ISSN: 0973-7510
E-ISSN: 2581-690X
The emergence of bacterial infections caused by resistant strains poses a threat to the development of new antibiotics. The majority of antibiotics being produced has been accelerated through the finding of newly reported natural products, especially those originated and produced by biological sources. Endophytic fungi residing in medicinal plants may be regarded as potential sources and encourage the exploration of more plant species for their antimicrobial activity. Our current study reports on the assemblage of endophytic fungi that colonize the rhizomes, using Globba patens a representative of Zingiberaceous species from North Sumatra. Twenty-six fungal morphotypes were obtained and differentiated by their morphological features. Each isolate was tested against human pathogenic bacteria namely Staphylococcus aureus ATCC® 29213™, Methicillin-resistant S. aureus (MRSA) ATCC® 43300™, Escherichia coli ATCC® 25922™, and Enteropathogenic E. coli (EPEC) K11 in a dual culture assay. The results revealed that the majority of fungal isolates were strong antagonists against S. aureus and E. coli but not against MRSA and EPEC. Isolate Gp07 was the most potential fungus with a wide range of antibacterial activities and was subjected to further species-level identification based on its morphological characteristics and DNA sequence in the ITS-rDNA region. The isolate Gp07 was identified as Colletotrichum siamense, yet the presence of C. siamense in the rhizome of G. patens is not fully understood while possibly being characterized as the antibiotics-producing agent in the future.
Antibiotic resistance, Biocontrol, Colletotrichum siamense, Endophytic fungi, ITS-rDNA
The emergence and rapidly increasing incidence of bacterial resistance limited the discovery and development of antibiotics for treating infections. During the past two decades, there have been very numerous reports of untreatable infections caused by multidrug-resistant bacteria such as Methicillin-resistant Staphylococcus aureus (MRSA) and its resistant strains as also of serious infections by diarrheagenic Escherichia coli strains1,2. Ninety-one years after the discovery of Penicillin in 1929, scientists from each region of the world continue to explore and investigate the many sources of new natural products. Natural products are significant sources that deserve investigation as alternative sources of antibiotics and are commonly derived or synthesized by plants, bacteria, and fungi. One study of bioactive compounds discovered between 1981 and 2010 reported that half of the molecules discovered originated from natural sources3.
In most cases, fungi are known as notable producers of natural products. Furthermore, half of the novel natural products derived from fungal sources was from endophytic origins, with the most studies on their antimicrobials in specific to antibacterial activities4. Endophytic fungi are new sources for the discovery of bioactive compounds with potential development in biotechnology, although their true potential may not be fully investigated due respect of their unknown and unlimited host species5,6. Interaction of endophytic fungi with medicinal plants as their hosts has revealed some beneficial features such as promotion of plant growth, increase of stress resistances, and increased bioactive accumulation of molecules by the plants7. The medicinal properties of directly consumed or consumption of processed medicinal plants may be studied thoroughly, particularly their endophytic fungal colonization. This may broaden the scale of the discovery of natural products, especially in the tropical regions that are rich with many plant species.
Zingiberaceae is one of the most studied families of pharmacological plants growing in the tropical region including Indonesia for its bioactive compounds and some of its endophytic associates. Globba spp. are examples and the third-largest genus of Zingiberaceae distributed in Southeast Asia. It comprises about 100 species in that region8. Members of the genus Globba have been used in many ethnobotanical practices by the local communities of India, Malaysia, and Indonesia9. Although most ginger species are known to exhibit antimicrobial activities, the reports in this respect on the species of Globba are still limited10. Furthermore, information regarding the microbial associates, especially endophytic fungi colonizing the rhizomes of Globba species is even more limited and, therefore, merit investigation.
In North Sumatra, four species of Globba are found, namely, Globba aurantiaca, G. paniculata, G. patens, and G. pendula11. Our preliminary investigation on endophytic fungi began with G. pendula rhizome and some of the fungal morphotypes found in it exhibited potential antibacterial activities against S. aureus ATCC 2921312. Our study was undertaken to isolate endophytic fungi from the rhizomes of Globba patens and use the dual culture assay to test them for their antagonistic activities against human pathogenic bacteria. Screening of endophytic fungi from G. patens for antibacterial activities will provide the basis for the discovery and development of new antibiotic agents.
Plant materials
Globba patens Miq. was collected during an exploration in Sibolangit Forest located in Deli Serdang District, North Sumatra, Indonesia. The asymptomatic G. patens plants were uprooted from the intact soil and stored in sterile plastic bags, then transported to the laboratory at Universitas Sumatera Utara (Fig. 1). The plant samples were kept in cold temperature until further analysis. Duplicate samples were submitted to Herbarium Medanese for species authentication. Fungal isolation was performed within 48 h of sample collection.
Fig. 1. (a) Leaf and (b) inflorescence part of Globba patens Miq. collected from Sibolangit forest, North Sumatra
Isolation of endophytic fungi
The procedure of isolating endophytic fungi was based on a modified surface sterilization method12. Rhizomes were gently cleaned under running tap water and cut into segments 10 cm long. The segments were surface sterilized in 75% EtOH for 2 min, 5.3% NaOCl for 5 min, and 75% EtOH for 1 min followed by three washings in sterile distilled water, each for about 1 min. The sterilized rhizome segments were cut into 1–2-cm fragments and placed on Petri dishes (90 mm) containing Potato Dextrose Agar (PDA) supplemented with Chloramphenicol (0.01%, w/v). Plates were incubated at 25°C ± 3°C and monitored daily for any fungal colonies emerging from the rhizome fragments. Individual fungal colonies were picked from the edge of growth and transferred to new PDA plates. Stock cultures were made by sub-culturing of fungal colonies for maintenance and further analysis. Each isolate was differentiated based on the features of their colonies such as form, elevation, margin, upper-lower color, texture, concentric rings (+/-), and radial line (+/-). Each isolate was assigned a code and maintained for further analysis.
Antagonistic activity of endophytic fungi against human pathogenic bacteria
Each isolate was tested against four representative human pathogenic bacteria, namely, Staphylococcus aureus ATCC® 29213™, Methicilin-resistant S. aureus ATCC® 43300™, Escherichia coli ATCC® 25922™, and Enteropathogenic E. coli K11 using a modified dual culture method12,14. Three 6-mm mycelial plugs were cut from active-growing fungal cultures and placed on top of PDA + 1% yeast extract (w/v) that were previously seeded with 1–2×108 CFU/mL of bacterial suspensions. Plates were incubated at 37°C for 2 days. Clear zones surrounding mycelial plugs indicated the antagonistic activities and measured in millimeters (mm) using digital calipers.
Molecular identification of the antagonistic fungus
The potential antagonistic fungal isolate, Gp07 was identified to the genus level based on the sequence of the internal transcribed spacer (ITS) rDNA. For extracting DNA, the manufacturer’s prescribed Protocol provided with Wizard® Genomic DNA Purification Kit (United States) was followed. Polymerase chain reaction (PCR) was performed using universal primers for fungi i.e ITS1 (5’-CTTGGTCATTTAGAGGAAGTAA-3)15 and ITS4 (5’-TCCTCCGCTTATTGATATGC-3’)16. Each reaction was performed in a 40 µL final volume containing 12 µL nuclease-free water (NFW), 20 µL GoTaq DNA Polymerase solution, 2 µL of each primer, and 4 µL DNA template solution in an Eppendorf tube. The PCR was conditioned as 95°C for 3 min, followed by 35 cycles of 95°C for 45 sec, 55°C for 45 sec, 72°C for 45 sec, and a final extension at 72°C for 7 min. The PCR products were visualized on 1% agarose gels and delivered for sequencing (Macrogen, Inc., Singapore). Multiple sequence alignments were generated from MUSCLE feature provided in MEGA X17. Phylogenetic analysis of the sequences retrieved from BLAST results (http://blast.ncbi.nlm.nih.gov/) and the Gp07 sequence was constructed into a dendrogram using a Neighbor-Joining (NJ) clustering method.
Twenty-six fungal isolates were successfully recovered from the rhizomes of G. patens and differentiated based on the characteristics of their colonies (Table 1). The number of isolates found is higher than the 16 (sixteen) fungal isolates derived in the previous study of G. pendula12. Also, the number of isolates was more than in the cases of other genera that have been studied, such as Amomum, Elettaria, Etlingera, and Hedychium from North Sumatra18-21. The results indicated that the possible habit of endophytic fungi colonizing the root parts may be heterotrophic eukaryons. The roots or rhizomes of Zingiberaceae play an important role as reservoirs of metabolites and nutrition for initiating and promoting the metabolism of endophytic fungi. The great diversity of endophytic fungi in a plant species may be explained by the differences in spatio-temporal distribution of host plants across regions or the wide range of colonization niches occupied by certain rhizospheric fungi22. Also, plants of a species at different stages of life-cycle may harbor distinct endophytic fungal communities over time. Younger parts of plants tend to accumulate a higher concentration of bioactive and anti-herbivory compounds which may reduce the opportunity for colonization by endophytic fungi. On the contrary, an endophytic study of Dioscorea zingiberensis showed that the number of endophytic fungal isolates may be accompanied by the higher concentration of saponins accumulated in the rhizome23. Another study on Alpinia officinarum using a metagenomic approach revealed that there was a connection between the colonization by endophytic fungal communities and two types of accumulated chemicals, namely, volatile oils and galangin which constructed the whole plant-microbe interaction in the rhizome24. However, no such link has so far been discussed for other Zingiberaceous species, including Globba spp., which may open the possibility of further investigation in the future due to the metabolite-rich rhizomes and their connection to endophytic fungi colonization.
Table (1):
Morphological features of 26 endophytic fungal isolates from Globba patens Miq.
| Isolate code |
Form | Elevation | Margin | Color | Texture | Concentric ring (+/–) |
Radial line (+/–) |
|
|---|---|---|---|---|---|---|---|---|
| Upper | Lower | |||||||
| Gp01 | Circular | Flat | Entire | Black | Black | Smooth | + | – |
| Gp02 | Irregular | Flat | Undulate | Grey | Black | Rough | – | – |
| Gp03 | Filamentous | Flat | Filiform | Black | Black | Rough | – | – |
| Gp04 | Circular | Raised | Undulate | White | White | Dull | – | – |
| Gp05 | Circular | Raised | Filiform | Brown | Dark Brown | Cottony | – | – |
| Gp06 | Irregular | Raised | Undulate | White | White | Powdery | – | – |
| Gp07 | Filamentous | Raised | Entire | Greyish Brown | Light Brown | Cottony | + | – |
| Gp08 | Circular | Raised | Filiform | White | White | Cottony | – | – |
| Gp09 | Irregular | Flat | Undulate | White | White | Cottony | – | – |
| Gp10 | Irregular | Raised | Entire | White | White | Smooth | – | – |
| Gp11 | Circular | Raised | Filiform | White | White | Cottony | – | + |
| Gp12 | Circular | Raised | Entire | White | White | Dull | – | – |
| Gp13 | Irregular | Flat | Undulate | Tangerine | Brownish Grey | Smooth | – | – |
| Gp14 | Rhizoid | Flat | Lobate | Black | Black | Dull | – | – |
| Gp15 | Filamentous | Raised | Filiform | Grey | Black | Rough | – | – |
| Gp16 | Irregular | Flat | Filiform | Black | Grey | Powdery | + | – |
| Gp17 | Filamentous | Umbonate | Filiform | Green | Brownish Red | Smooth | + | + |
| Gp18 | Irregular | Flat | Filiform | Pinkish White | Brownish Red | Smooth | + | – |
| Gp19 | Irregular | Flat | Filiform | Grey | Grey | Smooth | + | – |
| Gp20 | Filamentous | Flat | Filiform | White | White | Cottony | – | – |
| Gp21 | Irregular | Flat | Filiform | Green | Green | Powdery | – | – |
| Gp22 | Filamentous | Curvy | Filiform | Black | Black | Dull | – | – |
| Gp23 | Irregular | Flat | Filiform | White | White | Cottony | + | – |
| Gp24 | Filamentous | Flat | Filiform | Purple | Purple | Smooth | – | – |
| Gp25 | Filamentous | Flat | Filiform | White | White | Smooth | – | – |
| Gp26 | Filamentous | Curvy | Filiform | Pinkish White | Brown | Smooth | + | – |
The endophytic fungal isolates were subjected to antagonistic test against human pathogenic bacteria using dual culture assay. The majority of isolates were antagonists of S. aureus ATCC 29213 (96%), followed by E. coli ATCC 25922 (73%), Enteropathogenic E. coli K11 (15%), and Methicilin resistant S. aureus ATCC 43300 (3%). Gp07 stood out as the most potential isolate that exhibits a wide range of activities antagonistic to all the tested pathogenic bacteria based on the diameter of the clear zone (mm). Also, the isolate Gp07 was the only isolate which exhibited inhibitory activities towards the growth of MRSA, which was the only drug-resistant strain used in this study. The antibacterial activity demonstrated by each fungal isolate was attributable to certain metabolites diffusible in agar plugs and not the mycoparasitism of the isolates. These results may lead to the identification of the anti-MRSA compound and the significance of Gp07 for its production through fermentation. Meanwhile, the discovery of antibacterial compounds, particularly those with special potential as anti-MRSA, has been reported for endophytic fungi from other sources. Dothideomycete sp. isolated from a Thai medicinal plant species, Tiliacora triandra showed potential antibacterial activities against both S. aureus ATCC 25923 and MRSA ATCC 33591 by producing Dothideomycetide A with minimum inhibitory concentration (MIC) from 128 to 256 µg/mL25. Pestalotia sp. isolated from a mangrove medicinal plant species in Bangladesh, Heritiera fomes, was reported to produce oxysporone and xylitol that have the anti-MRSA properties with MIC from 32 to 128 µg/mL26. The major portion of antibacterial components produced by Gp07 may be identified in the future for the possibility of finding new compounds to combat bacterial infections.
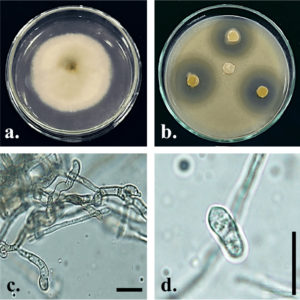
Fig. 2. Colletotrichum siamense Gp07. Upper (a) side of culture on PDA 7 days after inoculation; (b) antagonistic activity against S. aureus ATCC 29213 on PDA + 1% yeast extract (w/v) 2 days after incubation; (c) conidiogenous cell; (d) conidia; Bars = 10 µm.
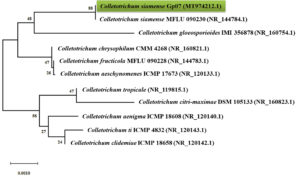
Fig. 3. Neighbor-joining (NJ) phylogram showing phylogenetic relationships between Colletotrichum siamense Gp07 and selected species of Colletotrichum based on ITS-rDNA sequences retrieved from GenBank. Phylogenetic tree was constructed based on the genetic distance using the Jukes-Cantor method with pairwise deletion of ambiguous positions and 1000× bootstrapping.
The identification of isolate Gp07 was based on its macroscopic and microscopic morphological features confirmed with a molecular identification in the region of ITS-rDNA (Fig. 2, 3). Based on the preliminary identification, the isolate Gp07 was preliminarily assigned as the member of Colletotrichum, for having the unique morphological characteristics, i.e, colony color, absence of setae, and cylindrical conidia27. However, some discrepancies in morphological features may exist due to the variation of the species and variations across the range of the host species. A phylogenetic tree was then constructed to reveal the species-level identification which assigned the Gp07 position as C. siamense, a fungal species described firstly from Thailand. However, no information is available so far about this species, especially the one extracted from Zingiberaceous species. Also, this is the first report from Indonesia on finding C. siamense as an endophytic fungus from G. patens. The ITS sequence of C. siamense Gp07 was then submitted to GenBank and allotted the accession number of MT974212.1. The majority of recent reports on this species are regarding its being found in new host species, mainly as a newly reported phytopathogenic fungus causing anthracnose on various horticultural plants grown in the Southeast Asia region28,29. Antibacterial or bioactive compound-producing Colletotrichum strains derived from a range of hosts and origins as endophytes have been documented. Colletotric acid produced by C. gloeosporioides from Artemisia mongolica in China was reported to inhibit a wide range of human pathogenic bacteria with MIC from 25 to 50 μg/mL30. Two novel compounds, apigenin-8-C-β-D-glucopyranoside, and 2-(hydroxymethylthio)ethanol were identified as produced by an endophytic Colletotrichum sp. from Chinese Ginkgo biloba through fermentation31. However, the presence of C. siamense in the rhizome of G. patens is not fully understood though it is, possibly, an antibiotics-producing agent of the future.
Table (2):
Antagonistic activity of endophytic fungi isolated from Globba patens Miq. rhizome.
| Isolate Code |
Antagonistic activity (mm ± S.D) | |||
|---|---|---|---|---|
| S. aureus ATCC 29213 |
MRSA ATCC 43300 |
E. coli ATCC 25922 |
Enteropathogenic E. coli K11 |
|
| Gp01 | 15.93 ± 2.19 | n.d. | 27.57 ± 1.72 | 15.09 ± 2.07 |
| Gp02 | 16.67 ± 2.03 | n.d. | 13.63 ± 0.4 | n.d. |
| Gp03 | 24.30 ± 0.9 | n.d. | 14.6 ± 1.93 | n.d. |
| Gp04 | 19.20 ± 2.78 | n.d. | 14.57 ± 3.19 | n.d. |
| Gp05 | 20.73 ± 2.73 | n.d. | n.d. | n.d. |
| Gp06 | 13.40 ± 2.52 | n.d. | 18.53 ± 0.25 | n.d. |
| Gp07 | 34 ± 1.22 | 12.83 ± 1.87 | 21.1 ± 0.85 | 18.87 ± 0.92 |
| Gp08 | 24.97 ± 1.69 | n.d. | 13.93 ± 0.81 | n.d. |
| Gp09 | 21.63 ± 0.99 | n.d. | 12.4 ± 3.63 | n.d. |
| Gp10 | 18.43 ± 0.91 | n.d. | 19.6 ± 0.61 | n.d. |
| Gp11 | 24.8 ± 2.72 | n.d. | 21.73 ± 0.65 | n.d. |
| Gp12 | 23.03 ± 5.96 | n.d. | 17.77 ± 3.41 | 18.1 ± 0.0 |
| Gp13 | 18.67 ± 3.19 | n.d. | 14.07 ± 0.93 | n.d. |
| Gp14 | 25.77 ± 4.31 | n.d. | n.d. | n.d. |
| Gp15 | 22.83 ± 5.82 | n.d. | 15.53 ± 3.19 | n.d. |
| Gp16 | 22.8 ± 2.72 | n.d. | 12.73 ± 1.47 | n.d. |
| Gp17 | 16.53 ± 4.22 | n.d. | 10.4 ± 1.11 | n.d. |
| Gp18 | 29.77 ± 0.49 | n.d. | 9.73 ± 0.5 | n.d. |
| Gp19 | 31.47 ± 0.55 | n.d. | 17.77 ± 0.59 | n.d. |
| Gp20 | 28.6 ± 2.6 | n.d. | 18.4 ± 1.31 | 14 ± 2.77 |
| Gp21 | 22.17 ± 2.99 | n.d. | n.d. | n.d. |
| Gp22 | 26.8 ± 3.54 | n.d. | n.d. | n.d. |
| Gp23 | 27.8 ± 0.56 | n.d. | 14.33 ± 3.07 | n.d. |
| Gp24 | 31.23 ± 4.7 | n.d. | n.d. | n.d. |
| Gp25 | 19.67 ± 5.48 | n.d. | n.d. | n.d. |
| Gp26 | n.d. | n.d. | n.d. | n.d. |
ACKNOWLEDGMENTS
None.
CONFLICT OF INTEREST
The authors declare that there is no conflict of interest.
AUTHORS’ CONTRIBUTION
AL: Concept of the main idea, Developed the supporting theory, Performed the experiment, Writing of manuscript. EM: Verified the analytical methods, Supervised the project. YY: Developed the theoretical formalism, Providing the laboratory necessity. MB: Supervised the findings of this project.
FUNDING
The research was funded by Ministry of Research, Technology and Higher Education, Republic of Indonesia under of PMDSU Research Grant (contract number: 11/E1/KP.PTNBH/2019).
ETHICS STATEMENT
This article does not contain any studies with human participants or animals performed by any of the authors.
AVAILABILITY OF DATA
All datasets generated and analyses are provided in this manuscript. The raw data may be requested upon acceptance by the authors.
- Russo TA, Johnson JR. Medical and economic impact of extraintestinal infections due to Escherichia coli: focus on an increasingly important endemic problem. Microbes Infect. 2003;5(5):449-456.
Crossref - Menichetti F. Current and emerging serious Gram-positive infections. Clin Microbiol Infect. 2005;11(Suppl 3):22-28.
Crossref - Newman DJ, Cragg GM. Natural products as sources of new drugs over the 30 years from 1981 to 2010. J Nat Prod. 2012;75(3):311-335.
Crossref - Deshmukh SK, Verekar SA, Bhave SV. Endophytic fungi: a reservoir of antibacterials. Front Microbiol. 2015;5:715.
Crossref - Newman DJ. Are microbial endophytes the ‘actual’ producers of bioactive antitumor agents? Trends Cancer. 2018;4(10):662-670.
Crossref - Strobel G. The emergence of endophytic microbes and their biological promise. J Fungi. 2018;4(2):57.
Crossref - Jia M, Chen L, Xin HL, et al. A friendly relationship between endophytic fungi and medicinal plants: a systematic review. Front Microbiol. 2016;7:906.
Crossref - Kress WJ, Prince LM, Williams KJ. The phylogeny and a new classification of the gingers (Zingiberaceae): evidence from molecular data. American J Bot. 2002;89(10):1682-1696.
Crossref - Aslam MS, Ahmad MS. Ethnobotanical uses of Globba species: a brief. BAOJ Pharm Sci. 2007;3:035.
- Rahmani AH, Al Shabrmi FM, Aly SM. Active ingredients of ginger as potential candidates in the prevention and treatment of diseases via modulation of biological activities. Int J Physiol Pathophysiol Pharmacol. 2014;6:125-136.
- Siregar ES, Pasaribu N. Inventarisasi jenis-jenis Zingiberaceae di Hutan Sibayak, Sumatera Utara. Journal Penelitian MIPA. 2008;2:22-24.
- Lutfia A, Munir E, Yurnaliza Y. Antagonistic endophytic fungi of Globba pendula Roxb. from Taman Hutan Raya, North Sumatra against Staphylococcus aureus ATCC® 29213™. IOP Conf Ser Earth Env Sci. 2019;305:012003.
Crossref - Lutfia A, Munir E, Yurnaliza Y. Molecular identification of endophytic fungi from Torch Ginger (Etlingera elatior) antagonist to phytopathogenic fungi. Biodiversitas. 2020;21(6):2681-2689.
Crossref - Balouiri M, Sadiki M, Ibnsouda SK. Methods for in vitro evaluating antimicrobial activity: a review. J Pharm Anal. 2016;6(2):71-79.
Crossref - Gardes M, Bruns TD. ITS primers with enhanced specificity for basidiomycetes – application to the identification of mycorrhizae and rusts. Mol Ecol. 1993;2(2):113-118.
Crossref - Manter DK, Vivanco JM. Use of the ITS primers, ITS1F and ITS4, to characterize fungal abundance and diversity in mixed-template samples by qPCR and length heterogeneity analysis. J Microbiol Methods. 2007;71(1):7-14.
Crossref - Kumar S, Stecher G, Li M, Knyaz C, Tamura K. MEGA X: Molecular Evolutionary Genetics Analysis across computing platforms. Mol Biol Evol. 2018;35(6):1547-1549.
Crossref - Lutfia A, Munir E, Yurnaliza Y. Antagonistic endophytic fungi of Hedychium coronarium J. Koenig from Hutan Sibayak and Taman Hutan Raya, North Sumatra against Staphylococcus aureus ATCC® 29213™. IOP Conf Ser Earth Env Sci. 2019;305:012002.
Crossref - Lutfia A, Munir E, Yurnaliza Y, Hartanto A. Antagonistic fungal endophytes colonizing rhizome of Amomum centrocephalum A. D. Poulsen from North Sumatera, Indonesia. Int J Adv Sci Eng Inf Tech. 2019;9(4):1427-1432.
Crossref - Lutfia A, Munir E, Yurnaliza, Basyuni M. Screening of antagonistic fungi from Etlingera littoralis (J. Konig) Giseke rhizome in Sibolangit Forest, North Sumatra. J Phys Conf Ser. 2019;1351: 012025.
Crossref - Munir E, Lutfia A, Yurnaliza Y. Records of culturable endophytic fungi inhabiting rhizome of Elettaria in Hutan Sibayak, North Sumatera. IOP Conf Ser Earth Env Sci. 2019;305:012004.
Crossref - Caruso G, Abdelhamid MT, Kalisz A, Sekara A. Linking endophytic fungi to medicinal plants therapeutic activity: a case study on Asteraceae. Agriculture. 2020;10:286.
- Liu T, Greenslade A, Yang S. Levels of rhizome endophytic fungi fluctuate in Paris polyphylla var. yuunanensis as plant age. Plant Divers. 2017;39(1):60-64.
Crossref - Shubin L, Juan H, Chao ZR, Ru XS, Xiao JY. Fungal endophytes of Alpinia officinarum rhizomes: Insights on diversity and variation across growth years, growth sites, and the inner active chemical concentration. PLoS ONE. 2014;9(12):e115289.
Crossref - Senadeera SP, Wiyakrutta S, Mahidol C, Ruchirawat S, Kittakoop P. A novel tricyclic polyketide and its biosynthetic precursor azaphilone derivatives from the endophytic fungus Dothideomycete sp. Org Biomol Chem. 2012;10(35):7220-7226.
Crossref - Nurunnabi TR, Nahar L, Al-Majmaie S, et al. Anti-MRSA activity of oxysporone and xylitol from the endophytic fungus Pestalotia sp. growing on the Sundarbans mangrove plant Heritiera fomes. Phytother Res. 2017;32(2):348-354.
Crossref - Prihastuti H, Cai L, Chen H, McKenzie EHC Hyde KD. Characterization of Colletotrichum species associated with coffee berries in northern Thailand. Fungal Divers. 2009;39:89-109.
- Salunkhe VN, Gawande SP, Gokte-Narkhedkar N, Nagrale DT, Hiremani NS, Waghmare VN. First report of Colletotrichum siamense causing leaf Anthracnose on cotton in India. Plant Dis. 2020;104:2015.
Crossref - Silva DD, Groenewald JZ, Crous PW, Ades PK, Nasruddin A, Mongkolporn O, Taylor PWJ. Identification, prevalence and pathogenicity of Colletotrichum species causing anthracnose of Capsicum annuum in Asia. IMA Fungus. 2019;10:8.
Crossref - Zou WX, Meng JC, Lu H, et al. Metabolites of Colletotrichum gloeosporioides, an endophytic fungus in Artemisia mongolica. J Nat Prod. 2000;63(11):1529-1530.
Crossref - Zhou SL, Zhou SL, Wang MX, Chen SL. Two compounds from the endophytic Colletotrichum sp. of Ginkgo biloba. Nat Prod Commun. 2011;6(8):1131-1132.
Crossref
© The Author(s) 2021. Open Access. This article is distributed under the terms of the Creative Commons Attribution 4.0 International License which permits unrestricted use, sharing, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.