibiotics. This review aimed to explore various antimicrobial resistance mechanisms in Proteus mirabilis and their impact on public health status.
Proteus mirabilis, Antibiotic Resistance, Beta-lactams, Cephalosporins, Fluoroquinolones, Tetracyclines, Public Health
Proteus mirabilis, belonging to the class Gammaproteobacteria and family Enterobacteriaceae, is a well-known rod-shaped Gram-negative bacteria that swarm across the agar plates to form characteristic bullseye-shaped motility.1 P. mirabilis strains representing 18 different species have been isolated from various geographical locations.2 P. mirabilis is found in multiple environments such as sewage, soil, water, and especially in the gastrointestinal tract of animals and humans.3 The patients having long-term indwelling catheters or complicated UTIs also suffer from the infection of this opportunistic pathogen.4 Several human infections are associated with this bacterium such as infections of the gastrointestinal tract, wounds, eyes, and UTIs especially catheter-associated urinary tract infections (CAUTI).5 Renal damage and the formation of kidney and bladder stones (urolithiasis) further complicate the P. mirabilis related UTIs and CAUTIs.6 In the urinary tract, P. mirabilis mainly forms two types of crystals including apatite and struvite (CaPO4 and MgNH3PO4), which prevent urine flow.7 The symptoms of P. mirabilis infections such as bacteriuria, acute pyelonephritis, catheter occlusion, and fever could further complicate into bacteremia and sepsis.8 CAUTI is quite common in nursing homes whereas bacteremia mostly occurs following CAUTI or UTI. P. mirabilis associated sepsis and bacteremia comparatively lead to a higher mortality rate than other infections.9,10
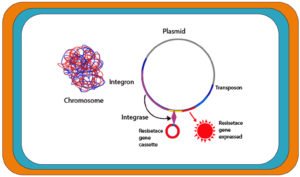
Antibiotics resistance exhibited in 48% P. mirabilis strains complicates the treatment of infections.11 The resistant strains are rising sharply and current therapies are becoming unable to cope with the situation. This scenario demands the urgent development of new antibiotic targets. Uropathogenic P. mirabilis might also be resistant to extended-spectrum beta-lactams, cephalosporins, fluoroquinolones, and aminoglycosides.11 P. mirabilis acquire genes encoding antimicrobial resistance via transferable plasmids, insertion sequences, transposons and integrons. Of these mobile genetic materials, integrons, which are not considered as transferable element, but are usually located on mobile plasmids, and play an important role in facilitating the horizontal gene transfer process of cassettes carrying resistance genes, i.e., integrons help incorporate gene cassettes encoding resistance to β-lactams, aminoglycoside and also plasmid-mediates quinolones resistance genes into recipient P. mirabilis cells (Figure 1).11 Integrons contain an integrase gene, attI (recombination site), and a promoter PC for the captured genes’ transcription.12 Integrons link with the mobile DNA elements (plasmids and transposons) to spread resistance determinants. The integrase gene sequence revealed five classes of integrons connected with resistance determinants.13 Integrons belonging to class 1 are mainly associated with MDR.14 Several antibiotic resistance determinants are present in Enterobacteriaceae strains, which are mediated by the integrons. Plasmid-mediated beta-lactamases gene coding and PMQR (quinolone resistance determinants) are complex integrons, which include ISCR1 and resistance genes through the duplication of the 3′ conserved region in addition to the variable part between 5′ and 3′ covered regions.15
Figure 1. Schematic diagram illustrating the role of integron in drug resistance acquisition in Proteus mirabilis
Antibiotic resistance mechanisms in Proteus mirabilis
Resistance to fluoroquinolones
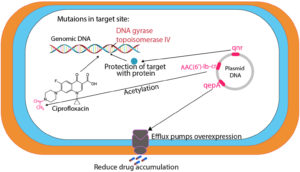
Fluoroquinolones are commonly used antibiotics in Western Europe, North America, and Japan to treat a broad range of infections including UTIs.16 European Antimicrobial Resistance Monitoring Network has reported significantly increased resistance to fluoroquinolones in Europe since 2001.17 Different fluoroquinolones resistance mechanisms have been identified including target enzyme modification in parC and parE encoded topoisomerase IV, gyrA and gyrB encoded DNA gyrase, and changes in the outer membrane to reduce drug accumulation through efflux pumps.18 Gram-negative organisms primarily target DNA gyrase whereas Gram-positive organisms target topoisomerase IV.19 gyrA is the essential target of fluoroquinolones in several Enterobacteriaceae species and its mutation is associated with fluoroquinolones resistance.20 Further mutations in DNA gyrase and topoisomerase IV cause higher resistance to fluoroquinolones.21 DNA sequence analyses have revealed the genetic characterization of mutations in clinical isolates. Quinolone resistance determining regions (QRDRs) have been reported to be extremely conserved.20 QRDRs linked with P. mirabilis resistance to fluoroquinolones exhibit substitutions in parC (S80) and gyrA (S83) whereas gyrB (S464) mutation could result in further higher fluoroquinolones resistance.22 QRDRs’ role in P. mirabilis resistance to fluoroquinolones is not well understood, which requires more data to elaborate its resistance mechanism. Levofloxacin-resistant P. mirabilis has been studied to investigate the fluoroquinolone resistance mechanism. The results depicted that parE (D420) and gyrA (E87) mutations are crucial for a higher resistance in P. mirabilis clinical isolates, which links ParE QRDRs and resistance to fluoroquinolones.18 Different spectroscopic techniques have been employed to identify new ciprofloxacin derivatives (hydroxamic acid, amide, and hydrazide) in addition to levofloxacin analogues. Some of these compounds exhibited significant efficacy against urease splitting P. mirabilis.23 Similar to the GyrA gene, the role of the ParC gene in ciprofloxacin resistance is also important. A couple of mutations in P. mirabilis GyrA and ParC genes could cause resistance to ciprofloxacin. Moreover, the percentage of quinolones resistance should be considered while aiming for other medical options. Therefore, drug susceptibility testing should be conducted for all patients with comparable infections before starting a specific medicine.24
The mutations in target enzymes (GyrB (Ser-464) and ParC (Ser-80) codons) and AcrAB efflux pump were investigated in relation to P. mirabilis resistance against fluoroquinolones. However, any relationship between mutation numbers in ParC, GyrA, and GyrB genes and the degree of P. mirabilis resistance to fluoroquinolone was not observed. The role of efflux pumps in fluoroquinolones resistance has been estimated by measuring the minimum inhibitory concentrations (MICs) through an efflux pump inhibitor CCCP. The CCCP (12.5 mM) was integrated with Mueller Hinton agar. Fifty isolates with uninfluenced fluoroquinolones susceptibility in response to CCCP were selected from a total of 100 isolates and characterized in terms of MICs and genotype for Levofloxacin (Figure 2).18
Resistance to tetracyclines
Several Gram-positive and Gram-negative bacterial infections are treated with tetracycline antibiotics but high tetracycline resistance rates in Enterobacteriaceae have been reported.25 Tetracycline resistance is presumed to be related to the efflux mechanism. The efflux resistance genes are often associated with the mobile elements such as the class A tetracycline resistance (tet) determinant that was the first to be identified from the RP1/Tn1721 system.26 Tigecycline (9-t-butylglycylamido derivative of minocycline) belongs to the novel class of tetracyclines that is used to treat Gram-negative bacteria.27 Klebsiella pneumoniae was the first tigecycline-resistant strain of Enterobacteriaceae with rpsJ mutation encoding Val57Leu on S10.28 Enterobacteriaceae tetracycline resistance is mostly considered to be linked with tet (A) to tet (E) gene determinants.29 P. mirabilis possesses a natural resistance against tetracycline that could be the main reason for its rising tolerance.30 The rise in acquired resistance of Enterobacteriaceae demands the development of new antibiotics to effectively treat bacterial infections.
AcrAB efflux pump, which is a member of the resistance-nodulation-division (RND) superfamily is found in Enterobacteriaceae. This efflux pump has been reported to be involved in P. mirabilis resistance to tigecycline.31 AcrAB provides intrinsic resistance to several structurally diverse lipophilic compounds, antibiotics, dyes, and inclusive detergents.32 P. mirabilis is a notable exemption to tigecycline activity, which normally exhibits 4 µg/ml MICs in tests. A typical clinical isolate was selected to identify the mechanism of decreased tigecycline sensitivity. Two independent transposon insertion mutants were isolated and inserted into the P. mirabilis chromosome. The results revealed a correlation between AcrRAB gene expression and observed MIC changes in various P. mirabilis strains. The classical tetracycline resistance determinants could not affect the tigecycline, however, AcrAB efflux pump identification in P. mirabilis explained its decreased susceptibility to tigecycline. Fortunately, the study did not report a direct threat of spreading tigecycline resistance.33 Nontoxic carbon nanoparticles could inhibit Gram-negative bacterial growth when integrated with tetracycline. This combination has generated tenfold higher activities against tetracycline-resistant bacteria as compared to solely tetracycline. The tetracycline-conjugated carbon nanoparticles could inhibit the efflux mechanism of bacteria. Tetracycline is supposed to direct nanoparticles into efflux pumps to block and inhibit their normal functioning. Qin et al.34 have conducted a study to acquire tigecycline, tetracycline, and colistin-resistant P. mirabilis for NDM-1 Plasmid and further characterized PM58 isolate. Molecular investigation elaborated that the PM58 chromosome contains a novel Salmonella genomic island 1 and conjugative NDM-1 plasmid.34,35
Resistance to β-lactams
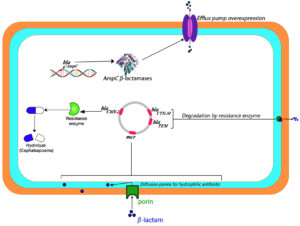
Lactamase genes are absent on the P. mirabilis chromosome whereas β-lactamase production includes AmpC β-lactamases and broad-spectrum β-lactamases.36 Gene cassette sequence analysis could not relate the resistance patterns and gene cassette content. The resistance patterns to beta-lactam antibiotics were more diverse than depicted by integrin-embedded cassettes. Gene screening revealed the presence of blaTEM genes in both genomes. blaTEM-2 encoding beta-lactamases are effective against early cephalosporins and penicillin. Thus, they could not be attributed to ESBL phenotype. Ye et al.37 have reported the involvement of another enzyme in ESBL-positive strains. P. mirabilis is known to possess CMY-2-like AmpC β-lactamases encoding genes, which facilitate to resist against cephamycins and cephalosporins. blaCMY sequence has been reported to conform with P. mirabilis blaCMY-15.
Ahn et al.38 have reported chromosome-borne genes coding for MY-15 in P. mirabilis strains in Poland. Colistin serves as a last-resort drug against MDR Gram-negative bacterial infections. P. mirabilis is naturally resistant to colistin due to the presence of the mcr genes, which are mediated by the plasmid. This bacterium can transmit these genes to other bacteria, which are susceptible to colistin.39 ESBL enzyme production confirms the wide-spectrum β-lactam antibiotic resistance. However, the presence of these genes does not necessarily generate phenotypical aspects of ESBLs as reported in several studies.40 Initially, the CTX-M gene appears in combination with the TEM gene but as the predominant gene spreads it replaces others. The selective pressure posed by the antibiotics misuse might provide a favorable environment for the diffusion of ESBLs among Enterobacteriaceae (Figure 3).41
Figure 3. Illustrative diagram of resistance mechanisms to β-lactams and cephalosporins exhibited by Proteus mirabilis
Resistance to cephalosporins
Cephalosporins are widely prescribed to treat respiratory, abdominal, and urinary infections. Such broad-scale utilization leads to significant selection pressure on Enterobacteriaceae members for resistance. Cephalosporins resistance is either associated with the higher chromosomal ‘AmpC’ b-lactamases production in Enterobacter spp. or transferable ESBLs.42 During a study in China, 2288 clinical isolates (non-repetitive) were collected from five laboratories in four cities to establish cefoselis epidemiological cut-off values (ECOFFs). Disc diffusion and broth micro-dilution methods based on European Committee on Antimicrobial Susceptibility Testing (EUCAST) guidelines were followed to determine MICs of Cefoselis and diameters of isolates inhibition zones. MIC ECOFFs were estimated through visual assessment and ECOF Finder software. Distributed cefoselis MICs ranged between 0.008 to >256 mg/L whereas MIC ECOFFs value was noted as 0.125 mg/L. P. mirabilis zone diameter ECOFF was observed as 26 mm.43 blaCTX, blaOXA-1, tetA, blaCTX-M, and sul1 genes were encoded for cephalosporins-resistance.44, 45
P. mirabilis isolates exhibited significant resistance (57.1%) to Cephalosporins (ceftazidime and cefotaxime).45 P. mirabilis strains are not frequently found in pneumonia patients but they can cause airborne acute infection of the lower airways (pneumonia) or infections that are transferred through the bloodstream from one body part to others.46 Cefepime, an antimicrobial agent that is administered to treat pneumonia patients, was found to be the most effective among six antibiotics used against severe Gram-negative bacterial infections.47 Several studies have confirmed the clinical efficacy of cefepime against drug-resistant organisms.48 However, the efficacy of ceftazidime has reduced over the past decade because of the extraordinary rise in microbial resistance.49 Cefotaxime is used against both types of bacteria (Gram-positive and Gram-negative) but P. mirabilis resistance to cefotaxime has been reported.50 Cefuroxime was found to display better efficiency against rod-shaped Gram-negative bacteria than cephalosporins (first-generation).51 have concluded that carbapenemase genes were not involved in the development of resistance in cephalosporin-resistant strains. The mutations in porin and protein of the outer membrane leading to low antibiotic permeability might have contributed to the resistance of cephalosporin-resistant strains.52 ESBL confirmatory tests revealed that 15 out of 50 cephalosporin-resistant Enterobacteriaceae were ESBL negative depicting that these strains might have acquired cephalosporin resistance via other mechanisms.53 The fosA gene mediated by plasmid could be transferred amidst Enterobacteriaceae species and fosA3 has been reported in 90% of E. coli isolates, which produces ESBL to resist fosfomycin (FOM).54,55 parC, gyrA, and fosA3 mutations might induce resistance to quinolone and further lead to high cross-resistance against fosfomycin (FOM), levofloxacin (LVX), and cephalosporin in UTI-causing bacteria. Ishii et al.56 have reported considerable gyrA and parC mutations based cross resistance of UTI causing bacteria to levofloxacin whereas the presence of fosA3 was linked to fosfomycin resistance. (Figure 3).56
Resistance to aminoglycosides
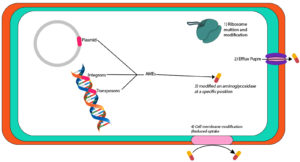
Broad-spectrum aminoglycosides antibiotics are primarily produced through Actinomyces species to treat Gram-negative and Gram-positive bacteria.57 Aminoglycosides have served as successful antibiotics but the resistance and toxicity aspects have hindered their application.58 However, they can still be efficiently used to counter MDR bacterial species.57 Mechanisms of aminoglycoside resistance and aminoglycoside-modifying enzymes (AMEs) have been detected frequently in bacteria.59 AMEs initiate resistance by changing aminoglycoside molecules at specific positions. Based on the modifications, these enzymes are known as aminoglycoside acetyltransferases (AACs), phosphotransferases (APHs), nucleotidyltransferases, and adenyltransferases (ANTs).60 The mobile agents such as plasmids, integrons, or transposons carry the AME coding genes, which often integrate with other resistance mechanisms.57 Recently, 16S ribosomal RNA (rRNA) methyltransferases have been used to code the aminoglycoside-resistance mechanism as these enzymes contain an aminoglycoside linking site in the ribosome to produce higher resistance against all clinically available aminoglycosides.61 Sometimes, the isolates already containing β-lactamases or Metallo-β-Lactamase (MBLs) carry 16S rRNA methyltransferases encoding genes.62 Alteration of membrane protein and ribosome, and raised efflux could be the other mechanisms of aminoglycoside resistance. However, these mechanisms are less spread as compared to AMEs.63 Plazomicin aminoglycoside (semi-synthetic) is obtained from sisomicin. The modifications in the plazomicin molecule structure make it resistant to AMEs-based alterations.64
Carbapenems are highly effective antimicrobial agents to cure hospital-acquired infections (HAIs). However, the development of carbapenemases (GES, VIM, KPC, IMP, OXA-48, and NDM) based resistance has limited their utility.65 Carbapenemases encoding genes are commonly found in plasmids and they might also contain AMEs encoding genes.66 AMEs-based enzymatic inefficiency is common aminoglycosides resistance mechanism followed by 16S rRNA methylation that also imparts significantly higher resistance against gentamicin, tobramycin, and amikacin.67 The studies have reported multiple isolates harbouring blaKPC-2, blaNDM-1, and AMEs encoding genes. The literature depicts the Klebsiella pneumoniae carbapenemase (KPC) insistence over the years that led to the emergence of a new carbapenemase known as NDM. The relationship of blaKPc-2 and blaNDM-1 genes and their association with AME genes in P. mirabilis isolates has been described (Figure 4). This association demonstrates a fast P. mirabilis evolution to obtain and preserve different genes, which urgently require further in-depth elaboration.68
Status of Proteus mirabilis resistance to various antimicrobial agents
The first report of ESBL-based resistance in Proteus species emerged in 1987.69 P. mirabilis isolates capable of producing ESBL are now more frequently detected in clinical settings. A study in France (1988 to 1990) revealed the presence of only 0.8% ESBL producing P. mirabilis strains, which has increased up to 6.9% and 9.5% in France and the USA, respectively.70-72 The isolation of ESBL-producing P. mirabilis strains reached 8.8% during 1997–1999.73 An Italian survey in 1999 ranked P. mirabilis as the second-highest ESBL producer in Enterobacteriaceae.74 In France, urine samples of 3340 patients were found positive for P. mirabilis from 1997 to 2002 whereas 45 (1.3%) patients were infected with extended-spectrum b-lactamases producing P. mirabilis.75 In Japan, 45.6% of the P. mirabilis strains were found to produce ESBL during 2009-2010.76 European Committee on Antimicrobial Susceptibility Testing revealed that74% of isolates were resistant to penicillin in 2010 whereas 1.23% of P. mirabilis strains were resistant to third-generation cephalosporins.77,78 In 2019, 8.4% of P. mirabilis isolates were noted to be resistant to various antibiotics such as ciprofloxacin, amoxicillin, gentamicin, amoxicillin/clavulanic acid, and cefotaxime. 28.6% of these isolates possessed ESBL genotype (blaCTX-M-2) whereas 71.4% had AmpC/ESBL genotype (blaCMY-2/blaTEM-1).79 Recently (i.e. 2020), 37% of strains produced ESBLs and all ESBLs-producing isolates contained blaTEM. These isolates were susceptible to cefotaxime/clavulanic, cefoxitin, and imipenem.80
Levofloxacine (LVX) resistance average has gradually increased between 2000 and 2005 and a continuous high prevalence (17.5%) has been reported in Europe and Japan since 2004.81,82 Similarly, a high spreading rate (37.0%) of cefotaxime (CTX)-resistant P. mirabilis strain was also noted in 2004. The rise in CTX-resistant P. mirabilis up to 45.6% is comparable to that reported in Japan between 2009 and 2010,76 whereas the spread of FQ-resistant P. mirabilis strain increased to 17.5% in Japan.18 In 2014, the ciprofloxacin resistance in uncomplicated UTIs in some European countries was reported as Germany (20.3%), France (4.8%), Sweden (7.3%), Spain (30.8%), and the UK (15.3%).83 P. mirabilis resistance rate against a novel antimicrobial agent glycylcycline reached up to 13% in Germany in 201647,51 whereas P. mirabilis resistance to imipenem (3.6%) and meropenem (4%) has also been reported in Iran.84 Similarly, decreased efficacy of imipenem (61.5%, 90.9) and ceftazidime-avibactam (72.7%, 93.8%) has been noticed in Canada for ESBL as compared to non-ESBL-producing Enterobacteriaceae in 2015. The situation has led to the lower response of complicated UTI patients to imipenem and ceftazidime-avibactam.85
Problems and Future Concerns
The ability of Proteus mirabilis to colonize and form crystalline multidrug-resistant (MDR) biofilms is a major reason for recurrent CAUTIs.86 Multidrug resistance (MDR) in the clinical isolates P. mirabilis is leading to public health anxiety and serious wildlife implications. Therefore, wildlife’s role in spreading resistance to antimicrobials has become a main topic of interest for researchers.87 Several studies have been conducted to understand the P. mirabilis ability to produce swarm cells but it remains unclarified. Peng et al.88 have reported that the swarming migration of the P. mirabilis strain is a rare feature.88 A recent study has revealed the appearance of infectious diseases and the mcr-1gene (colistin-resistant) in MDR Enterobacteriaceae in the Syrian refugee camps’ sewage water.89 These findings further elevate concerns about the health and sanitary conditions in Syrian camps. Similarly, mcr-1 gene has been detected in P. mirabilis samples collected from sewer and domestic waters of Lebanon’s war refugee camps. These results are alarming as P. mirabilis association with healthcare and community infections has already been established. Furthermore, the mcr gene encodes colistin resistance that serves as a last-resort antibiotic to treat complex Gram-negative bacterial infections.39
ACKNOWLEDGMENTS
None.
CONFLICT OF INTEREST
The authors declare that there is no conflict of interest.
AUTHORS’ CONTRIBUTION
All authors listed have made a substantial, direct, and intellectual contribution to the work, and approved it for publication.
FUNDING
None.
DATA AVAILABILITY
All datasets generated or analyzed during this study are included in the manuscript.
ETHICS STATEMENT
This article does not contain any studies with human participants or animals performed by any of the authors.
- Adeolu M, Alnajar S, Naushad S, Gupta RS. Genome-based phylogeny and taxonomy of the ‘Enterobacteriales‘: proposal for Enterobacterales ord. nov. divided into the families Enterobacteriaceae, Erwiniaceae fam. nov., Pectobacteriaceae fam. nov., Yersiniaceae fam. nov., Hafniaceae fam. nov., Morganellaceae fam. nov., and Budviciaceae fam. nov. Int J Syst Evol Microbiol. 2016;66(12):5575-5599.
Crossref - Zloch M, Maslak E, Kupczyk W, Jackowski M, Pomastowski P, Buszewski B. Culturomics Approach to Identify Diabetic Foot Infection Bacteria. Int J Molec Sci. 2021;22(17):9574.
Crossref - Drzewiecka D. Significance and roles of Proteus spp. bacteria in natural environments. Microb Ecol. 2016;72:741-758.
Crossref - Yuan F, Huang Z, Yang T, et al. Pathogenesis of Proteus mirabilis in Catheter-Associated Urinary Tract Infections. Urol Int. 2021;105:354-361.
Crossref - Armbruster CE, Prenovost K, Mobley HL, Mody L. How often do clinically diagnosed catheter-associated urinary tract infections in nursing homes meet standardized criteria? J Am Geriat Soc. 2017;65(2):395-401.
Crossref - Wasfi R, Hamed SM, Amer MA, Fahmy LI. Proteus mirabilis biofilm: development and therapeutic strategies. Front Cell Infect Microbiol. 2020;10:414.
Crossref - Durgadevi R, Abirami G, Alexpandi R, et al. Explication of the potential of 2-hydroxy-4-methoxybenzaldehyde in hampering uropathogenic Proteus mirabilis crystalline biofilm and virulence. Front Microbiol. 2019;2804.
Crossref - Schaffer JN, Pearson MM. Proteus mirabilis and urinary tract infections. In: Mulvey MA, Klummp DJ, Stapleton AE (eds.), Urinary Tract Infections: Molecular Pathogenesis and Clinical Management 2nd edition, Wiley, New York. 2017;17:383-433.
Crossref - Daniels KR, Lee GC, Frei CR. Trends in catheter-associated urinary tract infections among a national cohort of hospitalized adults, 2001-2010. Am J Infect Control. 2014;42(1):17-22.
Crossref - Armbruster CE, Mobley HL, Pearson MM. Pathogenesis of Proteus mirabilis infection. EcoSal Plus. 2018;8(1).
https://doi.org/10.1128/ecosalplus.ESP-0009-2017 - Girlich D, Bonnin RA, Dortet L, Naas T. Genetics of acquired antibiotic resistance genes in Proteus spp. Front Microbiol. 2020;11:256.
Crossref - Jove T, Da Re S, Tabesse A, Gassama-Sow A, Ploy MC. Gene expression in class 2 integrons is SOS-independent and involves two Pc promoters. Front Microbiol. 2017;8:1499.
Crossref - Partridge SR, Kwong SM, Firth N, Jensen SO. Mobile genetic elements associated with antimicrobial resistance. Clin Microbiol Rev. 2018;31(4):e00088-17.
Crossref - Kargar M, Mohammadalipour Z, Doosti A, Lorzadeh S, Japoni-Nejad A. High prevalence of class 1 to 3 integrons among multidrug-resistant diarrheagenic Escherichia coli in southwest of Iran. Osong Public Health Res Perspect. 2014;5(4):193-198.
Crossref - Szabo O, Gulyas D, Szabo N, Kristof K, Kocsis B, Szabo D. Plasmid-mediated quinolone resistance determinants in Enterobacteriaceae from urine clinical samples. Acta Microbiol Immunol Hung. 2018;65(3):255-265.
Crossref - Versporten A, Zarb P, Caniaux I, et al. Antimicrobial consumption and resistance in adult hospital inpatients in 53 countries: results of an internet-based global point prevalence survey. Lancet Glob Health. 2018;6(6):e619-e629.
Crossref - EARS-Net. Antimicrobial Resistance Surveillance in Europe 2017. Annual Report of the European Antimicrobial Resistance Surveillance Network (EARS-Net), ECDC, Stockholm. 2018.
- Nakano R, Nakano A, Abe M, et al. Prevalence and mechanism of fluoroquinolone resistance in clinical isolates of Proteus mirabilis in Japan. Heliyon. 2019;5(3):e01291.
Crossref - Yimin H, Houguang S, Mingwei Z, et al. Discovery of pyrido [2, 3-b] indole derivatives with Gram-negative activity targeting both DNA gyrase and topoisomerase IV. J Med Chem. 2020;63(17):9623-9649.
Crossref - Varughese LR, Rajpoot M, Goyal S, Mehra R, Chhokar V, Beniwal V. Analytical profiling of mutations in quinolone resistance determining region of gyrA gene among UPEC. PLoS One. 2018;13:e0190729.
Crossref - Khademi F, Maarofi K, Arzanlou M, Peeri-Dogaheh H, Sahebkar A. Which missense mutations associated with DNA gyrase and topoisomerase IV are involved in Pseudomonas aeruginosa clinical isolates resistance to ciprofloxacin in Ardabil? Gene Rep. 2021;24:101211.
Crossref - Piekarska K, Wolkowicz T, Zacharczuk K, et al. Co-existence of plasmid-mediated quinolone resistance determinants and mutations in gyrA and parC among fluoroquinolone-resistant clinical Enterobacteriaceae isolated in a tertiary hospital in Warsaw, Poland. Int J Antimicrob Agents. 2015;45(3):238-243.
Crossref - Abdullah MA, Abuo-Rahma GEDA, Abdelhafez ESM, Hassan HA, Abd El-Baky RM. Design, synthesis, molecular docking, anti-Proteus mirabilis and urease inhibition of new fluoroquinolone carboxylic acid derivatives. Bioorg Chem. 2017;70:1-11.
Crossref - Abdelkreem RH, Yousuf AM, Elmekki MA, Elhassan MM. DNA gyrase and topoisomerase IV mutations and their effect on quinolones resistant Proteus mirabilis among UTIs patients. Pak J Med Sci. 2020;36(6):1234.
Crossref - Grossman TH. Tetracycline antibiotics and resistance. Cold Spring Harb Perspect Med. 2016;6(4):a025387.
Crossref - Wang S, Gao X, Gao Y,et al. Tetracycline resistance genes identified from distinct soil environments in China by functional metagenomics. Front microbial. 2017;8:1406.
Crossref - Zhang R, Dong N, Shen Z, et al. Epidemiological and phylogenetic analysis reveals Flavobacteriaceae as potential ancestral source of tigecycline resistance gene tet (X). Nat Commun. 2020;11(1):1-13.
Crossref - Villa L, Feudi C, Fortini D, Garcia-Fernandez A, Carattoli A. Genomics of KPC-producing Klebsiella pneumoniae sequence type 512 clone highlights the role of RamR and ribosomal S10 protein mutations in conferring tigecycline resistance. Antimicrob Agents Chemother. 2014;58(3):1707-1712.
Crossref - Kuleshov KV, Pavlova AS, Shedko ED, et al. Mobile colistin resistance genetic determinants of non-typhoid Salmonella enterica isolates from Russia. Microorganisms. 2021;9(12):2515.
Crossref - Mirzaei A, Esfahani BN, Raz A, Ghanadian M, Moghim S. From the Urinary Catheter to the Prevalence of Three Classes of Integrons, β-Lactamase Genes, and Differences in Antimicrobial Susceptibility of Proteus mirabilis and Clonal Relatedness with Rep-PCR. Biomed Res Int. 2021;2021:9952769.
Crossref - Lv L, Wan M, Wang C, et al. Emergence of a plasmid-encoded resistance-nodulation-division efflux pump conferring resistance to multiple drugs, including tigecycline, in Klebsiella pneumoniae. MBio. 2020;11(2):e02930-19.
Crossref - Zhaowei J. A new high-throughput assay for quantification of antibiotic penetration in gram-negative bacterial cells. Syracuse University. 2020. https://surface.syr.edu/thesis/439
- van der Putten BC, Remondini D, Pasquini G, Janes VA, Matamoros S, Schultsz C. Quantifying the contribution of four resistance mechanisms to ciprofloxacin MIC in Escherichia coli: a systematic review. J Antimicrob Chemother. 2019;74(2):298-310.
Crossref - Qin S, Qi H, Zhang Q, et al. Emergence of extensively drug-resistant Proteus mirabilis harboring a conjugative NDM-1 plasmid and a novel Salmonella genomic island 1 variant, SGI1-Z. Antimicrob Agents Chemother. 2015;59(10):6601-6604.
Crossref - Kim TH, Raiz A, Unni AD, et al. Combating antibiotic-resistant gram-negative bacteria strains with tetracycline-conjugated carbon nanoparticles. Adv Biosyst. 2020;4(9):2000074.
Crossref - Yang JH, Sheng WH, Hsueh PR. Antimicrobial susceptibility and distribution of extended-spectrum β-lactamases, AmpC β-lactamases and carbapenemases among Proteus, Providencia and Morganella isolated from global hospitalised patients with intra-abdominal and urinary tract infections: Results of the Study for Monitoring Antimicrobial Resistance Trends (SMART), 2008-2011. J Glob Antimicrob Resist. 2020;22:398-407.
Crossref - Ye Q, Wu Q, Zhang S, et al. Characterization of extended-spectrum β-lactamase-producing Enterobacteriaceae from retail food in China. Front Microbiol. 2018;9:1709.
Crossref - Ahn JY, Ann HW, Jeon Y, et al. The impact of production of extended-spectrum β-lactamases on the 28-day mortality rate of patients with Proteus mirabilis bacteremia in Korea. BMC Infect Dis. 2017;17(1):1-10.
Crossref - Hmede Z, Kassem II. First report of the plasmid-borne colistin resistance gene (mcr-1) in Proteus mirabilis isolated from a toddler in non-clinical settings. ID Cases. 2019;18:e00651.
Crossref - Tseng BS, Zhang W, Harrison JJ, et al. Parsek MR. The extracellular matrix protects Pseudomonas aeruginosa biofilms by limiting the penetration of tobramycin. Environ Microbiol. 2013;15(10):2865-2878.
Crossref - Musa HA, Osman MA, Abdelaziz YH, Mohamed S, Ibrahim-Saeed M. Distribution of extended-spectrum beta-lactamase TEM and CTX-M resistance genes among Proteus species isolated in Sudan. Vacci Monitor. 2019:28(2):80-84.
- Ku YH, Lee MF, Chuang YC, Yu WL. Detection of plasmid-mediated β-lactamase genes and emergence of a novel AmpC (CMH-1) in Enterobacter cloacae at a medical center in Southern Taiwan. J Clin Med. 2019;8(1):8.
Crossref - Li X, Jia P, Zhu Y, et al. Establishment of epidemiological cut-off values for cefoselis, a new fourth-generation cephalosporin, against Escherichia coli, Klebsiella pneumoniae, Enterobacter cloacae, Proteus mirabilis and Pseudomonas aeruginosa. J Antimicrob Chemother. 2021;76(10):2593-2599.
Crossref - Shi X, Lin Y, Qiu Y, et al. Comparative screening of digestion tract toxic genes in Proteus mirabilis. PLoS One. 2016;11(3):e0151873.
Crossref - Algammal AM, Hashem HR, Alfifi KJ, et al. atpD gene sequencing, multidrug resistance traits, virulence-determinants, and antimicrobial resistance genes of emerging XDR and MDR-Proteus mirabilis. Sci Rep. 2021;11(1):9476.
Crossref - Zhang J, Hoedt EC, Liu Q, et al. Elucidation of Proteus mirabilis as a key bacterium in Crohn’s disease inflammation. Gastroenterol. 2021;160(1):317-330.
Crossref - Yayan J, Ghebremedhin B, Rasche K. Cefepime shows good efficacy and no antibiotic resistance in pneumonia caused by Serratia marcescens and Proteus mirabilis-an observational study. BMC Pharmacol Toxicol. 2016;17:10.
Crossref - Thomson KS, AbdelGhani S, Snyder JW, Thomson GK. Activity of cefepime-zidebactam against multidrug-resistant (MDR) Gram-negative pathogens. Antibiotics (Basel). 2019;8(1):32.
Crossref - Hatfull GF, Dedrick RM, Schooley RT. Phage therapy for antibiotic-resistant bacterial infections. Ann Rev Med. 2022;73:197-211.
Crossref - Morris S, Cerceo E. Trends, epidemiology, and management of multi-drug resistant gram-negative bacterial infections in the hospitalized setting. Antibiotics. 2020;9(4):196.
Crossref - Bui T, Preuss CV. Cephalosporin. National Library for Medicine, Bookshelf ID: NBM551517. StatPearls Publishing LLC. 2022. https://www.ncbi.nlm.nih.gov/books/NBK551517/
- Ye Y, Xu L, Han Y, Chen Z, Liu C, Ming L. Mechanism for carbapenem resistance of clinical Enterobacteriaceae isolates. Exper Therapeut Med. 2018;15(1):1143-1149.
Crossref - Lau CHF, DeJong EN, Dussault F, et al. A penicillin-binding protein that can promote advanced-generation cephalosporin resistance and genome adaptation in the opportunistic pathogen Pseudomonas aeruginosa. Int J Antimicrob Agents. 2020;55(1):105896.
Crossref - Klontz EH, Tomich AD, Gunther S, et al. Structure and dynamics of FosA-mediated fosfomycin resistance in Klebsiella pneumoniae and Escherichia coli. Antimicrob Agents Chemother. 2017;61(11):e01572-17.X
Crossref - Cao XL, Shen H, Xu YY, et al. High prevalence of fosfomycin resistance gene fosA3 in blaCTX-M-harbouring Escherichia coli from urine in a Chinese tertiary hospital during 2010-2014. Epidemiol Infect. 2017;145(4):818-824.
Crossref - Ishii A, Shigemura K, Kitagawa K, et al. Cross-Resistance and the Mechanisms of Cephalosporin-Resistant Bacteria in Urinary Tract Infections Isolated in Indonesia. Curr Microbiol. 2021;78(5):1771-1777.
Crossref - Castanheira M, Deshpande LM, Woosley LN, Serio AW, Krause KM, Flamm RK. Activity of plazomicin compared with other aminoglycosides against isolates from European and adjacent countries, including Enterobacteriaceae molecularly characterized for aminoglycoside-modifying enzymes and other resistance mechanisms. J Antimicrob Chemother. 2018;73(12):3346-3354.
Crossref - Goodlet KJ, Benhalima FZ, Nailor MD. A systematic review of single-dose aminoglycoside therapy for urinary tract infection: is it time to resurrect an old strategy? Antimicrob Agents Chemother. 2019;63(1):e02165-18.
Crossref - Kloos, J. M. Horizontal transfer, selection and maintenance of antibiotic resistance determinants. The Arctic University of Norway, 2021, PhD Thesis.
- Zarate SG, De la Cruz Claure ML, Benito-Arenas R, Revuelta J, Santana AG, Bastida A. Overcoming aminoglycoside enzymatic resistance: design of novel antibiotics and inhibitors. Molecules. 2018;23(2):284.
Crossref - Doi Y, Wachino JI, Arakawa Y. Aminoglycoside resistance: the emergence of acquired 16S ribosomal RNA methyltransferases. Infect Dis Clin. 2016;30(2):523-537.
Crossref - Shrestha S, Tada T, Sherchan JB, et al. Highly multidrug-resistant Morganella morganii clinical isolates from Nepal co-producing NDM-type metallo-β-lactamases and the 16S rRNA methylase ArmA. J Med Microbiol. 2020;69(4):572-575.
Crossref - Peterson E, Kaur P. Antibiotic resistance mechanisms in bacteria: relationships between resistance determinants of antibiotic producers, environmental bacteria, and clinical pathogens. Front Microbiol. 2018;9:2928.
Crossref - Abdul-Mutakabbir JC, Kebriaei R, Jorgensen SC, Rybak MJ. Teaching an old class new tricks: a novel semi-synthetic aminoglycoside, plazomicin. Infect Dis Ther. 2019;8(2):155-170.
Crossref - Codjoe FS, Donkor ES. Carbapenem resistance: a review. Med Sci. 2018;6(1):1.
Crossref - Aires CAM, Pereira PS, de Araujo CFM, et al. Multiclonal expansion of Klebsiella pneumoniae isolates producing NDM-1 in Rio de Janeiro, Brazil. Antimicrob Agents Chemother. 2017;61(4):e01048-16.
Crossref - Kawai A, Suzuki M, Tsukamoto K, Minato Y, Doi Y. Functional and structural characterization of acquired 16s rRNA methyltransferase npmb1 conferring pan-aminoglycoside resistance. Antimicrob Agents Chemother. 2021;65(10):e01009-21.
Crossref - Firmo EF, Beltrao EMB, da Silva FRF, Alves LC, Brayner FA, Veras DL, Lopes ACS. Association of blaNDM-1 with blaKPC-2 and aminoglycoside-modifying enzyme genes among Klebsiella pneumoniae, Proteus mirabilis and Serratia marcescens clinical isolates in Brazil. J Glob Antimicrob Resist. 2020;21:255-261.
Crossref - Uzunovic S, Ibrahimagic A, Hodzic D, Bedenic B. Molecular epidemiology and antimicrobial susceptibility of AmpC-and/or extended-spectrum (ESBL) b-lactamase-producing Proteus spp. clinical isolates in Zenica-Doboj Canton, Bosnia and Herzegovina. Med Glas (Zenica). 2016;13(2):103-112.
Crossref - Sirot DL, Goldstein FW, Soussy CJ, et al. Resistance to cefotaxime and seven other beta-lactams in members of the family Enterobacteriaceae: a 3-year survey in France. Antimicrob Agents Chemother. 1992;36(8):1677-1681.
Crossref - De Champs C, Bonnet R, Sirot D, Chanal C, Sirot J. Clinical relevance of Proteus mirabilis in hospital patients: a two year survey. J Antimicrob Chemother. 2000;45(4):537-539.
Crossref - Saurina G, Quale JM, Manikal VM, Oydna E, Landman D. Antimicrobial resistance in Enterobacteriaceae in Brooklyn, NY: epidemiology and relation to antibiotic usage patterns. J Antimicrobial Chemother. 2000;45(6):895-898.
Crossref - Luzzaro F, Perilli M, Amicosante G, et al. Properties of multidrug-resistant, ESBL-producing Proteus mirabilis isolates and possible role of β-lactam/β-lactamase inhibitor combinations. Int J Antimicrob Agents. 2001;17(2):131-135.
Crossref - Spanu T, Luzzaro F, Perilli M, Toniolo A, Amicosante G, Fadda G. Resistance profile of Enterobacteriaceae showing synergy between clavulanate and β-lactams: an Italian survey. 10th European Congress of Clinical Microbiology and Infectious Diseases, Stockholm, Sweden. 2000.
- Biendo M, Thomas D, Laurans G, et al. Molecular diversity of Proteus mirabilis isolates producing extended-spectrum β-lactamases in a French university hospital. Clin Microbiol Infect. 2005;11(5):395-401.
Crossref - Kanayama A, Kobayashi I, Shibuya K. Distribution and antimicrobial susceptibility profile of extended-spectrum β-lactamase-producing Proteus mirabilis strains recently isolated in Japan. Int J Antimicrob Agents. 2015;45(2):113-118.
Crossref - European Committee on Antimicrobial Susceptibility Testing. Breakpoint tables for interpretation of MICs and zones diameters. EUCAST; 2010. http://www.eucast. org
- Mestrovic T, Lukic-Grlic A, Bogdan M, et al. Molecular Epidemiology of Cephalosporinases and Extended Spectrum β-Lactamases (ESBLs) in Proteus mirabilis Isolates From Croatia: Following the Spread of Resistance Determinants Between Long-Term Care Facilities and the Community. Open Forum Infect Dis. 2018;5(Suppl 1):S363.
Crossref - Boudjemaa H, Allem R, Fonkou MDM, el Houda Khennouchi NC, Kerkoud M. Molecular drivers of emerging multidrug resistance in Proteus mirabilis clinical isolates from Algeria. J Glob Antimicrob Resist. 2019;18:249-256.
Crossref - Han P, Luo L, Shahid M, et al. Prevalence, Genetic Diversity and Antimicrobial Resistance of Proteus mirabilis Isolated from Dogs Hospitalized in Beijing. Pak Vet J. 2020;40(1):61-66.
Crossref - Muratani T, Matsumoto T. Urinary tract infection caused by fluoroquinolone-and cephem-resistant Enterobacteriaceae. Int J Antimicrob Agents. 2006;28(Suppl 1):10-13.
Crossref - Sedlakova MH, Urbanek K, Vojtova V, Suchankova H, Imwensi P, Kolar M. Antibiotic consumption and its influence on the resistance in Enterobacteriaceae. BMC Res Notes. 2014;7:1-10.
Crossref - Kahlmeter G, Ahman J, Matuschek E. Antimicrobial resistance of Escherichia coli causing uncomplicated urinary tract infections: a European update for 2014 and comparison with 2000 and 2008. Infect Dis Ther. 2015;4(4):417-423.
Crossref - Sheykhsaran E, Baghi HB, Soroush MH, Ghotaslou R. An overview of tetracyclines and related resistance mechanisms. Rev Med Microbiol. 2019;30(1):69-75.
Crossref - Mendes RE, Castanheira M, Gasink L, et al. β-Lactamase characterization of Gram-negative pathogens recovered from patients enrolled in the phase 2 trials for ceftazidime-avibactam: clinical efficacies analyzed against subsets of molecularly characterized isolates. Antimicrob Agents Chemother. 2015;60(3):1328-1335.
Crossref - Esmael A, Abo-Elmaaty SA, Khafaga EM, Abdelrahman S, Hassan MG. Efficacy of three lytic bacteriophages for eradicating biofilms of multidrug-resistant Proteus mirabilis. Arch Virol. 2021;166(12):3311-3322.
Crossref - Kang Q, Wang X, Zhao J, et al. Multidrug-resistant Proteus mirabilis isolates carrying blaOXA-1 and blaNDM-1 from wildlife in China: increasing public health risk. Integr Zool. 2021;16(6):798-809.
Crossref - Peng L, Chen DQ, Jiang GM, et al. Transcriptome Analysis of Two Strains of Proteus mirabilis with Swarming Migration Deficiency Isolated from Patients with Urinary Tract Infection. Curr Microbiol. 2020;77(8):1381-1389.
Crossref - Isenring E, Fehr J, Gultekin N, Schlagenhauf P. Infectious disease profiles of Syrian and Eritrean migrants presenting in Europe: a systematic review. Travel Med Infect Dis. 2018:25:65-76.
Crossref
© The Author(s) 2022. Open Access. This article is distributed under the terms of the Creative Commons Attribution 4.0 International License which permits unrestricted use, sharing, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.